Journal of Biomedical Science and Engineering
Vol.08 No.12(2015), Article ID:61961,10 pages
10.4236/jbise.2015.812077
An Assay Study of Molecular Recognition of Amino Acids in Water: Covalent Imprinting of Cysteine
Harsha Vardhan Reddy Burri, Donghong Yu*
Department of Chemistry and Bioscience, Aalborg University, Aalborg, Denmark

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 20 August 2015; accepted 14 December 2015; published 17 December 2015

ABSTRACT
A novel synthetic N-(9-fluorenyl methoxy carbonyl)-L-Cysteine (Fmoc-Cys(SH)-OH) receptor was pre- pared by co-polymerizing (9-fluorenyl methoxy carbonyl)-S-(1-propene-2-thiol)-L-Cysteine (Fmoc- Cys(SCH2CHCH2)-OH) and a non-imprinted polymer prepared from 1-propene-1-thiol photo-chemi- cally 15 h at room temperature and additional 3 h thermally at 80˚C. Subsequently, disulfides were reduced with lithium aluminum hydride (LiAlH4) from imprinted polymers. The imprinted polymers selectively recognized Fmoc-Cys(SH)-OH with high binding constants in aqueous and protic solvents by thiol-disulfide exchange reactions. In order to estimate the covalent rebinding, particles were further extracted and disulfides reduced were estimated with the non-covalent recognized and covalently bounded analytes. From rebinding studies that were conducted, we observed that proved polymer particles could be reproducible and contain constant binding strengths and recognition properties. Furthermore, we proved that short incubation periods resulted in fast and efficient thiol-disulfide interchange reactions.
Keywords:
Synthetic Receptor-Like Structure, Molecularly Imprinted Polymers, Thiol-Disulfide Exchange Based Recognition, Covalent Recognition

1. Introduction
The dynamic nature of receptor recognizes the variety of ligands with multiple non-covalent interactions, and plays an important role in many biological processes. Natural receptors are highly selective and have strong binding capacity to corresponding ligands such as proteins, small molecules, neurotransmitters, toxins, hormones, drugs, microbes or viruses. With the availability of natural receptors being limited and cost effective, the man made mimics of artificial receptors are promising alternatives to natural receptors, further employing functional groups at preferred sites which are not found in natural receptors [1] -[6] . In the recent-past, numerous techniques have been reported to make artificial receptors such as cyclodextrins [7] , crown ethers [8] , cyclophanes [9] , coordination complexes [10] and molecularly imprinted polymers (MIP) [11] -[13] . Among these techniques template mediating molecularly imprinting technique is more attractive. Since, MIPs serve as an attractive receptor due to simple and inexpensive, additionally resulted polymers are stable and versatile with dense, rigid and predetermine chiral selective molecular recognition sites. The state-of-art template mediated functional monomers co-polymerize with the cross linker, with subsequent removal of template producing template specific recognition sites also called MIPs [14] -[16] . So far MIPs have been successfully applied in separation science [17] , catalysts [18] , artificial antibodies [19] and sensors [20] . The recognition pattern of MIPs is reversible bond formation of guest molecule with the binding sites, either covalent or non-covalent bonds. Many of the non-covalent bonds such as hydrogen bonding and electrostatic interactions facilitate reversible interactions in recognition, nevertheless binding strengths are weak and the choice of the solvents is limited [21] -[23] . To vary these, covalent bonds show specific strong binding strengths in polar and non-polar solvents, while few of the reversible covalent bonds are available such as boronic acid esters [24] , cyclo-additions [25] , schiff bases [26] and disulfide bonds [27] -[29] .
Till date, a wide variety of MIPs were synthesized, and successfully used in recognizing the various guest molecules such as carbohydrates [30] , lipids [31] , nucleotide bases [32] , amino acids [33] and small peptides [12] [34] -[37] . The recognition of proteins is a fascinating area due to an interest to study their key role in biological pathways. A wide variety of synthetic peptide receptors like oxytocin [12] , macrotricyclic [37] , synthetic tripeptide [35] , ATP receptors [34] , and microcontact imprinting of proteins [37] have been reported.
In the protein structures amide, hydroxyl, amine and thiol groups are available and ideally interact with the MIPs; therefore this recognition in water or polar solvents is poor. On the other hand, covalent recognition in polar solvents is efficient since reversible covalent bonds are limited. Cysteine plays an important role in protein folding and function even as proteins with at least one cysteine in its structure can be recognized by the thiol- disulfide exchange reaction. In the current study, we have employed a covalent imprinting method to synthesize cystein receptor and recognition carried out covalently by thiol-disulfide exchange reaction. Further, we focus on imprinting and recognition carried out through disulfide interactions. The typical molecular imprinting procedure schematically represented in Scheme 1.
2. Experimental Section
2.1. Materials and Instruments
All commercially available solvents were used without purification and distillation. Tetrahydrofuran (THF), methanol (MeOH), trifluoroacetic acid (TFA), N,N-Diisopropylethylamine (DIPEA) and piperidine were purchased from Sigma-Aldrich GmBH (Germany); 1-propene-2-thiol, 2,2’-azo-bis(iso-butyronitrile) (AIBN) and N,N-dimethyl formamide (DMF) from Fluka analytical (Buchs, Switzerland); 2M Lithium aluminum hydride solution in THF, ethylene glycol dimethacrylate (EGDMA) and triisopropylsilane (TIS) purchased from Aldrich chemie (Germany). Acetonitril (ACN), ethyl acetate and N-(9-fluorenyl methoxycarbonyl)-L-phenyl alanine (Fmoc-Phe-OH) were obtained from Fisher-Scientific (U.K), Merck KGaAGmBH (Germany) and Iris Biotech GmBH (Germany) respectively. N-Hydroxybenzotriazole monohydrate (HOBt) and (Benzotriazol-1-yl)-N,N,N',N'- tetramethyluroniumhexafluorophosphate (HBTU) were acquired from Advanced Chemtech (USA). N-(9-fluo- renyl methoxy carbonyl)-S-tert-butylthio-L-Cysteine (Fmoc-Cys(StBu)-OH) and N-(9-fluorenyl methoxy carbonyl)-S-sulfonic-L-Cysteine disodium salt (Fmoc-Cys(SO3H)-OH) obtained from Bachem AG (Switzerland). Deuterated methanol (MeOH-D4) and dimethyl suloxide (DMSO) were procured from Cambridge Isotope Laboratories, Inc. (USA). Nuclear magnetic resonance (NMR) spectra’s recorded on a Bruker DRX 600 spectrophotometer with 14.1 T field strength and 5 mm xyz gradient TXI (H/C/N) probe was used. Topspin 1.3 and 1.2 software were used to collect and analyze data. High performance liquid chromatography (HPLC) performed on Dionex chromatography interfaces model UCI-50 was connected with the ASI-10U model auto sampler and UVD 170U model UV/VIS-detector. Chromolen 6.60 version software was used for control, processing and analyzing the spectra on UV-280 nm channel. A phenomenex, kromasil C-18 (5 µm) column (150 × 4.6 mm) at 25˚C eluted with two eluents systems. The Milliq water was used as a HPLC eluent collected from Barnstednano pure diamond U.V™ D11911 ultra-pure water system and Barnsted D3750 gamma irradiated hollow fiber
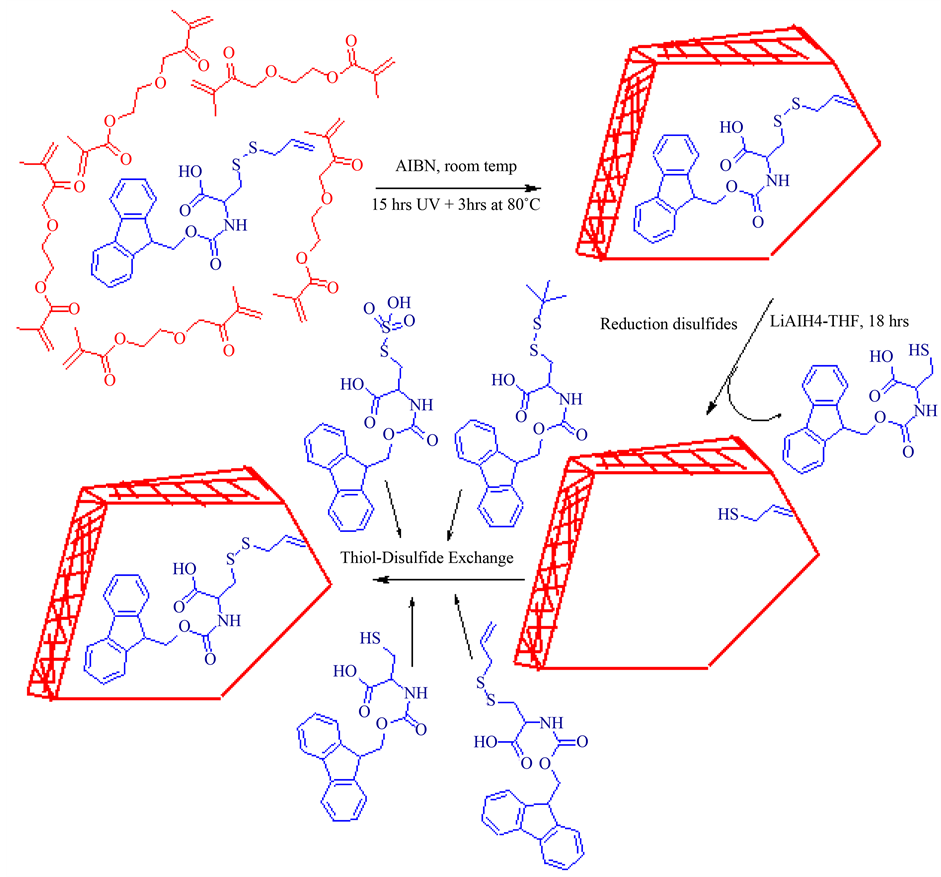
Scheme 1. Schematic representation of molecule imprinting procedure step-by step. The Fmoc-Cys(SCH2CHCH2)-OH copolymerized with cross linker EGDMA. Further disulfides were reductively cleaved from polymer particles. The obtained Fmoc-Cys(SH)-OH MIPs were recognized template covalently (thiol-disulfide exchange) and non-covalently hydrogen bonding).
filters. The column runs at Ultraviolet lamp model 3040, 400 W power, MP from Photo chemical reactors Ltd (U.K) was used for the photo chemical radical polymerization reaction.
2.2. Preparation of Print Molecule (II)
A 25-ml round bottomed flask equipped with a drooping funnel and mechanical stirrer was charged with 2.5 mmol of sodium hydroxide (100 mg) in 3 mL water. 2.5 mmol of 1-propene-2-thiol were added drop wise and vigorous stirring upon 3 hrs of time at room temperature. The viscous solution of 1-propene-2-sodiumthiolate (I) were obtained and diluted with 3 mL water and kept on ice bath (0˚C). A 250 mL three necked round bottomed flask equipped with a mechanical stirrer, a thermometer and dropping funnel was charged with the 2.5 mmol of Fmoc-Cys(SSO3H)-OH. Disodium salt (1.17 g) in 50 ml water was cooled to 0˚C on ice bath. The cold 1-Propene-2-sodiumthiolate solution (I) were added rapidly, with vigorous stirring for 30 minutes resulted white thick precipitation washed with dilute HCl and extracted with ethyl acetate (3 × 150 mL). The organic layers were combined and dried briefly over anhydrous CaCO3 and then filtered. The filtrate was evaporated under rotary evaporator gave 2.15 mmol of (9-fluorenyl methoxy carbonyl)-S-(1-propene-2-thiol)-L-Cysteine (Fmoc- Cys(SCH2CHCH2)-OH) (II) as a color less oil. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.8 (2H, Ph-H, Fmoc); 7.7 (2H, Ph-H, Fmoc); 7.4 (2H, Ph-H, Fmoc); 7.3 (2H, Ph-H, Fmoc); 5.7 (1H, ethylene); 5.2 (2H, ethylene); 4.3 (4H, CH2); 4.02 (1H, CH); 3.3 (2H, CH2); 3.1 (1H, CHαCys); 2.9 (1H, CHαCys). 13C NMR (100 HZ, DMSO-d6) δ (ppm): 156.4 (C=O); 144.2 (Ph); 141.1 (Ph); 133.9 (1-ethylene); 128 (Ph); 125.5 (Ph); 119.1 (1-ethylene); 66.1 (CH2), 55.3 (Cα); 47 (CH); 41.2 (CH2); 40.3 (CH2).
2.3. Preparation of Dipeptide (III)
Fmoc-Phe-Cys(StBu)-CONH2 have been synthesized on batch wise manual solid phase peptide synthesizer. The synthesis was carried out in a 15 mL reactor loaded with the resin (0.65 mmol/g loading capacity) and washed with the DMF. The resin swollen in the DMF, subsequently Fmoc group of the resin deprotected with the 25% pipaeridine in DMF at 20 mints. 2.6 mmol of Fmoc-Cys(StBu)-OH (1.12 g) carboxylic group was activated with the 0.5 M HBTU/HOAt and 1M DIPEA. The activated amino acid coupled to the resin over 120 minutes on vigorous shaking and followed by washing the resin. The deprotection and coupling steps were repeated with the Fmoc-Phe-OH except the final fmoc cleavage. The corresponding dipeptide cleaved from the resin with the mixture of TFA-TIS-H2O in 95:2.5:2.5 was gently shaked for over 2.5 h. The resin was filtered off and filtrate lyophilized gave 0.4 mmol of Fmoc-Phe-Cys(StBu)-CONH2 as a white powder.
2.4. Preparation of Polymers
The polymers were prepared following general free radical polymerization procedure. In a 25 mL glass tube, 1.3 mmol of Fmoc-Cys(SCH2CHCH2)-OH (0.54 g) was dissolved in 26 mmol of DMF (2 mL) and 26 mmol of EGDMA (5.2 mL), 0.4 mmol of AIBN (65 mg) and 260 mmol of THF (21 mL) were added. The solution was sonicated in short period of time and purged with nitrogen gas for 5 min and then irradiated under UV lamp for 15 h at room temperature, additional 3 h at 80˚C. The resulted polymers grinded and dried overnight at 40˚C in vacuum incubator and named as MIPs. The non-imprinted polymers (NIP’s) were prepared through the same manner without template molecule and 1-propene-2-thiol used as a functional monomer.
2.5. Disulfide Reduction
In order to estimate the amount of un-reacted Fmoc-Cys(SCH2CHCH2)-OH, the corresponding polymer particles were extracted with 100 mL of MeOH by Soxhlet extraction for 12 h at 65˚C and extractives examined on analytical HPLC. Further polymer particles were suspended in 2 M LiAlH4 in THF and stirred reaction mixture over 18 h at room temperature, dilute H2SO4 and MeOH was added drop wise, then particles were filtered, supernatant was analyzed on HPLC.
2.6. Binding and Selectivity Studies
The binding and selectivity experiments of MIPs and NIPs were performed with batch-wise incubations and MeOH used as a solvent (unless water used) for Fmoc-Cys(SO3H)-OH. In order to examine the covalent recognition and binding strengths, in 50 ml centrifuge tubes, MIPs (0.65 g) or NIPs (0.625 g) were incubated with 10, 20, 30 and 40 µmoles of the Fmoc-Cys(SSO3H)-OH and Fmoc-Cys(StBu)-OH for 18 h at room temperature on shaking table. Then tubes were centrifuged and supernatant was filtered off and analyzed on HPLC. The free amount (F) of Fmoc-groups under 280 nm UV channel was calculated. To study the non-covalent recognition Fmoc-Leu-OH and for selectivity Fmoc-Phe-Cys(StBu)-OH, it was further incubated in the same procedure mentioned above.
2.7. HPLC Conditions
All samples were employed on analytical HPLC system with two eluent systems (Eluent A is 0.1% TFA in ACN and Eluent B is 0.1% TFA in milliq water). The elution rate is 1 ml/min over 35 min, starting conditions are eluent A (15%) and B (85%) and gradient reaching at 18 min to eluent A (85%) and B (15%). FROM 18 - 20 mints system reaches to initial state. The detection carried out at 280 nm UV channel.
3. Results and Discussion
3.1. Binding Sites
During the imprinting procedure stoichiometry of template and functional monomer plays an important role in formation of homogenous binding sites. Within self-assembly approach, only non-covalent interactions takes place and excess of functional monomer uses to shift equilibrium towards complex formation further leading to the formation of heterogeneous binding throughout polymeric networks. However in some extent advantageous reversible forming covalent imprinting such as disulfide imprinting template and functional monomer coupled 1:1 molar ratio results in reduce the heterogeneous binding sites. We have employed a disulfide bond between Fmoc-Cys(SH)-OH and 1-propene-2-thiol yielded the product (III), which is used as a precursor molecule for making synthetic receptor like structure for Fmoc-Cys(SH)-OH. To complete polymerization depends on various factors for instance amount of cross linker, reaction time, temperature and source of irradiation. So, we have set optimal conditions from the available sources and print molecule co-polymerized with the 20 times EGDMA under UV lamp for 15 h and additional 3 h at 80˚C. We subsequently focused on investigating the un-reacted print molecules, corresponding MIPs were extracted with MeOH and quantified 60% of the print molecule was not involved in reaction with remaining 40% of reacted print molecule (0.52 mmol) available in MIPs, i.e. maximum binding sites (Bmax) can be 40 µmoles/g. Furthermore, reductively disulfide bonds were cleaved by strong reducing agent like LiAlH4 in THF mixture. In order to examine the released product, supernatant was applied on HPLC and observed Fmoc groups under 280 nm UV channel.
3.2. Rebinding Studies
The key to the success of dynamic recognition of templates depends on reversible formation bonds ability to the binding sites. The nature of thiol-disulfide interconversion is more attractive and can be controlled at certain pH conditions, at pH ≥ 7 disulfides rapidly undergoes interconversions with thiols and at pH < 5 disulfides are stable. Although in our present studies, recognition was carried out in water and MeOH at neutral pH and room temperature, to study the recognition properties of polymers concentration, independent batch rebinding studies were performed. At neutral pH and room temperature, polymer particles of MIPs and NIPs were incubated with Fmoc-Cys(SO3H)-OH and Fmoc-Cys(StBu)-OH for 18 h on shaking table. The binding isotherms (Figure 1) shows that these two template molecules were recognized with high binding affinities in MIPs because of these templates involving thiol-disulfide exchange with the thiolic functional groups properly located in binding sites of imprinted polymers. These binding curves (as seen in Figure 1) are constantly saturated and reaches 100% binding affinities at each point, which corresponds that a predetermined number of binding sites created via imprinting process results in strong covalent recognition and is concentration independent with same binding affinities in organic and inorganic solvents. This is the more attractive to compare with non-covalent recognitions, many of the non-covalent recognitions based on hydrogen interactions, it can be achieved in aprotic organic
Figure 1. Showing the recognition properties of MIPs towards structurally related templates and NIP’s were showing poor recognition properties since NIPs were prepared without templates.
solvents and binding constants are varies in different concentrations. The work of Takeda et al. (2009) is taken into consideration and compared, they have been covalently imprinted a disulfide template as a print molecule with styrene and divinyl benzene, further reduced disulfides yielded thiolic containing binding sites were modified in to sulfonic acid form via post-imprinting modification in order to recognize the templates through non-covalently with hydrogen bonding and they have been reported binding curves were saturated in post-im- printing modified particles than the unmodified particles [29] . Their work can be compared with our present work where in they used Ethylene Glycol. Dimethacrylate (EGDMA) and covalently thiol-disulfide template recognition without post-imprinting modification.
Although the relative imprinting effect examined on binding curves of MIPs are comparable with the NIPs (Figure 2), the guest molecules show very poor binding with the NIPs even though NIPs contain the same functional group in linking to template recognition via thiol-disulfide exchange, which means that the imprinting process created structurally related shape specific cavities correspond to template molecules.
3.3. Covalent vs Non-Covalent Recognition
The results, so far have proved that imprinted MIPs recognize the guest molecules covalently via thiol-disulfide exchange. However, in connection with our present work, other range of non-covalent interactions involves in recognition process along with covalent interactions such as hydrogen bonding, vander-walls forces, electrostatic interactions etc. Due to the corresponding guest molecules that contain amide and hydroxyl groups are able to form hydrogen interactions with the thiol groups located in binding sites of MIPs and NIPs. Earlier, Mukawa et al. (2003) reported thiol groups in binding sites were able to form hydrogen-bonding with the hydroxyl groups containing guest molecules [38] . So, due to this effect, the entire bounded amount (B) is not only with the covalent interactions but also non-covalent interactions. In order to examine this effect, the template bounded MIP particles after batch rebinding experiments were extracted with MeOH and extractives are analyzed on HPLC. The 1% - 5% template only released from the total bounded MIP indicates that 95% - 99% of amount covalently bounded (Bcov) and remaining small amount 1% - 5% is non-covalently bounded (Bnon-cov). Further to conclude the covalent recognition (Bcov), disulfides were reduced with LiAlH4 from the corresponded MIPs and batch rebinding studies were performed same as mentioned above. The repeated batch rebinding studies also show the same binding curves and total bounded amount in repeated batch experiments (Brep) were compared with the first rebinding results (Figure 3).
3.4. Selectivity Studies
The versatile applications of MIPs are varied in chiral selectivity, separation science, synthetic receptors and sensors etc. So, relative selectivity of imprinted MIPs was investigated with template and structurally analogues molecules were compared with the NIPs. Furthermore, Fmoc-Leu-OH and Fmoc-Phe-Cys(tBu)-OH molecules were incubated with the MIPs and NIPs as above mentioned way. The relative binding strengths of recognition
Figure 2. Showing the MIPs template recognition through covalently Fmoc- Cys(SO3H)-OH of towards structurally related templates and NIPs were showing poor recognition properties since NIPs were prepared without templates.
of MIPs with various structurally related templates were found to be in Fmoc-Leu-OH and Fmoc-Phe- Cys(tBu)-OH (Figure 4). Although Fmoc-Leu-OH shows considerable amounts of binding affinities towards MIPs not in the NIPs, perhaps due to amide and hydroxyl groups in Fmoc-Leu-OH interacting with the hydrogen bonding with the thiol group binding sites containing structural similarity, the imprinted template has a missing the thiol functional group. Ourresults suggest that this covalently imprinted MIPs can be used in recognition of the corresponding templates covalently as well recognition of structurally related templates non-cova- lently. In addition, Takeda et al. (2009) and Takeuchi et al. (2006) reported that formed thiolic binding sites can be tuned with post-imprinting modification to obtained desirable recognition of templates without losing molecular memory. From Figure 4, it is clear that Fmoc-Phe-Cys(StBu)-OH shows very low binding affinities with MIPs as well in NIPs. These results strongly support the fact that MIPs are not only sensitive to thiol-disulfide interconversions but also sensitive to structurally related templates. The non-specific binding affinities of dipeptide are similar to MIPs and NIPs.
Figure 3. Flow chart of MIPs reproducibility with Fmoc-Cys(SO3H)-OH. Illustrates the amounts of templates, bounded amounts, template removals and rebounded amounts.
Figure 4. Showing relative binding strengths of MIPs recognition with various structurally related templates.
4. Conclusions and Future Perspectives
A wide range of imprinting solvents can be used depending on components solubility and type of interactions involved. In this work, we focused on imprinting and recognition carried out through disulfide interactions, excluding a range of non-covalent interactions such as functional groups amide and carboxyl groups present in the print molecule.
This work demonstrates the disulfide based covalent imprinting and feasibility of molecular recognition covalently (thiol-disulfide interconversion) and non-covalently (hydrogen bonding) in organic and inorganic solvents. The results proved covalent recognition showed higher binding affinities than non-covalent recognitions. The dynamic nature of reversible forming bonds in imprinted MIPs was reused multiple times by removing templates without loss of molecular memory and the same binding affinities were maintained. Further studies will be conducted with these MIPs as synthetic cystein receptors in ligand based binding assays in biological fluids to recognize the cystein and structurally related amino acids. To extent this peptidic receptors will be prepared by this disulfide based imprinting and used as analytical reagents to recognize the corresponding peptides in biological fluids to evaluate the diseases. Such site-directed thiol-disulfide exchange recognition model could be used in predicting the active sites of biological receptors and designing new drugs for receptor modulation.
Acknowledgements
HVRB would like to thank Kim Lambertsen Larsen and Reinhard Wimmer for useful discussions. Funding from Danish Council for Independent Research: Technology and Production Sciences―FTP Grant with File No. 09-064172 and Case No. 274-07-0405 is gratefully acknowledged.
Cite this paper
Harsha Vardhan ReddyBurri,DonghongYu, (2015) An Assay Study of Molecular Recognition of Amino Acids in Water: Covalent Imprinting of Cysteine. Journal of Biomedical Science and Engineering,08,805-814. doi: 10.4236/jbise.2015.812077
References
- 1. Dutt, S., Wilch, C. and Schrader, T. (2011) Artificial Synthetic Receptors as Regulators of Protein Activity. Chemical Communications, 47, 5376-5383.
http://dx.doi.org/10.1039/c0cc05812b - 2. Bongrand, P. (1999) Ligand-Receptor Interactions. Reports on Progress in Physics, 62, 921-968.
http://dx.doi.org/10.1088/0034-4885/62/6/202 - 3. Song, S., Qin, Y., He, Y., Huang, Q., Fan, C. and Chen, H.Y. (2010) Functional Nanoprobes for Ultrasensitive Detection of Biomolecules. Chemical Society Reviews, 39, 4234-4243.
http://dx.doi.org/10.1039/c000682n - 4. Hoshino, Y., Koide, H., Urakami, T., Kanazawa, H., Kodama, T., Oku, N. and Shea, K.J. (2010) Recognition, Neutrallzation, and Clearance of Target Ppetides in the Bloodstream of Living Mice by Molecularly Imprinted Polymer Nanoparticles: A Plastic Antibody. Journal of American Chemical Society, 132, 6644-6645.
http://dx.doi.org/10.1021/ja102148f - 5. Hoshino, Y., Kodama, T., Okahata, Y. and Shea, K.J. (2008) Peptide Imprinted Polymer Nanoparticles: A Plastic Antibody. Journal of American Chemical Society, 130, 15242-15243.
http://dx.doi.org/10.1021/ja8062875 - 6. Annamma, K.M. and Mathew, B. (2011) Design of 2,4-Dichlorophenoxyacetic Acid Imprinted Polymer with High Specificity and Selectivity. Materials Sciences and Applications, 2, 131-140.
http://dx.doi.org/10.4236/msa.2011.23017 - 7. Lee, K.P., Choi, S.H., Ryu, E.N., Ryoo, J.J., Park, J.H., Kim, Y. and Hyun, M.H. (2002) Preparation and Characterization of Cyclodextrin Polymer and Its High-Performance Liquid-Chromatography Stationary Phase. Analytical Sciences, 18, 31-34.
http://dx.doi.org/10.2116/analsci.18.31 - 8. Zhang, Y.M., Ren, H.X., Zhou, Y.Q., Luo, R., Xu, W.X. and Wei, T.B. (2007) Studies on the Anion Recognition Properties of Synthesized Receptors III: a Novel Thiourea-Based Receptor Constructed by Benzo-15-Crown-5 for Sensing Anions in a Strong Polar Solvent. Turkish Journal of Chemistry, 31, 327-334.
- 9. Verboom, W., Rudkevich, D.M. and Reinhoudt, D.N. (1994) Molecular Recognition by Artificial Receptors. Pure & Applied Chemistry, 66, 679-686.
http://dx.doi.org/10.1351/pac199466040679 - 10. Chin, J., Lee, S.S., Lee, K.J., Park, S. and Kim, D.H. (1999) A Metal Complex That Binds α-Amino Acids with High and Predictable Stereospecificity. Nature, 401, 254-257.
http://dx.doi.org/10.1038/45751 - 11. Scorrano, S., Mergola, L., Sole, R.D. and Vasapollo, G. (2011) Synthesis of Molecularly Imprinted Polymers for Amino Acid Derivates by Using Different Functional Monomers. International Journal of Molecular Sciences, 12, 1735-1743.
http://dx.doi.org/10.3390/ijms12031735 - 12. Kempe, M. (2000) Oxytocin Receptor Mimetics Prepared by Molecular Imprinting. Letters in Peptide Science, 7, 27- 33.
http://dx.doi.org/10.1007/BF02443559 - 13. Ramstrom, O., Nicholls, I.A. and Mosbach, K. (1994) Synthetic Peptide Receptor Mimics: Highly Stereoselective Recognition in Non-Covalent Molecularly Imprinted Polymers. Tetrahedron: Asymmetry, 5, 649-656.
http://dx.doi.org/10.1016/0957-4166(94)80027-8 - 14. Kirk, C., Jensen, M., Kjaer, C.N., Smedskjaer, M.M., Larsen, K.L., Wimmer, R. and Yu, D. (2009) Aqueous Batch Rebinding and Selectivity Studies on Sucrose Imprinted Polymers. Biosensors and Bioelectronics, 25, 623-628.
http://dx.doi.org/10.1016/j.bios.2009.01.021 - 15. Takeuchi, T., Murase, N., Maki, H., Mukawa, T. and Shinmori, H. (2006) Dopamine Selective Molecularly Imprinted Polymers via Post-Imprinting Modification. Organic and Biomolecular Chemistry, 4, 565-568.
http://dx.doi.org/10.1039/b514432a - 16. Shea, K.J. and Dougherty, T.K. (1986) Molecular Recognition on Synthetic Amorphous Surfaces. The Influence of Functional Group Positioning on the Effectiveness of Molecular Recognition. Journal of the American Chemical Society, 108, 1091-1093.
http://dx.doi.org/10.1021/ja00265a046 - 17. Sellergren, B. (1990) Molecular Recognition in Macroporous Polymers Prepared by a Substrate Analogue Imprinting Strategy. The Journal of Organic Chemistry, 55, 3381-3383.
http://dx.doi.org/10.1021/jo00297a074 - 18. Muratsugu, S. and Tada, M. (2013) Molecularly Imprinted Ru Complex Catalysts Integrated on Oxide Surfaces. Accounts of Chemical Research, 46, 300-311.
http://dx.doi.org/10.1021/ar300142p - 19. Haupt, K. (2010) Plastic Antibodies. Nature Materials, 9, 612-614.
http://dx.doi.org/10.1038/nmat2818 - 20. Kandimalla, V.B. and Ju, H. (2004) Molecular Imprinting: A Dynamic Technique for Diverse Applications in Analytical Chemistry. Analytical and Bioanalytical Chemistry, 380, 587-605.
http://dx.doi.org/10.1007/s00216-004-2793-9 - 21. Umpleby, II., Bode, M., Robert, J. and Shimizu, K.D. (2000) Measurement of the Continuous Distribution of Binding Sites in Molecularly Imprinted Polymers. Analyst, 125, 1261-1265.
http://dx.doi.org/10.1039/b002354j - 22. Turiel, E. and Esteban, A.M. (2004) Molecularly Imprinted Polymers: Towards Highly Selective Stationary Phases in Liquid Chromatography and Capillary Electrophoresis. Analytical and Bioanalytical Chemistry, 378, 1876-1886.
http://dx.doi.org/10.1007/s00216-003-2331-1 - 23. Chandler, D. (2005) Interfaces and the Driving Force of Hydrophobic Assembly. Nature, 437, 640-647.
http://dx.doi.org/10.1038/nature04162 - 24. Rajkumar, R., Warsinke, A., Mohwald, H., Scheller, F.W. and Katterle, M. (2007) Development of Fructosyl Valine Binding Polymers by Covalent Imprinting. Biosensors and Bioelectronics, 22, 3318-3325.
http://dx.doi.org/10.1016/j.bios.2007.03.001 - 25. White, S.R., Sottos, N.R., Geubelle, P.H., Moore, J.S., Sriram, S.R., Brown, E.N. and Viswanathan, S. (2001) Autonomic Healing Polymer Composites. Nature, 409, 794-797.
http://dx.doi.org/10.1038/35057232 - 26. Wulff, G., Heide, B. and Helfmeier, G. (1986) Molecular Recognition through the Exact Placement of Functional Groups on Rigid Matrices via a Template Approach. Journal of the American Chemical Society, 108, 1089-1091.
http://dx.doi.org/10.1021/ja00265a045 - 27. Otto, S., Furlan, R.L.E. and Sanders, J.K.M. (2002) Recent Developments in Dynamic Combinatorial Chemistry. Current Opinion in Chemical Biology, 6, 321-327.
http://dx.doi.org/10.1016/S1367-5931(02)00331-9 - 28. Otto, S., Furlan, R.L.E. and Sanders, J.K.M. (2000) Dynamic Combinatorial Libraries of Macrocyclic Disulfides in Water. Journal of the American Chemical Society, 122, 12063-12064.
http://dx.doi.org/10.1021/ja005507o - 29. Takeda, K., Kuwahara, A., Ohmori, K. and Taukechi, T. (2009) Molecularly Imprinted Tunable Binding Sites Based on Conjugated Prosthetic Groups and Ion-Paired Cofactors. Journal of the American Chemical Society, 131, 8833- 8838.
http://dx.doi.org/10.1021/ja9004317 - 30. Ramstrom, O. and Lehn, J.M. (2000) In Situ Generation and Screening of a Dynamic Combinatorial Carbohydrate Library against Concanavalin A. ChemBioChem, 1, 41-48.
http://dx.doi.org/10.1002/1439-7633(20000703)1:1<41::AID-CBIC41>3.0.CO;2-L - 31. Ogawa, K.I., Hyuga, M., Okada, T. and Minoura, N. (2012) Development of Lipid A-Imprinted Polymer Hydrogels That Selectively Recognize Lipopolysaccharides. Biosensors and Bioelectronics, 38, 215-219.
http://dx.doi.org/10.1016/j.bios.2012.05.028 - 32. Yano, K., Tanabe, K., Takeuchi, T., Matsu, J., Ikebukuro, K. and Karube, I. (1998) Molcularly Imprinted Polymers which Mimic Multiple Hydrogen Bonds between Nucleotide Bases. Analytica Chimica Acta, 363, 111-117.
http://dx.doi.org/10.1016/S0003-2670(98)00082-8 - 33. Wu, L., Gao, Y. and Wang, J. (2007) Synthesis, Application, and Molecular Recognition Mechanism Study of Phenylalanine Molecularly Imprinted Polymer. Analytical Letters, 40, 3129-3147.
http://dx.doi.org/10.1080/00032710701603835 - 34. Matsui, J., Nagano, J., Miyoshi, D., Tamaki, K. and Sugimoto, N. (2009) An Approach to Peptide-Based ATP Receptors by a Combination of Random Selection, Rational Design, and Molecular Imprinting. Biosensors and Bioelectronics, 25, 563-567.
http://dx.doi.org/10.1016/j.bios.2009.01.031 - 35. Hioki, H. and Still, W.C. (1998) Chemical Evolution: A Model System That Selects and Amplifies a Receptor for the Tripeptide (D)Pro(L)Val(D)Val. The Journal of Organic Chemistry, 63, 904-905.
http://dx.doi.org/10.1021/jo971782q - 36. Hong, J.I., Namgoong, S.K., Bernardi, A. and Still, W.C. (1991) Highly Selective Binding of Simple Peptides by a C3 Macrotricyclic Receptor. Journal of the American Chemical Society, 113, 5111-5112.
http://dx.doi.org/10.1021/ja00013a084 - 37. Lin, H.Y., Hsu, C.Y., Thomas, J.L., Wang, S.E., Chen, H.C. and Chou, T.C. (2006) The Microcontact Imprinting of Proteins: The Effect of Cross-Linking Monomers for Lysozyme, Ribonuclease A and Myoglobin. Biosensors and Bioelectronics, 22, 534-543.
http://dx.doi.org/10.1016/j.bios.2006.07.038 - 38. Mukawa, T., Goto, T., Nariai, H., Aoki, Y., Imamura, A. and Takeuchi, T. (2003) Novel Strategy for Molecular Imprinting of Phenolic Compounds Utilizing Disulfide Templates. Journal of Pharmaceutical and Biomedical Analysis, 30, 1943-1947.
http://dx.doi.org/10.1016/S0731-7085(02)00538-1
NOTES
*Corresponding author.






