Food and Nutrition Sciences
Vol. 2 No. 4 (2011) , Article ID: 5447 , 10 pages DOI:10.4236/fns.2011.24053
Uptake and Organ Distribution of Feed Introduced Plasmid DNA in Growing or Pregnant Rats*
![]()
1GenØk - Centre for Biosafety, Tromsø, Norway; 2Department of Biochemistry, Faculty of Health Sciences, University of Tromsø, Tromsø, Norway; 3Department of Pharmacy, Faculty of Health Sciences, University of Tromsø, Tromsø, Norway; 4Centre for Agricultural Sciences and Engineering, University of Debrecen, Debrecen, Hungary.
Email: terje.traavik@genok.org
Received January 12th, 2011; revised March 25th, 2011; accepted April 2nd, 2011.
Keywords: DNA Uptake, DNA Persistence, DNA Degradation, Rodents, Feeding Trial
ABSTRACT
Fragments of DNA present in food and feed are taken up by the gastrointestinal tract (GIT) of mammals. The extent of uptake varies according to organism, study design and DNA source. This study explores the hypothesis that actively growing, as well as pregnant rats, are more likely to take up DNA from the GIT than mature animals due to the high demand for nutrients for tissue and organ development. Plasmid DNA (pDNA) was added to standard feed for growing, and pregnant rats. The young rats received one pDNA (50 μg) containing meal by gavage. Blood, organ and tissue samples were harvested at 2 h to 3 days post feeding (p.f). The pregnant females were fed pellets containing pDNA (100 μg) daily, starting at day 5 after established pregnancy. Females and foeti were killed at days 7 and 14 of gestation, and pups at the time of weaning. Genomic DNA was analyzed by PCR followed by Southern blot and real-time PCR. A 201 bp target sequence was detected in mesenteric lymph nodes, spleen, liver and pancreas from growing rats 2 h p.f. At 6 h, target DNA was detectable in the kidneys, and at three days p.f. in the liver. Target DNA was not detected in samples from pregnant rats, their foeti or pups. In conclusion, low level of feed introduced DNA could be transiently detected in organs of young, growing rats. However, indications of increased DNA uptake levels in the GIT of growing rats were not found.
1. Introduction
Several published studies now suggest that minor portions of feed introduced DNA is taken up by the gastrointestinal tract (GIT) of mammals. For instance, the digestive fate of the recombinant portion of DNA from GM crops has been investigated in mice, rats, fish, cattle, poultry and pigs [1-35]. Food is often processed by different treatments prior to consumption that fragment DNA such as high temperatures and acidic conditions [36-39]. The extent of DNA fragmentation will, however, also be determined by other protective factors as well as the storage conditions [38-39].
The first site of possible degradation in the GIT is the oral cavity. Studies by Duggan et al. [10], and Mercer et al. [40-42] demonstrated that plasmid DNA survived for a short time in human saliva and was able to transform Escherichia coli or Streptococcus gordonii; implying that DNA released from the diet may survive and provide a source of transforming DNA for bacteria present in the oral cavity. Feeding experiments in adult mice performed by Walter Doerfler’s group in Cologne, Germany, have shown that most foreign DNA is substantially degraded during passage through the lower GIT [1-3,5]. However, the persistence of fragments up to about 1700 bp in length was demonstrated in the caecum and large intestine. Fragments were subsequently also detected in leucocytes, spleen, liver and kidney. Moreover, these studies indicated that fragments of phage DNA added to the feed had become covalently linked to chromosomal DNA after transplacental transfer [1-3,5]. Other studies have also reported detection of feed introduced DNA in mammals. For instance, fragments of plant DNA have been observed in muscle, liver, spleen, and kidney of chicken after feeding with corn [17-20]. In pigs, sheep and cattle, fragments of ingested DNA from corn have been detected in GIT contents as well as in tissues and organs [7-9,12,15,21,31,33]. In contrast, a study in fallow deer fed on GM plants concluded that no recombinant DNA fragments were detected in GIT contents or the visceral tissues [23].
The cellular processes governing the degradation, timelimited survival or cellular uptake of feed-introduced DNA remain poorly understood and characterized despite the many observational studies available. Schubbert et al. [2] suggested that extracellular DNA enters the organism via the gut mucosa and then reaches the different organs via the blood stream. Small amounts of feed introduced DNA are therefore expected to circulate in human blood after feed intake and to be cleared from circulation by the liver [43,44].
The high demand for efficient nutrient uptake from the GIT (to supply energy and nutrients, including DNA metabolites) for tissue and organ development in growing individuals suggest the frequency of uptake of feed introduced DNA may differ from those recorded in previous studies of adult individuals. Only few studies have examined the fate of feed introduced DNA in rapidly growing/developing animals. In this study the hypothesis that young actively growing, as well as pregnant rats, are more susceptible to and more likely to take up and integrate DNA from the GIT than mature animals was explored. A plasmid that contained a gfp (green fluorescent protein) gene under the control of the 35S cauliflower mosaic virus (CaMV) promoter was added to the standard, semi-synthetic DNA void, rat feed source to determine if: 1) feed introduced DNA was taken up from the GIT and distributed to internal organs, 2) there was a difference in DNA uptake efficiency between young, actively growing and adult, pregnant female rats, 3) feed introduced DNA sequences crossed the placental “barrier” and could be detected in foeti and pups, and if 4) there was a difference in uptake frequency between linear and circular pDNA.
2. Materials and Methods
2.1. Preparation of DNA for the Rat Feeding Experiments
The plasmid phrGFP-CaMV [45] carries the gene for the green fluorescent protein (GFP) under control of the 35S Cauliflower Mosaic virus (CaMV) promoter. Plasmid DNA was isolated from E. coli strain DH5α and was solubilized in sterilized water according to the Qiagen Plasmid Maxi Kit protocol (Qiagen, Germany). The DNA concentration was quantified spectrophotometrically at OD260 with a SmartSpec TM Plus (Bio-Rad, USA). Linear plasmids were obtained by incubation with the EcoRI enzyme (New England BioLabs, USA) at 37˚C according to the manufacturer’s instructions. The restriction reaction was terminated with thermal inactivation of the enzyme at 65˚C for 20 minutes.
2.2. Preparation of Rat Feed
The basic feed for the young, actively growing rats was a breast-milk substitute (NAN, AS Nestle, Norway), which had a composition closely related to the fully balanced semi-synthetic lactalbumin based diet (LA; 150 g protein/ kg diet) [46]. The pregnant rats were fed rat chow AIN- 93G recommended for growth, pregnancy and lactation and AIN-93M, recommended for maintenance after pregnancy [47] (Scanbur BK AS, Norway). All feedlots used in these experiments were tested for the absence of the target PCR DNA. Food pellets were made by mixing 80 ml sterile, autoclaved water with 200 g rat feed- -powder. The solution was poured into a box, lined with aluminum foil, and left at 4˚C overnight. The solidified material was then cut into feed pellets (1.5 × 1.5 × 1.5 cm pellets) and left on a tray covered with baking paper overnight to dry at room temperature. This work was performed in a sterile laminar airflow cabinet. Pellets were then frozen at –20˚C until they were used.
2.3. DNA Stability in Feed
A total of 50 mg plasmid DNA pKTG from E. coli strain DH5a [48] was mixed with the feed pellets in 3 parallels and left at room temperature for 0, 1, 12, 36 or 72 hours. The pellets were frozen at –20˚C until DNA extraction. Frozen pellet samples (0.5 g) were crushed to a fine powder using a mortar. The DNA was extracted using the FastDNA SPIN kit (Q-Biogene, Heidelberg, Germany). Absorbance at OD260 was used to determine the DNA concentrations. To test whether the isolated DNA remained functionally intact over the incubation period, a transformation experiment was performed using the naturally competent Acinetobacter baylyi strain BD413 [49]. For the filter-based transformation assay, 1 µg of DNA isolated from each rat feed pellet was mixed with 80 µl of bacterial cells and the filter transformation, with appropriate controls, was carried out as described by Ray and Nielsen [50]. The transformation frequency was determined as the number of transformants (expressing the pKTG harbored nptII kanamycin resistance gene) divided by the total number of recipient BD413 cells.
2.4. Feeding Experiments
Wistar rats, Mol:WIST Han, M&B Denmark, bred at the rat facility of the Department of Comparative Medicine of the University of Tromsø, Norway, were used for the feeding experiments. The animal experiments and housing procedures were approved by The National Animal Research Authority, Norway. Two series of experiments were conducted:
Experiment series 1: Young actively growing rats (80 ± 3 grams) were divided into three different groups. Each group consisted of 6 rats: 3 females and 3 males. The target meal contained the circular plasmid DNA (pDNA) for group one, the linear pDNA for group two, and no pDNA (negative control group) for group three. For the target meal, which was administered by gavaging, a total of 50 mg pDNA was mixed with the feed to group one and two. Two rats from each group were killed in a CO2 chamber 2 h, 6 h and 3 days after receiving the target meal.
Experiment series 2: Six pregnant rats were separated into two groups. Group one did not receive any pDNA (control group). Group two were given pDNA mixed with their food daily from day 5 of established pregnancy. pDNA consisted of 50% circular and 50% linearized pDNA in a total of 100 µg added (pipetted into the core region of the pellets) to 3 feed pellets that were given in the morning. In the afternoon, and after finishing the 3 feed pellets with DNA, the rats were given the same feed but without pDNA. The pregnant rats did not receive any pDNA the day they were killed. The adult rats and their foeti were killed (one rat from each group) in a CO2 chamber at days 7 and 14 of gestation, and at weaning. The pups were killed by decapitation at weaning.
2.5. Rat Organs
Organs from both experiments were sampled and dissected as described by Pusztai et al. [46] and frozen in liquid nitrogen within 5 minutes post mortem. The following organs were sampled: mesenteric lymph nodes, pancreas, spleen, liver, kidney, thymus, heart, ovaries/ uterus, brain, lung, gastronomic muscle and blood. The fetuses and placentas were collected from the pregnant rats.
2.6. DNA Isolation
The different rat organs were weighed and homogenized in a digestion buffer (10 mM Tris/HCl pH 8.0, 100 mM NaCl, 25 mM EDTA pH 8.0, 0.5% SDS) using an Ultra Turax T25 Basic homogenizer (Ika-Labotechnik, Germany). DNA was isolated using the QIAamp DNA isolation kit (Qiagen) according to the manufacturer’s instructions. For mesenteric lymph nodes and spleen, 10 mg tissue was used for DNA isolation per sample (according to recommendations given in the instructions of the QIAamp DNA isolation kit) as they have a very high cell number for a given mass of tissue. For all other organs, 25 mg tissue was used. The DNA was isolated from baby rat organs, fetuses, rat feed and gastrointestinal contents using the FastPrep (FP120) shaker (Q-Biogene,) and the FastDNA kit (Q-Biogene). The DNA was isolated from blood using the QIAamp DNA blood kit (Qiagen). The quantity and purity of DNA were measured spectrophotometrically (BioRad SmartSpec TM Plus) at OD260/280.
2.7. PCR of a Rat Housekeeping Gene
Extracted DNA was analyzed for the presence of inhibitors by PCR amplification of a 1277 bp part of the rat Bradykinin B2 receptor gene [51,52]. The reactions were performed in three parallels in a total volume of 50 µl containing the following: 1.3 ml of primers (forward: 5- GTC CTT CGT TTT GAG TCT GG -3, reverse: 5- GGT TCT GTG TTG TAG GGA GT -3) at 10 µM, 25 µl DYNAzyme™ II PCR Master Mix (Finnzymes Oy, Finland), 17.4 µl water and 5 µl DNA template (200 ng). PCR was performed with the following thermal cycling conditions: 94˚C for 4 min; 30 cycles of 94˚C for 30 sec, 55˚C for 30 sec and 74˚C for 1 min and 30 sec, final extension at 74˚C for 3 min. The PCR products were run on agarose gels for visualization.
2.8. PCR and Southern Hybridization of PhrGFPCaMV Amplicons
DNA isolated from the various tissues was analyzed for the presence of plasmid sequences. The PCR was designed to amplify a 391 bp (primers CaMV08F and CaMV08R- 2) and a 922 bp (primers CaMV08F and CaMV08R) fragment, including the 35S CaMV promoter of the plasmid phrGFPCaMV (Figure 2). The reactions were performed in a total volume of 50 µl containing the following: 1.0 ml of primers (CaMV08F: 5-GAG TCG ACC TCG AGA CCA T-3, CaMV08R-2: 5-GCG GTA CAC GAA CAT CTC CT-3, CaMV08R: 5-AAA TGA GCC TTG GGA CTA TG) at 10 µM, 25 µl DYNAzyme™ II PCR Master Mix (Finnzymes Oy, Finland), 18 µl water and 5 µl DNA template (200 ng). Negative PCR controls (no template), negative rat controls (no DNA in feed), commercial rat genomic DNA (BD Bioscience) and positive controls (plasmid dilution series) were always included in each set of PCR analysis. PCR for the 391 bp sequence was performed with the following thermal cycling conditions: 94˚C for 5 min; 35 cycles of 94˚C for 30 sec, 64˚C for 30 sec, and 72˚C for 40 sec, final extension at 72˚C for 7 min. and for the 922 bp sequence: 94˚C for 5 min; 35 cycles of 94˚C for 30 sec, 65˚C for 30 sec, and 72˚C for 1 min, final extension at 72˚C for 7 min. The PCR products were run on 1% agarose gels and blotted onto Nytran N membranes (Schleicher & Schoell, Germany) with the TurboBlotter system (Schleicher & Schoell) before chemoluminescence detection using sequence specific dig-labeled oligonucleotide probes for the detection of the corresponding region in the CaMV promoter region. Signals were developed using the substrate CDP- -Star (Roche, Germany) and visualized using a Lumi-Imager F1 (Roche).
2.9. Real time PCR Standard Curves, Amplification Efficiencies and Statistics
To assess the quantification range of the real time PCR system, a 10-fold dilution of the plasmid phrGFPCaMV ranging from 23 100 to 2.3 copies was prepared and analyzed by real time PCR as described below. The standard curve for phrGFPCaMV resulting from Ct values of each reaction plotted against the log of estimated copy numbers is presented in Figure 1(a). The curve is linear in the range tested by triplicate reactions. The slope of the standard curve in Figure 1(a) is –3.35 and the square regression coefficient after the linear regression (R2) is 0.99. The R2 indicated a good correlation between Ct value of the amplification and the amount of template (DNA). The 10-fold diluted phrGFPCaMV samples were also spiked with the amount of DNA used (200 ng).
2.10. Real Time PCR Analysis
The DNA samples were analyzed for the presence of a 201 bp region within the 35S-CaMV promoter of the phrGFPCaMV plasmid using real-time PCR. Primers and probes were designed using the Primer Express program (Primer Express Software V. 2.0, Applied Biosystems, US). The real time PCR was performed on an ABI 7900 HT Fast Real Time PCR system (Applied Biosystems) with the primers 5-TGC CAT CAT TGC GAT AAA GG-3 and 5-GGA AGG GTC TTG CGA AGG AT-3 (Eurogentec, Belgium). The amplified PCR fragments were detected by a TaqMan probe with a 5´-FAM fluorescent dye and 3´-Dark Quencher label (5-ATC GTT GAA GAT GCC TCT GCC GAC A-3) (Eurogentec). The real time PCR included denaturation at 95˚C for 10 sec., and 60˚C for 1 min for 50 cycles. For amplification, the qPCR Mastermix Plus reagent (Eurogentec) was used. All samples were analyzed in triplicate, including positive (isolated plasmid DNA), negative control samples (no DNA in feed) and no template controls (dH2O). A total of 200 ng DNA was analyzed in each well. This corresponds to approximately 28,500 cells as 1 mammalian cell in average contains 7 pg DNA [53]. For blood and sera, the DNA concentrations were lower due to overall lower DNA content and difficulties in extracting enough DNA.
3. Results
3.1. DNA Stability in Feed Pellets
Plasmid DNA was as added to rat feed pellets and incubated over time (0 to 72 hours) to determine whether this treatment would result in quantifiable pDNA degradation. The added DNA showed no signs of degradation over the 72 h incubation period at room temperature (Table 1). A bacterial transformation assay was subsequently performed to verify that the feed-incubated pDNA retained its biological functionality if acquired by competent cells. As seen in Table 2, no significant reduction in the transformation frequencies could be seen when the bacteria were exposed to DNA extracted from feed pellets after up to 72 hours of incubation.
3.2. Target DNA Isolation, Housekeeping Gene PCR, and Real-Time PCR Standard Curves
The DNA was isolated from mesenteric lymph nodes, pancreas, spleen, liver, kidney, thymus, heart, ovaries, uterus, brain, lung, gastronomic muscle and blood. The % sample weight of each organ is given in Table 3. For each analysis 200 ng total DNA was used. Prior to regular PCR and real time PCR, a rat BK2-targeted PCR was run to detect any inhibition of the PCR amplification and no indication of the presence of inhibitors was found. The standard curve used in the real-time PCR was spiked with rat genomic DNA (200 ng) to check whether the PCR efficiency was affected. The Ct values and R2 for these runs indicated that the amount of total DNA present have no effect on the amplification efficiency for any of the plasmid dilutions (Figure 1(b)).
3.3. Detection of Target DNA in Rat Organ and Tissue Samples
To detect the presence of feed introduced pDNA in the organ and tissue samples, a regular PCR amplification and subsequent Southern blotting were performed with primers and probes targeting part of the CaMV promoter. However, pDNA was not detected in organs, blood/sera or foeti of the pregnant rats and their offspring.
The real-time PCR quantification was based on detecting a 201 bp sequence inside the 35S CaMV promoter, and calculating copy numbers according to the standard curve based on serial dilutions of the pDNA (described in Materials and Methods) (Figure 1(a)). The mean value of estimated copy numbers was >2.3 if considered positive (see Material and Methods). All negative
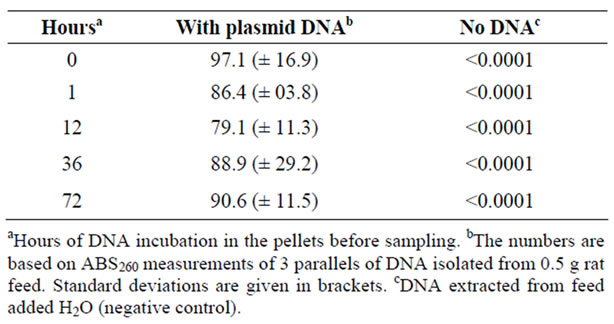
Table 1. Quantification of DNA isolated from feed pellets.
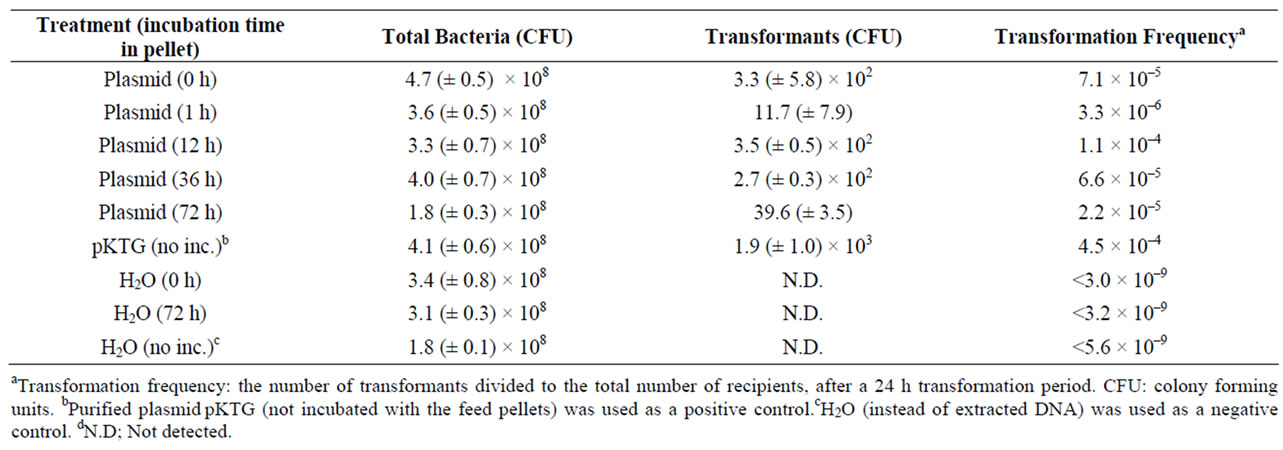
Table 2. Natural transformation assay of Acinetobacter baylyi by plasmid DNA isolated from rat feed pellets.
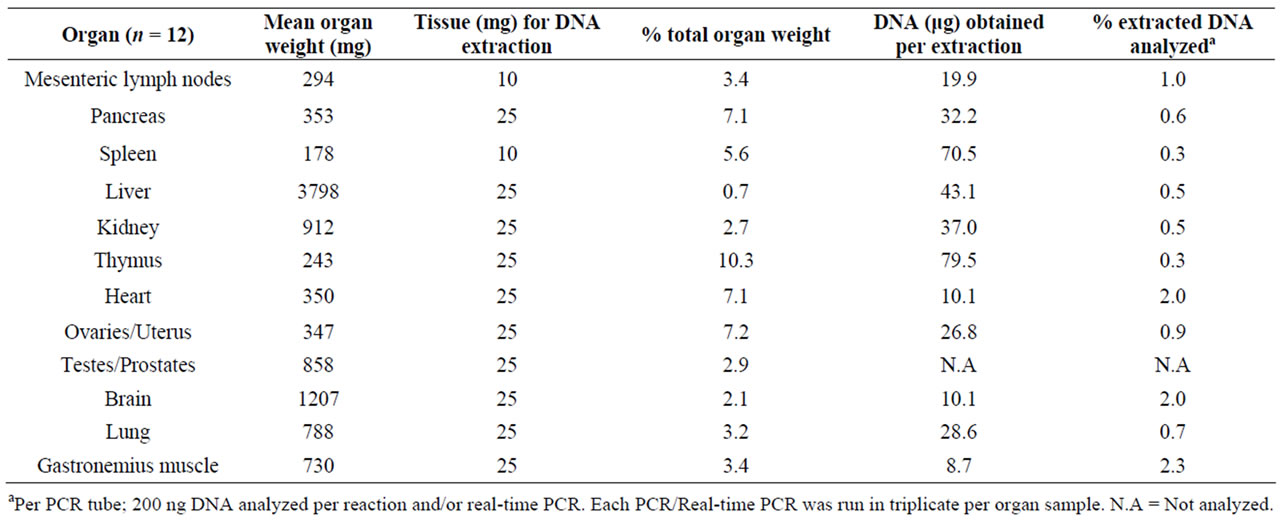
Table 3. Amount of rat tissue used for DNA extraction and PCR analyses.
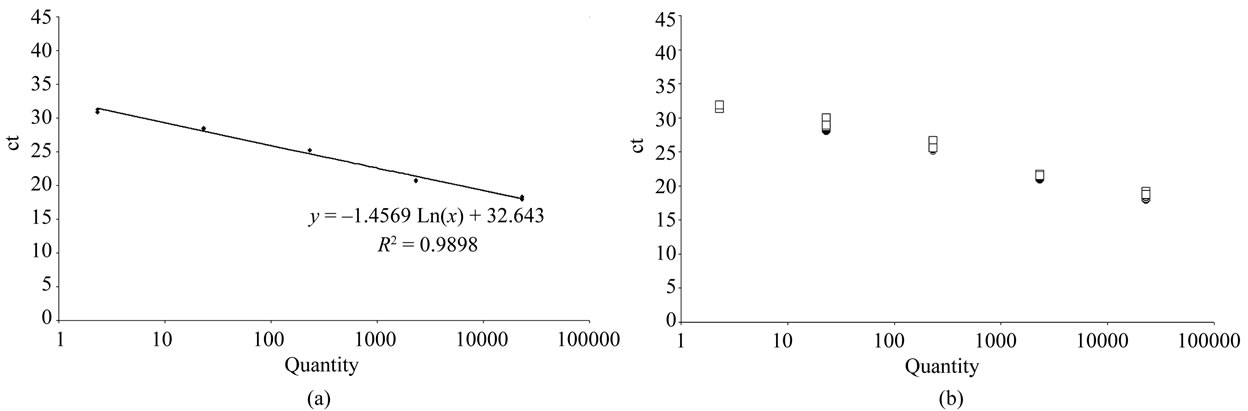
Figure 1. (a) Standard curve for real time PCR. Ten-fold dilutions of the plasmid phrGFPCaMV ranging from 100 fg to 0.01 fg (23 100 to 2.3 copies) were used and each dilution was run in triplicate. The slope and correlation coefficients were calculated for each run. The Ct value is the threshold cycle at which the system starts to detect the increase in signal associated with an exponential growth of the PCR product during the log-linear phase. (b) Standard curve for real time PCR with added rat DNA. The plasmid dilutions, ranging from 23,100 to 2.3 copies, were also in parallel added 200 ng genomic rat DNA. Squared open points; not added genomic DNA, and filled black circles; added genomic DNA.
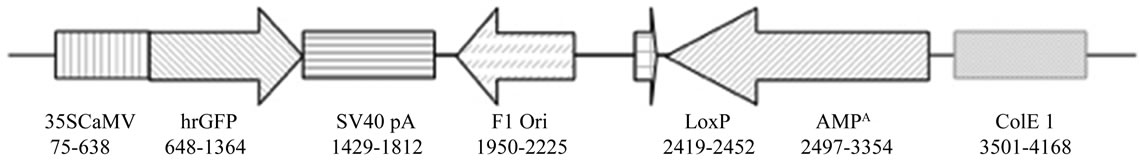
Figure 2. Linear map of the phrGFP-CaMV plasmid (4220 bp)(not to scale).
sample controls (rats fed with no DNA) were negative under these conditions. The results for young, actively growing rats are summarized below. The results are given as estimated plasmid copy numbers using the standard curve. The pDNA targets detected were below 64 copies per analyzed sample. Collectively, through the 3 days sampling period after the pDNA feeding, target pDNA was detected in 4 out of 12 rats (three female and one male). Of these four, three had received linear pDNA and one received circular pDNA. It is noteworthy that pDNA was still detectable in the liver of one rat 3 days after the single pDNA-containing meal. More specifically, pDNA was detected in the mesenteric lymph node (11 copies), the spleen (5 copies), the liver (3 copies) and the pancreas (63 copies) of one female rat fed linear plasmid DNA two hours after feeding, (Rat 4). In another female rat fed linear DNA and sampled after two hours, pDNA was detected in the pancreas only (6 copies) (Rat 3). The pDNA was detected in the kidney (6 copies) of one female rat (Rat 6) after 6 hours. After 3 days, 2 pDNA copies were detected in the liver of one female rat fed linear DNA (Rat 12). No pDNA detections were made in the samples from the 6 control rats (numbers 13 - 18) that received a meal without pDNA. Tissue samples from pregnant rats, foeti, and offspring pups were also investigated by real-time PCR. However, no pDNA from organs, blood/sera and foeti samples could be detected (detection limit >2 copies per sample).
4. Discussion
The data presented here demonstrate uptake and intraorganismal spread of feed-introduced DNA. They also extend the observations made in previous feeding studies [1-35] as only few studies published so far have examined the fate of feed introduced DNA in rapidly growing/developing animals. This study examined the hypothesis that the frequency of uptake of feed introduced DNA from the GIT may differ in rapidly growing individuals from those recorded in previous studies of adult individuals due to the high demand for nutrient uptake during growth. However, although pDNA was more readily detected in the organs of young actively growing rats that the pregnant adults, no indications of a substantially elevated uptake rate in young, growing rats as compared to adults were found. Additional novel observations reported here include that feed introduced DNA is present in the pancreas. This has not been shown in previous feeding studies. Interestingly, three of the four rats with detectable pDNA in tissues were feed linear DNA, possibly indicating that linear DNA may be more easily taken up from the GIT or persist longer than circular DNA. Two previous studies in mice have also indicated that linearized polyomavirus DNA is more infectious than circular versions [54,55].
The current study also showed several differences to previous reports. Importantly, our study was not able to detect feed introduced DNA in any organs or in offspring foeti/pups of the continuously pDNA exposed pregnant rats as previously demonstrated by Schubbert et al. [3]. The amount of pDNA used in our study (50 µg plasmid DNA per rat) corresponds to approximately 1013 phrGFP plasmid molecules. Schubbert et al. [3] detected lowlevel DNA transfer from gut to liver and spleen after feeding 1013 plasmids molecules per mouse. This latter feeding design therefore represents a higher dose relative to body weight, when compared to the current study. However, both studies are within the normal range of DNA exposures as rodents ingest milligram quantities of foreign DNA with their daily food intake [53].
Our study did not detect pDNA in blood, although DNA has previously been reported to persist in blood samples from mammals, including humans and mice [24, 27,56,57]. The reason for this is unclear. However, the different feeding regimes used in the various published studies might have influenced the time-dependent distribution of feed-introduced DNA and caused the differences seen. For instance, gavage feeding avoids initial DNA degradation in the oral cavity, and the amount of DNA that reaches the GIT will be higher compared to the experimental designs using orally ingested pDNA. Duggan et al. [10] demonstrated that approximately 70% of pDNA was extensively degraded after incubation for one minute in vivo in the ovine oral cavity. The results that differ from other reports may also be due to variations in sampling in relation to a time-dependent DNA uptake, clearance and degradation of DNA in different rodent species.
In summary, although it is known that the penetration/ transfer of macromolecules over the GIT barrier and their further distribution/clearance is influenced by age and that mammals are especially susceptible to macromolecules uptake in the neonatal period [58,59], no evidence was found suggesting actively growing rats are more susceptible to take up feed introduced DNA. Other novel findings include the observation of feed DNA in the pancreas, and possibly increased uptake rates of linear DNA as compared to circular DNA.
The presented and previous studies have all focused on the mechanistic aspects of DNA uptake and distribution. Few studies have examined the biological outcome of rare uptake and integration events of feed-derived DNA in mammalian cells [28,29,60]. However, alimentary administration of DNA vaccines provide evidence that DNA, if taken up from the GIT, can be transcribed, translated, and presented to the immune system as a functional protein [61-64]. Therefore, further studies on the biological effect and physical/functional state of food-ingested DNA in mammals’ GIT should be undertaken.
5. Acknowledgements
We thank the Department of Comparative Medicine of the University of Tromsø, for technical assistance and support. We thank Elisabeth Olsen, Julia Eggert and Martin Hammerø for technical assistance and Joachim Karlsen for advice on the real-time PCR. The authors I. M. Grønsberg and L. Nordgård contributed equally to this work.
REFERENCES
- R. Schubbert, C. Lettmann and W. Doerfler, “Ingested Foreign (Phage M13) DNA Survives Transiently in the Gastrointestinal Tract and Enters the Bloodstream of Mice,” Molecular and General Genetics, Vol. 242, No. 5, 1994, pp. 495-504. doi:10.1007/BF00285273
- R. Schubbert, D. Renz, B. Schmitz and W. Doerfler, “Foreign (M13) DNA Ingested by Mice Reaches Peripheral Leukocytes, Spleen, and Liver via the Intestinal Wall Mucosa and Can be Covalently Linked to Mouse DNA,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 94, No. 3, 1997, pp. 961-966. doi:10.1073/pnas.94.3.961
- R. Schubbert, U. Hohlweg, D. Renz and W. Doerfler, “On the Fate of Orally Ingested Feed-Derived DNA in Mice: Chromosomal Association and Placental Transmission to the Fetus,” Molecular and General Genetics, Vol. 259, No. 6, 1998, pp. 569-576. doi:10.1007/s004380050850
- W. Doerfler, R. Schubbert, H. Heller, J. Hertz, R. Remus, J. Schröder, C. Kämmer, K. Hilger-Eversheim, U. Gerhardt, B. Schmidt, D. Renz and G. Schell, “Foreign DNA in Mammalian Systems,” Acta Pathologica, Microbiologica, et Immunologica, Scandinavica, Supplement, Vol. 106, No. 84, 1998, pp. 62-68.
- U. Hohlweg and W. Doerfler, “On the Fate of Plant or Other Foreign Genes upon the Uptake in Food or after Intramuscular Injection in Mice,” Molecular Genetics and Genomics, Vol. 265, No. 2, 2001, pp. 225-233. doi:10.1007/s004380100450
- P. A. Chambers, P. S. Duggan, J. Heritage and M. J. Forbes, “The Fate of Antibiotic Resistance Marker Genes in Transgenic Plant Feed Material Fed to Chickens,” Journal of Antimicrobial Chemotherapy, Vol. 49, No. 1, 2002, pp. 161-164. doi:10.1093/jac/49.1.161
- E. H. Chowdhury, O. Mikami, Y. Nakajima, A. Hino, H. Kuribara, K. Suga, M. Hanazumi and C. Yomemochi, “Detection of Genetically Modified Maize DNA Fragments in the Intestinal Contents of Pigs Fed StarLink CBH351,” Veterinary and Human Toxicology, Vol. 45, No. 2, 2003, pp. 95-96.
- E. H. Chowdhury, H. Kuribara, A. Hino, P. Sultana, O. Mikami, N. Shimada, K. S. Guruge, M. Saito and Y. Nakajama, “Detection of Corn Intrinsic and Recombinant DNA Fragments and Cry1Ab Protein in the Gastrointestinal Contents of Pigs Fed Genetically Modified Corn Bt11,” Journal of Animal Science, Vol. 81, No. 10, 2003, pp. 2546-2551.
- E. H. Chowdhury, O. Mikami, H. Murata, P. Sultana, N. Shimada, M. Yoshioka, K. S. Guruge, S. Yamamoto, S. Miyazaki, N. Yamanaka and Y. J. Nakajima, “Fate of Maize Intrinsic and Recombinant Genes in Calves Fed Genetically Modified Maize Bt11,” Journal of Food Protection, Vol. 67, No. 2, 2004, pp. 365-370.
- P. S. Duggan, P. A Chambers, J. Heritage and M. J. Forbes, “Fate of Genetically Modified Maize DNA in the Oral Cavity and Rumen of Sheep,” British Journal of Nutrition, Vol. 89, No. 2, 2003, pp. 159-166. doi:10.1079/BJN2002764
- R. H. Phipps, E. R. Deaville and B. C. Maddison, “Detection of Transgenic and Endogenous Plant DNA in Rumen Fluid, Duodenal Digesta, Milk, Blood, and Feces of Lactating Dairy Cows,” Journal of Dairy Science, Vol. 86, No. 12, 2003, pp. 4070-4078. doi:10.3168/jds.S0022-0302(03)74019-3
- T. Reuter and K. Aulrich, “Investigations on Genetically Modified Maize (Bt-Maize) in Pig Nutrition: Fate of Feed-Ingested Feed-Derived DNA in Pig Bodies,” European Food Research and Technology, Vol. 216, No. 3, 2003, pp. 185-192.
- E. R. Deaville and B. C. Maddison, “Detection of Transgenic and Endogenous Plant DNA Fragments in the Blood, Tissues, and Digesta of Broilers,” Journal of Agricultural and Food Chemistry, Vol. 53, No. 26, 2005, pp. 10268-10275. doi:10.1021/jf051652f
- C. R. Nielsen, K. G. Berdal, A. M. Bakke-McKellup and A. Holst-Jensen, “Dietary DNA in Blood and Organs of Atlantic Salmon (Salmo Salar L.),” European Food Research and Technology, Vol. 221, No. 1-2, 2005, pp. 1-8. doi:10.1007/s00217-005-1160-1
- R. Sharma, D. Damgaard, T. W. Alexander, M. E. R. Dugan, J. L. Aalhus, K. Stanford and T. A. McAllister, “Detection of Transgenic and Endogenous Plant DNA in Digesta and Tissues of Sheep and Pigs Fed Roundup Ready Canola Meal,” Journal of Agricultural and Food Chemistry, Vol. 54, No. 5, 2006, pp. 1699-1709. doi:10.1021/jf052459o
- G. Flachowsky, K. Aulrich, H. B'ohme and I. Halle, “Animal Nutrition with Feeds from Genetically Modified Plants,” Animal Feed Science and Technology, Vol. 133, No. 1-2, 2007, pp. 2-30. doi:10.1016/j.anifeedsci.2006.08.002
- R. Einspanier, A Klotz, J. Kraft, K. Aulrich, R. Poser, F. Schwägele, G. Jahreis and G. Flachowsky, “The Fate of Forage Plant DNA in Farm Animals: A Collaborative Case-Study Investigating Cattle and Chicken Fed Recombinant Plant Material,” European Food Research and Technology, Vol. 212, No. 2, 2001, pp. 129-134. doi:10.1007/s002170000248
- K. Aeschbacher, R. Messikommer, L. Meile and C. Wenk, “Bt176 Corn in Poultry Nutrition: Physiological Characteristics and Fate of Recombinant Plant DNA in Chickens,” Poultry Science, Vol. 84, No. 3, 2005, pp. 385-394.
- M. A. Tony, A. Butschke, H. Broll, L. Grohmann, J. Zagon, I. Halle, S. Dänike, M. Schauzu, H. M. Hafez and G. Flachowsky, “Safety Assessment of Bt176 Maize in Broiler Nutrition: Degradation of Maize-DNA and Its Metabolic Fate,” Archives of Animal Nutrition, Vol. 57, No. 4, 2003, pp. 235-252. doi:10.1080/00039420310001594397
- F. Rossi, M. Morlacchini, G. Fusconi, A. Pietri, R. Mazza and G. Piva, “Effect of Bt Corn on Broiler Growth Performance and Fate of Feed-Derived DNA in the Digestive Tract,” Poultry Science, Vol. 84, No. 7, 2005, pp. 1022- 1030.
- A. Klotz and R. Einspanier, “Detection of ‘Novel-Feed’ in Animals? Injury of Consumers of Meat or Milk is Not Expected,” In German, Mais, Vol. 3, 1998, pp. 109-111.
- R. Sharma, D. Damgaard, T. W. Alexander, M. E. R. Dugan, J. L. Aalhus, K. Stanford and T. A. McAllister, “Detection of Transgenic and Endogenous Plant DNA in Digesta and Tissues of Sheep and Pigs Fed Roundup Ready Canola Meal,” Journal of Agricultural and Food Chemistry, Vol. 54, No. 5, 2006, pp. 1699-1709. doi:10.1021/jf052459o
- R. Guertler, B. Lutz, R. Kuehn, H. H. D. Meyer, R. Einspanier, B. Killermann and C. Albrecht, “Fate of Recombinant DNA and Cry1Ab Protein after Ingestion and Dispersal of Genetically Modified Maize in Comparison to Rapeseed by Fallow Deer (Dama Dama),” European Journal of Wildlife Research, Vol. 54, No. 1, 2007, pp. 36-43. doi:10.1007/s10344-007-0104-4
- A. Forsman, D. Ushameckis, A. Bindra, Z. Yun and J. Blomberg, “Uptake of Amplifiable Fragments of Retrotransposon DNA from the Human Alimentary Tract,” Molecular Genetic and Genomics, Vol. 270, No. 4, 2003, pp. 362-368. doi:10.1007/s00438-003-0930-3
- M. Palka-Santini, B. Schwarz-Herzke, M. Hösel, D. Renz, S. Auerochs, H. Brondke and W. Doerfler, “The Gastrointestinal Tract as the Portal of Entry for Foreign Macromolecules: Fate of DNA and Proteins,” Molecular Genetics and Genomics, Vol. 270, No. 3, 2003, pp. 201- 215. doi:10.1007/s00438-003-0907-2
- A. Wilcks, A. H. A. M. Hoek, R. G. Joosten, B. B. L. Jacobsen and H. J. M. Aarts, “Persistence of DNA Studied in Different ex Vivo and in Vivo Rat Models Simulating the Human Gut Situation,” Food and Chemical Toxicology, Vol. 42, No. 3, 2004, pp. 493-502. doi:10.1016/j.fct.2003.10.013
- W. Doerfler and R. Schubbert, “Uptake of Feed-Derived DNA from the Environment: The Gastrointestinal Tract and the Placenta as Portals of Entry,” Wiener Klinische Wochenschrift, Vol. 110, No. 2, 1998, pp. 40-44.
- W. Doerfler, R. Schubbert, H. Heller, C. Kämmer, K. Hilger-Eversheim, M. Knoblauch and R. Remus, “Integration of Foreign DNA and Its Consequences in Mammalian Systems,” Trends in Biotechnology, Vol. 15, No. 8, 1997, pp. 297-301. doi:10.1016/S0167-7799(97)01061-5
- W. Doerfler, R. Schubbert, H. Heller, J. Hertz, R. Remus, J. Schröer, C. Kämmer, K. Hilger-Eversheim, U. Gerhardt, B. Schmitz, D. Renz and G. Schell, “Foreign DNA in Mammalian Systems,” APMIS Supplement, Vol. 106, No 84, 1998, pp. 62-68.
- S. Swiatkiewicz, M. Twardowska, J. Markowsi, M. Mazur, Z. Sieradzki and K. Kwiatek, “Fate of Transgenic DNA from Bt Corn and Roundup Ready Soybean Meal in Broilers Fed GMO Feed,” Bulletin of the Veterinary Institute in Pulawy, Vol. 54, No. 2, 2010, pp. 237-242.
- R. Tudisco, V. Mastellone, M. I. Cutrignelli, P. Lombardim, F. Bovera, N. Mirabella, G. Piccolo, S. Calabro, S. Avallone and F. Infascelli, “Fate of Transgenic DNA and Evaluation of Metabolic Effects in Goats Fed Genetically Modified Soybean and Their Offsprings,” Animal, Vol. 4, No. 10, 2010, pp. 1662-1671. doi:10.1017/S1751731110000728
- J. A. Heinemann, “Report on Animals Exposed to GM Ingredients in Animal Feed,” Prepared for the Commerce Commission of New Zealand, 2009.
- R. Tudisco, M. I. Cutrignelli, S. Calabro and F. Infascelli, “Investigation on Genetically Modified Soybean (RoundUp Ready) in Goat Nutrition: DNA Detection in Suckling Kids,” Italian Journal of Animal Science, Vol. 6, No. 1, 2007, pp. 380-382.
- R. Tudisco, F. Infascelli, M. I. Cutrignelli, F. Bovera, C. Morcia, P. Faccioli and V. Terzi, “Fate of Feed Plant DNA Monitored in Water Buffalo (Bubalus Bubalis) and Rabbit (Orytolagus Cuniucus),” Livestock Science, Vol. 105, No. 1, 2006, pp. 12-18. doi:10.1016/j.livsci.2006.04.036
- R. Tudisco, et al., “Genetically Modified Soya Bean in Rabbit Feeding: Detection of DNA Fragments and Evaluation of Metabolic Effects by Enzymatic Analysis,” Animal Science, Vol. 82, No. 2, 2006, pp. 193-199. doi:10.1079/ASC200530
- R. Adam and B. H. Zimm, “Shear Degradation of DNA,” Nucleic Acid Research, Vol. 4, No. 5, 1977, pp. 1513- 1537. doi:10.1093/nar/4.5.1513
- A. Chiter, M. Forbes and E. G. Blair, “DNA Stability in Plant Tissues: Implications for the Possible Transfer of Genes from Genetically Modified Food,” FEBS Letters, Vol. 481, No. 2, 2000, pp. 164-168. doi:10.1016/S0014-5793(00)01986-4
- T. Bauer, P. Weller, W. P. Hammes and C. Hertel, “The Effect of Processing Parameters on DNA Degradation in Food,” European Food Research and Technology, Vol. 217, No. 4, 2003, pp. 338-343. doi:10.1007/s00217-003-0743-y
- M. Kharazmi, S. Sczesny, M. Blaut, W. P. Hammes and C. Hertel, “Effect of Food Processing on the Fate of DNA with Regard to Degradation and Transformation Capability in Bacillus Subtilis,” Applied and Environmental Microbiology, Vol. 69, No. 10, 2003, pp. 6121-6127. doi:10.1128/AEM.69.10.6121-6127.2003
- D. K. Mercer, C. M. Melville, K. P. Scott and H. J. Flint, “Natural Genetic Transformation in the Rumen Bacterium Streptococcus Bovis JB1,” FEMS Microbiology Letters, Vol. 179, No. 2, 1999, pp. 485-490. doi:10.1111/j.1574-6968.1999.tb08767.x
- D. K. Mercer, K. P. Scott, W. A. Bruce-Johnson, A. L. Glover and H. J. Flint, “Fate of Free DNA and Transformation of the Oral Bacterium Streptococcus Gordonii DL1 by Plasmid DNA in Human Saliva,” Applied and Environmental Microbiology, Vol. 65, No. 1, 1999, pp. 6- 10.
- D. K. Mercer, K. P. Scott, C. M. Melville, L. A. Glover and H. J. Flint, “Transformation of an Oral Bacterium via Chromosomal Integration of Free DNA in the Presence of Human Saliva,” FEMS Microbiology Letters, Vol. 200, No. 2, 2001, pp. 163-167. doi:10.1111/j.1574-6968.2001.tb10709.x
- G. I. Nedredal, K. H. Elvevold, L. M. Ytrebø, R. Olsen, A. Revhaug and B. Smedsrød, “Liver Sinusoidal Endothelial Cells Represents an Important Blood Clearance System in Pigs,” Comparative Hepatology, Vol. 2, No. 1, 2003. doi:10.1186/1476-5926-2-1
- J. Hisazumi, N. Kobayashi, M. Nishikawa and Y. Takakura, “Significant Role of Liver Sinusoidal Endothelial Cells in Hepatic Uptake and Degradation of Naked Plasmid DNA after Intravenous Injection,” Pharmaceutical Research, Vol. 21, No. 7, 2004, pp. 1223-1228. i:10.1023/B:PHAM.0000033009.17594.e5
- M. Myhre, K. A. Fenton, J. Eggert, K. M. Nielsen and T. Traavik, “The 35 S CaMV Plant Virus Promoter is Active in Human Enterocyte-Like Cells,” European Food Research and Technology, Vol. 222, No. 1-2, 2006, pp. 185-193.
- A. Pusztai, S. Bardocz, R. Alonso, M. J. Chrispeels, H. E. Schroeder, L. M. Tabe and T. J. V. Higgins, “Expression of the Insecticidal Bean Alpha-Amylase Inhibitor Transgene Has Minimal Detrimental Effect on the Nutritional Value of Peas Fed to Rats at 30% of the Diet,” Journal of Nutrition, Vol. 129, No. 8, 1999, pp. 1597-1603.
- P. G. Reeves, “Components of the AIN-93 Diets as Improvements in the AIN-76A Diet,” Journal of Nutrition, Vol. 127, No. 5, 1997, pp. 838S-841S.
- K. M. Nielsen, A. M. Bones and J. D. van Elsas, “Induced Natural Transformation of Acinetobacter Calcoaceticus in Soil Microcosms,” Applied and Environmental Microbiology, Vol. 63, No. 10, 1997, pp. 3972-3977.
- K. M. Nielsen, K. Smalla and J. D. van Elsas, “Transformation of Acinetobacter sp. Strain BD413 (pFG4- DeltanptII) with Transgenic Plant DNA in Soil Microcosms and Effects of Kanamycin on Selection of Transformants,” Applied and Environmental Microbiology, Vol. 66, No. 3, 2000, pp. 1237-1242. doi:10.1128/AEM.66.3.1237-1242.2000
- J. L. Ray and K. M. Nielsen, “Experimental Methods for Assaying Natural Transformation and Inferring Horizontal Gene Transfer,” Methods in Enzymology, Vol. 395, 2005, pp. 491-520.
- J. B. Pesquero, C. J. Lindsey, K. Zeh, C. M. Paiva, D. Ganten and M. Bader, “Molecular Structure and Expression of Rat Bradykinin B2 Receptor Gene. Evidence for Alternative Splicing,” The Journal of Biological Chemistry, Vol. 269, No. 43, 1994, pp. 26920-26925.
- Z. Saifudeen, H. Du, S. Dipp and S. S El-Dahr, “The Bradykinin Type 2 Receptor is a Target for p53-Mediated Transcriptional Activation,” The Journal of Biological Chemistry, Vol. 275, No. 20, 2000, pp. 15557-15562. doi:10.1074/jbc.M909810199
- G. M. Lee, J. T. Thornthwaite and E. M. Rasch, “Picogram per Cell Determination of DNA by Flow Cytofluorometry,” Analytical Biochemistry, Vol. 137, No. 1, 1984, pp. 221-226. doi:10.1016/0003-2697(84)90374-9
- D. B. Lowrie, “DNA Vaccination Exploits Normal Biology,” Nature Medicine, Vol. 4, No. 2, 1998, pp. 147-148. doi:10.1038/nm0298-147
- A. Israel, H. W. Chan, W. P. Rowe and M. A. Martin, “Molecular Cloning of Polyoma Virus DNA in Escherichia Coli: Oncogenicity Testing in Hamsters,” Science, Vol. 14, No. 205, 1979, pp. 1140-1142. doi:10.1126/science.224458
- K. Kawabata, Y. Takakura and M. Hashida, “The Fate of Plasmid DNA after Intravenous Injection in Mice: Involvement of Scavenger Receptors in Its Hepatic Uptake,” Pharmaceutical Research, Vol. 12, No. 6, 1995, pp. 825-830. doi:10.1023/A:1016248701505
- J. P. Shaw, K. Kent, J. Bird, J. Fishback and B. Froehler, “Modified Deoxyoligonucleotides Stable to Exonuclease Degradation in Serum,” Nucleic Acids Research, Vol. 19, No. 4, 1991, pp. 747-750. doi:10.1093/nar/19.4.747
- J. N. Udall and W. A. Walker, “The Physiologic and Pathologic Basis for the Transport of Macromolecules across the Intestinal Tract,” Journal of Pediatric Gastroenterology and Nutrition, Vol. 1, No. 3, 1982, pp. 295- 301. doi:10.1097/00005176-198201030-00004
- L. Vellenga, J. M. V. M. Mouwen, J. E. van Dijk and H. J. Breukink, “Biological and Pathological Aspects of the Mammalian Small Intestinal Permeability to Macromolecules,” The Veterinary Quarterly, Vol. 7, No. 4, 1985, pp. 322-332.
- J. M. Hertz, G. Schell and W. Doerfler, “Factors Affecting de Novo Methylation of Foreign DNA in Mouse Embryonic Stem Cells,” The Journal of Biological Chemistry, Vol. 274, No. 34, 1999, pp. 24232-24240. doi:10.1074/jbc.274.34.24232
- T. C. Tonheim, J. Leirvik, M. Løvoll, A. I. Myhr, J. Bøgwald and R. A. Dalmo, “Detection of Supercoiled Plasmid DNA and Luciferase Expression in Atlantic Salmon (Salmo Salar L.) 535 Days after Injection,” Fish and Shellfish Immunology, Vol. 23, No. 4, 2007, pp. 867- 876. doi:10.1016/j.fsi.2007.03.015
- T. C. Tonheim, R. A. Dalmo, J. Bøgwald and T. Seternes, “What Happens to the DNA Vaccine in Fish? A Review of Current Knowledge,” Fish and Shellfish Immunology, Vol. 25, No. 1-2, 2008, pp. 1-18. doi:10.1016/j.fsi.2008.03.007
- K. A. Garver, C. M. Conway, D. G. Elliot and G. Kurath, “Analysis of DNA-Vaccinated Fish Reveals Viral Antigen in Muscle, Kidney and Thymus, and Transient Histopathologic Changes,” Marine Biotechnology, Vol. 7, No. 5, 2005, pp. 540-553. doi:10.1007/s10126-004-5129-z
- N. P. Restifo, H. Ying, L. Hwang and W. W. Leitner, “The Promise of Nucleic Acid Vaccines,” Gene Therapy, Vol. 7, No. 2, 2000, pp. 89-92. doi:10.1038/sj.gt.3301117
NOTES
*The work was funded by the Research Council of Norway (148704).

