Journal of Cancer Therapy
Vol. 4 No. 5A (2013) , Article ID: 31441 , 7 pages DOI:10.4236/jct.2013.45A002
Equivocal Differential Effect of NDRG1 in Human Ovarian Cancer Cells*
![]()
1Department of Human Biology, Faculty of Natural Sciences, University of Haifa, Haifa, Israel; 2Department of Molecular Genetics, Carmel Medical Center, Haifa, Israel; 3Pediatric Gastroenterology and Nutrition Unit, Carmel Medical Center, Haifa, Israel; 4Bruce Rappaport Faculty of Medicine, Technion-Israel Institute of Technology, Haifa, Israel.
Email: #ffares@sci.haifa.ac.il
Copyright © 2013 Tina Napso et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received March 5th, 2013; revised April 16th, 2013; accepted April 14th, 2013
Keywords: NDRG1; Human Ovarian Cancer; p21
ABSTRACT
Inactivation of tumor suppressor genes is a key factor in cancer regulation. N-myc downstream regulated gene 1 (NDRG1) is a tumor suppressor gene well known to be involved in carcinogenesis of numerous cancer types. The present study was designed to investigate the role of NDRG1 in human ovarian cancer, using SKOV-3 and SW626 (moderately and well differentiated cancer cells, respectively). Our results revealed that over-expressed NDRG1 significantly up-regulated the differentiation marker p21, in the ovarian cancer cell lines. This regulation led to decrease in cell viability and DNA synthesis rates in SW626 cells (83% and 89.5%, respectively). However, no effect on viability or on DNA synthesis was observed in SKOV-3 NDRG1-transfected cells. These findings prove that NDRG1 is indubitably functional in human ovarian cancer cells, as it up-regulated p21 expression. Nevertheless, this regulation showed differential effect on cell viability and DNA formation thus promoting the perception that downstream regulation of p21 could be inefficient in some cancer cells, a concept that needs to be further explored in order to understand its disability to play as regulator of cell cycle progression.
1. Introduction
N-myc downstream regulated gene-1 (NDRG1) has been identified in several independent in vitro studies of human cell lines and is known by various names such as Drg-1 [1], RTP [2], Cap43 [3], rit42 [4], and PROXY-1 [5]. The gene is known to be involved in cellular differentiation [6,7], proliferation and growth arrest [6], heavy metal response [3,8], DNA damage [4], hypoxia response [8], tumor progression and metastasis [9,10]. The role of NDRG1 in tumor progression, invasion and metastasis remains somewhat enigmatic and controversial. Based on the analytic interactome of NDRG1 performed on an androgen-responsive prostatic cell line, LNCaP, it can be argued that NDRG1 is functionally linked to the formation of the E-cadherin-β-catenin complex, a key player in cell adhesion, cytoskeleton and metastatic spreading [11].
Furthermore, NDRG1 is directly involved in endoplasmic reticulum stress response and nuclear transcription factor activation, consequently modulating the expression of multiple genes [11]. The role of NDRG1 expression in normal and cancer tissue remains controversial. Studies have shown down-regulation of NDRG1 in tumors of colon [12], breast [13] and prostate [4,9]. In contrast, NDRG1 up-regulation has been reported in hepatocellular carcinoma [14], renal cell carcinoma [15,16], and endometrial cancer [17]. NDRG1’s function and regulation in cancerous human ovary is not clear. We undertook this task and investigated two distinctly differentiated human ovarian cancer cell lines: SKOV-3 (moderately differentiated) and SW626 (well differentiated). It is shown that NDRG1 is expressed and is functional in these cells, as it was able to induce p21 expression. However, unlike in SW626 cells, NDRG1 failed to reduce cell viability and DNA synthesis in SKOV-3 cells. The present study sheds new light on NDRG1’s equivocal function in human ovarian cancer cells.
2. Methods
2.1. Chemical and Biological Reagents
Cell culture media and reagents were purchased from Biological Industries (Beit Haemek, Israel). The expression vector pcDNA3.1(+) was obtained from Invitrogen (San Diego, CA) and TransIT®-LT1 transfection reagent was purchased from Mirus Bio (Madison, WI). AntiNDRG1 and anti-Actin goat polyclonal antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and secondary antibody peroxidase-conjugated rabbit anti-goat IgG was obtained from Jackson ImmunoResearch (West Grove, PA). Additional chemicals were purchased from Sigma (St. Louis, MO).
2.2. Cell Culture
The human ovary cancer cell lines; SKOV-3 and SW626 were purchased from American Tissue Culture Collection (ATCC, Bethesda, MD). Cells were cultured at 37˚C in a humidified 5% CO2 atmosphere in McCoy’s 5A medium (SKOV-3) or in Leibowitz L-15 medium (SW626), supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin and 10% fetal calf serum. As the Leibowitz L-15 medium used for the SW626 cell line was devised for use in a free gas with atmospheric air, flasks containing this cell line were kept closed in order to prevent a mixture of CO2 and air, which would be detrimental to the cells.
2.3. Cloning of NDRG1
To create mammalian expression plasmid, total mRNA was isolated from normal human placenta tissue using RNAqueous®-4PCR Kit (Ambion, Austin, TX) according to manufacturer’s instructions. Briefly, tissue was mechanically disrupted and lysed with lysis buffer and lysate was applied to silica filter that selectively binds mRNA. Bound RNA was washed and eluted in nucleasefree water. Extracted RNA was reverse transcribed for cDNA preparation. cDNA of NDRG1 was amplified by PCR reaction using forward primer (5’-TTAGGCAGGTACCAGCAGGGACATG-3’), which harbored a KpnΙ restriction site, and reverse primer (5’- GAGGAGGGGGCCACTCGAGAGATCAGAGTC-3’), that included an XhoΙ restriction site. The PCR amplification was conducted with initial denaturation at 95˚C for 2 min, followed by 40 cycles of: 95˚C for 30 sec, 58˚C for 1 min and 72˚C for 3 min, and ended with a final synthesis step at 72˚C for 10 min. The cDNA of NDRG1 was cloned into eukaryotic expression vector pcDNA3.1(+) which contains the CMV promoter. Using KpnΙ and XhoΙ sites, the NDRG1 insert was fully sequenced to ensure that no mutation existed in the the cloned gene.
2.4. NDRG1 Transfection
The NDRG1 expression plasmid or the empty vector were transfected into human ovarian cancer cell lines using TransIT®-LT1 Transfection Reagent (Mirus Bio, Madison, WI) according to the manufacturer’s protocol. The expression of NDRG1 in transfected cells was tested 72 h following transfection by reverse transcription-PCR (RT-PCR), in order to confirm the presence of NDRG1 mRNA.
2.5. RT-PCR Analysis
Total RNA from transfected cells was extracted using RNAqueous®-4PCR Kit (Ambion, Austin, TX) according to manufacturer’s instructions. The quality of extracted RNA was examined by electrophoresis on a denaturing 1% agarose gel. Extracted RNA was subjected to a twostep RT-PCR protocol. For the creation of first strand, cDNA EZ-first Strand cDNA Isolation Kit (Biological Industries, Beit Haemek, Israel) was used. The cDNA products were then amplified by PCR. In brief, PCR amplification mixture contained cDNA, Taq Polymerase and PCR Buffer ×10 (Sigma, St. Louis, MO), dNTP’s (Promega, Madison, WI), ddH2O and primer pairs. The primers were obtained from Invitrogen (San Diego, CA). The thermal cycling procedure was identical to that described before: denaturation at 95˚C for 2 min, followed by 40 cycles of: 95˚C for 30 sec, 58˚C for 1 min, 72˚C for 1 min and an end-up synthesis step at 72˚C for 10 min.
2.6. Western Blot
Samples were resolved by SDS-PAGE on a 10% polyacrylamide gel, and blotted onto nitrocellulose membrane (Biological Industries, Beit Haemek, Israel). The membrane was blocked with 5% non-fat dry milk in TBST for 1 h at RT, followed by overnight incubation with primary antibody solution, diluted 1:2000 with 5% BSA in TBST. The membrane was washed three times in TBST and incubated for 1 h at RT with horseradish peroxidaseconjugated secondary antibody solution, diluted 1:10,000 with 1% non-fat dry milk in TBST. Following three consecutive washes in TBST, the antigen antibody complex was detected by the ECL procedure (Biological Industries, Beit Haemek, Israel) according to manufacturer’s instructions. The proteins were visualized using the ChemiDocTM XRS Gel Documentation System (BioRad, Hercules, CA).
2.7. Cell Viability
Transfected cells were seeded on 96-well plates at a cell density of 10,000 cells per well, when assayed 72 h and 96 h after transfection, and at a cell density of 4000 cells per well, when performed 120 h after transfection. Cell viability was determined using Cell Proliferation Assay kit (XTT) (Biological Industries, Beit Haemek, Israel). The absorbance of each sample was measured using an ELISA reader (Tecan, Spectra) at a wavelength of 450 nm with a reference absorbance of 620 nm.
2.8. DNA Synthesis
DNA synthesis was examined using the 5-Bromo-2’- deoxy-uridine Labeling and Detection Kit III (Roche, Manheim, Germany) which is based on the incorporation of BrdU into proliferating cells. Cells were seeded onto 96-well plates as described in the previous section. Cells were incubated with 10 µM BrdU for 4 h and fixed with 0.5 M ethanol/HCl, followed by incubation with nucleases to partially digest the DNA. Incorporated BrdU was detected with peroxidase labeled anti-BrdU-POD, followed by the addition of peroxidase substrate ABTS which produces a colored reaction product. The absorbance of each sample was measured using an ELISA reader at a wavelength of 405 nm with a reference absorbance of 492 nm.
2.9. Statistical Analysis
Each experiment was performed at least three times unless otherwise stated, and results are represented as means ± SD. Statistical analysis of data presented were performed using SPSS software. For the protein expression, cell viability and DNA synthesis analysis, twotailed student t-test was used in which significance was determined at p < 0.05.
3. Results
3.1. NDRG1 Exhibit Basal Expression Levels in SKOV-3 and SW626 Cells
Untreated cells revealed basal levels of NDRG1 expression at the mRNA level of both cell lines tested by RT-PCR (Figure 1). However, basal protein levels of NDRG1 were not detected in these cells, indicating a translation inhibition control that prevented protein formation (Figure 2). Over-expression of NDRG1, using the constructed plasmid pcDNA3.1/NDRG1, resulted in significantly increased levels of NDRG1’s mRNA and protein in the two cell lines. SKOV-3 cells showed six times fold of increase in mRNA, which was mirrored in protein expression, and SW626 cells exhibited 2.9 fold of increase in the mRNA level that comparably led to protein expression in these cells (Figures 1 and 2).
3.2. NDRG1 Up-Regulates p21 Expression in the Ovarian Cancer Cells
As NDRG1 is considered to be a differentiating factor in
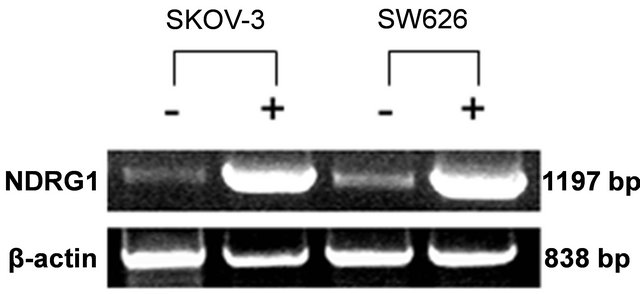
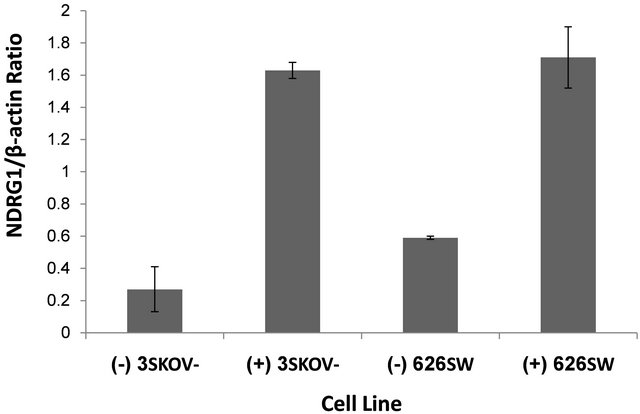
Figure 1. mRNA levels of NDRG1 in transfected ovarian cancer cells. pcDNA3.1+/NDRG1 were transfected into the SKOV-3 and SW626 cells and mRNA expression levels of the gene were detected by RT-PCR and quantified by densitometer analysis. Cells which transfected with pcDNA/ NDRG1 marked with (+) and control cells that were transfected with empty pcDNA3.1 marked with (−). Results are presented as average of three independent experiments (mean ± SD) and statistical significance was determined by two-tailed student’s t-test. Significance was determined as *p < 0.05.
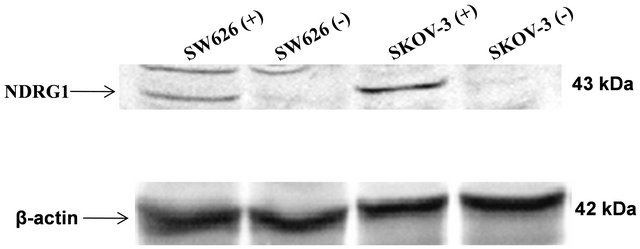
Figure 2. NDRG1 protein expression in transfected ovarian cell lines. Protein samples (80 µg) were separated on SDS-PAGE. Western blots were probed with anti-NDRG1 antibody or anti-β-actin antibody. Figure shown is representative of three experiments. For protein amount equalization, mouse anti-human β-actin monoclonal antibody was used. Bands detected were subtracted from background noise and density values calculated as percentage of β-actin serving as control. Statistical significance determined by a two-tailed student’s t-test. Significance was determined as *p < 0.05.
numerous cancer cell types, it was logical to explore the regulation of other differentiation factors in ovarian cancer cell lines. p21 and cytokeratine 8/18, both playing key elements in cell cycle progression, alteration and cell differentiation, were selected to be further analyzed following NDRG1 transfection. The mRNA expression levels of these two markers were examined 72 h post-transfection. Interestingly, SW626 cells, well-differentiated cancer cells by definition, were able to show basal levels of p21 expression (Figure 3), whereas SKOV-3 cells failed to demonstrate any basal expression of p21. While mRNA levels of p21 were found to be increased by NDRG1 over-expression in the cell, no differences were observed in mRNA levels of the c8/18 in pcDNA3.1/ NDRG1 transfected cells as compared to mock-transfection (data not shown), in both tested cell lines. It is worth noting that the fold of increase in p21 mRNA of the SKOV-3 cells was high and reached almost 700% fold of increase, whilst in SW626, which already showed high levels of p21 expression, the fold of increase was set on approximately 35% (Figure 3).
3.3. Effect of NDRG1 on Cell Viability and DNA Synthesis
Cell viability was measured using the XTT method at three time intervals post transfection (72, 96, 120 h). SKOV-3 cells, which showed increased levels of NDRG1
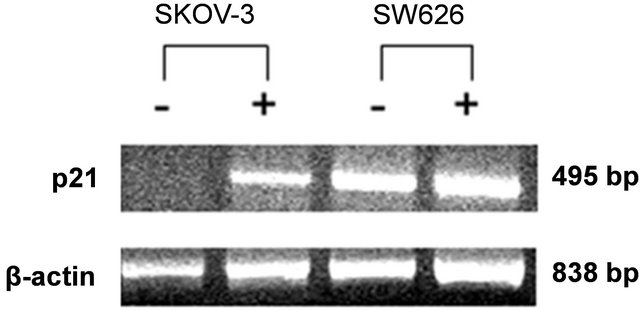
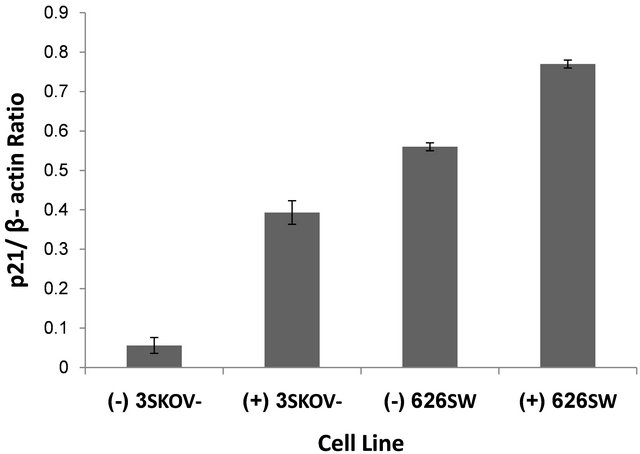
Figure 3. Effect of NDRG1 on the expression levels of p21. mRNA expression levels of the p21 gene following NDRG1 transfection were detected by RT-PCR and further quantified by densitometer analysis. Cells which transfected with pcDNA/NDRG1 marked with (+) and control cells that were transfected with empty pcDNA3.1 marked with (−). Results presented as average of three independent experiments (mean ± SD) and statistical significance determined by a two-tailed student’s t-test. Significance was determined as *p < 0.05.
expression alongside high increase in p21 expression, failed to demonstrate significant decrease neither in cell proliferation (Figure 4) nor in DNA synthesis (Figure 5). Alternatively, over-expressed NDRG1 in SW626 cells, with slight increase in p21 expression, was accompanied with significant reduction in cell proliferation and DNA synthesis levels, 96 h post transfection, (83% and 89.5%, respectively).
4. Discussion
In the present study, new basic aspects are enlightened concerning NDRG1 expression and function in two diversely differentiated human ovarian cancer cell lines. Basal amounts of NDRG1 mRNA exist in both the mod-
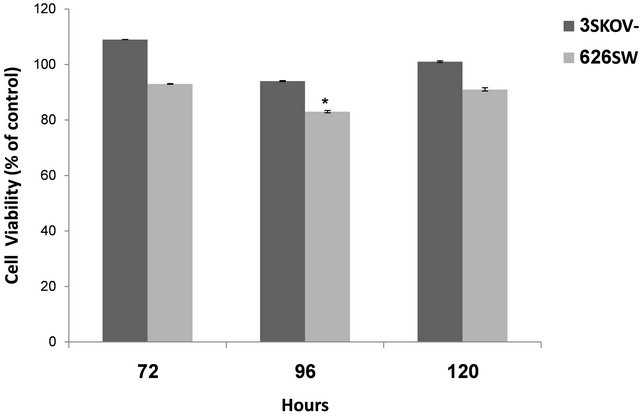
Figure 4. Cell viability following over-expression of NDRG1 in SKOV-3 and SW626 cells. Cell viability detected using the XTT method. Data presented are average of three experiments, conducted in four replicates (mean ± SD) and are expressed as percentages of the respective control. Statistical significance was determined by a two-tailed student’s t-test (*p < 0.05).
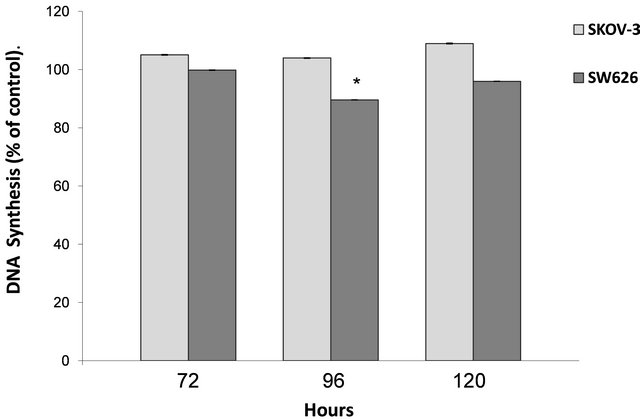
Figure 5. Effect of NDRG1 over-expression on DNA synthesis. DNA synthesis was detected by BrdU method. Data presented are averages of three independent experiments each conducted in four replicates (mean ± SD), and are expressed as percentages of respective control. Statistical significance was determined by a two-tailed student’s t-test (*p < 0.05).
erately and well differentiated cells (SKOV-3 and SW626, respectively). However, no basal protein was detected in the control cells. Following transfection, protein bands of NDRG1 at the expected molecular weight of 43 kDa were observed in both cell lines tested. Furthermore, the NDRG1 protein was proven to be functional in the transfected cells, as evidenced by p21 induction following over-expression of NDRG1. In contrast to the significant increase in NDRG1 mRNA expression following transfection, the surge in protein expression was much less than expected. The discrepancy between transcription and translation of the NDRG1 gene was reported earlier in normal ovarian human tissue [18]. It is proposed that there is a probability of an inhibitory effect existing on the translational pathway of NDRG1, which inhibits NDRG1 protein translation in ovarian cancer cells. This result might explain the failure of NDRG1 to operate as tumor suppressor gene along with ovarian cancer progression.
In addition, our findings illustrate that over-expression of NDRG1 induced p21 expression in transfected cells without affecting cell differentiation in SKOV-3, while causing only small amount of cell viability reduction in SW626 cells. A parallel phenomenon was shown in human ovarian cancer biopsies in which in 40% of the cases, p21 was expressed at a higher level than in normal ovarian epithelial cells, without any association to cellular proliferation inhibition [19]. Several reports also indicated that down-regulated p21 was related to cancerogenesis and survival rates of ovarian cancer [20,21]. For this reason, down-regulated or nonfunctional p21, as seen in the present study, would probably drive towards the same consequences. The loss of activity of p21 in ovarian cancer cells could be explained in several ways: 1) High mRNA levels of p21 might not be necessarily accompanied by p21 protein translation, since a mechanism of gene silencing such as miRNA may be involved; 2) The p21 mRNA levels might not be high enough for the p21 functionality, as it was proven earlier that p21’s cell cycle activity is dose dependent. While lower levels of p21 promote CDK/cyclin complex assembly, higher levels have an inhibitory effect [19]. On the other hand, as suggested by Barboule et al. [19], over-expressed cdk2 and cyclinA observed in ovarian IGROV1 cells allow the cells to escape p21 inhibitory activity; 3) p21 mutations might explain the loss of activity although this is quite unlikely, since mutations in p21 are rare and no such mutations were depicted in human ovarian adenocarcinoma [22]; 4) Various mechanisms are involved in the inactivation of p21, in which the inhibitory effect of p21 may be overcome by activated oncogenes or mutations in tumor suppressor genes in those cells; 5) An epigenetic phenomena such as DNA methylation and covalent modification of histones that alter the expression of p21, could be considered, since hyper-methylation of the CpG islands, hypoacetylated and hypermethylated histons and silencing of several promoters of tumor suppressor genes (p33ING1b, ARHI , PEG3, BRCA1, p16, RASSF1A), all were described earlier in human ovarian cancer progression [23-26]; 6) Finally, mutations in PTEN and β catenin, which were reported earlier [27-29], may influence p21 activity.
Most recently, a new relation between p21 and stem cell biology was unraveled [30,31]; p21 protects stem cells from acute genotoxic stress by preventing inappropriate cycling of acutely damaged cells. Moreover, oncogene expression induces DNA damage, which leads to reversible cell-cycle arrest and DNA repair in a p21-dependent cellular response. This defense mechanism is not operative in ovarian cancer cell lines, as first demonstrated in the present study.
5. Conclusions
The results of this study reveal that NDRG1 is functional in human ovarian cancer cell lines; however, it has an ambivalent effect on cell growth progression, making the pathway downstream NDRG1 occasionally non-functional. Further studies are needed in order to clarify the role of NDRG1 in human ovarian cancer as well as the molecular pathway of NDRG1 function.
With regard to p21 role, epigenetic and gene silencing mechanisms should be examined in order to reveal possible effects on p21 function. Elucidation of the functional molecular cascade operating downstream NDRG1, especially below the p21 block, can stimulate development of new diagnostic and therapeutic strategies in the fight against human ovarian cancer.
REFERENCES
- N. van Belzen, W. N. Dinjens, M. P. Diesveld, N. A. Groen, A. C. van der Made, Y. Nozawa, et al., “A Novel Gene Which Is Up-Regulated during Colon Epithelial Cell Differentiation and Down-Regulated in Colorectal Neoplasms,” Laboratory Investigation, Vol. 77, No. 1, 1997, pp. 85-92.
- K. Kokame, H. Kato and T. Miyata, “Homocysteine-Respondent Genes in Vascular Endothelial Cells Identified by Differential Display Analysis. GRP78/BiP and Novel Genes,” The Journal of Biological Chemistry, Vol. 271, No. 47, 1996, pp. 29659-29665. doi:10.1074/jbc.271.47.29659
- D. Zhou, K. Salnikow and M. Costa, “Cap43, a Novel Gene Specifically Induced by Ni2+ Compounds,” Cancer Research, Vol. 58, No. 10, 1998, pp. 2182-2189.
- S. K. Kurdistani, P. Arizti, C. L. Reimer, M. M. Sugrue, S. A. Aaronson and S. W. Lee, “Inhibition of Tumor Cell Growth by RTP/Rit42 and Its Responsiveness to p53 and DNA Damage,” Cancer Research, Vol. 58, No. 19, 1998, pp. 4439-4444.
- H. Park, M. A. Adams, P. Lachat, F. Bosman, S. C. Pang and C. H. Graham, “Hypoxia Induces the Expression of a 43-kDa Protein (PROXY-1) in Normal and Malignant Cells,” Biochemical and Biophysical Research Communications, Vol. 276, No. 1, 2000, pp. 321-328. doi:10.1006/bbrc.2000.3475
- D. Piquemal, D. Joulia, P. Balaguer, A. Basset, J. Marti and T. Commes, “Differential Expression of the RTP/ Drg1/Ndr1 Gene Product in Proliferating and Growth Arrested Cells,” Biochimica et Biophysica Acta, Vol. 1450, No. 3, 1999, pp. 364-373. doi:10.1016/S0167-4889(99)00056-7
- Y. Taketomi, T. Sugiki, T. Saito, S. Ishii, M. Hisada, T. Suzuki-Nishimura, et al., “Identification of NDRG1 as an Early Inducible Gene during in Vitro Maturation of Cultured Mast Cells,” Biochemical and Biophysical Research Communications, Vol. 306, No. 2, 2003, pp. 339-346. doi:10.1016/S0006-291X(03)00942-2
- K. Salnikow, M. V. Blagosklonny, H. Ryan, R. Johnson and M. Costa, “Carcinogenic Nickel Induces Genes Involved with Hypoxic Stress,” Cancer Research, Vol. 60, No. 1, 2000, pp. 38-41.
- S. Bandyopadhyay, S. K. Pai, S. C. Gross, S. Hirota, S. Hosobe, K. Miura, et al., “The Drg-1 Gene Suppresses Tumor Metastasis in Prostate Cancer,” Cancer Research, Vol. 63, No. 8, 2003, pp. 1731-1736.
- S. Bandyopadhyay, S. K. Pai, S. Hirota, S. Hosobe, Y. Takano, K. Saito, et al., “Role of the Putative Tumor Metastasis Suppressor Gene Drg-1 in Breast Cancer Progression,” Oncogene, Vol. 23, No. 33, 2004 , pp. 5675- 5681. doi:10.1038/sj.onc.1207734
- L. C. Tu, X. Yan, L. Hood and B. Lin, “Proteomics Analysis of the Interactome of N-MYC Downstream Regulated Gene 1 and Its Interactions with the Androgen Response Program in Prostate Cancer Cells,” Molecular & Cellular Proteomics, Vol. 6, No. 4, 2007, pp. 575-588. doi:10.1074/mcp.M600249-MCP200
- R. J. Guan, H. L. Ford, Y. Fu, Y. Li, L. M. Shaw and A. B. Pardee, “Drg-1 as a Differentiation-Related, Putative Metastatic Suppressor Gene in Human Colon Cancer,” Cancer Research, Vol. 60, No. 3, 2000, pp. 749-755.
- S. Bandyopadhyay, S. K. Pai, S. Hirota, S. Hosobe, T. Tsukada, K. Miura, et al., “PTEN Up-Regulates the Tumor Metastasis Suppressor Gene Drg-1 in Prostate and Breast Cancer,” Cancer Research, Vol. 64, No. 21, 2004, pp. 7655-7660. doi:10.1158/0008-5472.CAN-04-1623
- M. S. Chua, H. Sun, S. T. Cheung, V. Mason, J. Higgins, D. T. Ross, et al., “Overexpression of NDRG1 is an Indicator of Poor Prognosis in Hepatocellular Carcinoma,” Modern Pathology, Vol. 20, No. 1, 2007, pp. 76-83. doi:10.1038/modpathol.3800711
- K. Masuda, M. Ono, M. Okamoto, W. Morikawa, M. Otsubo, T. Migita, et al., “Downregulation of Cap43 Gene by von Hippel-Lindau Tumor Suppressor Protein in Human Renal Cancer Cells,” International Journal of Cancer, Vol. 105, No. 6, 2003, pp. 803-810.
- A. Nishie, K. Masuda, M. Otsubo, T. Migita, M. Tsuneyoshi, K. Kohno, et al., “High Expression of the Cap43 Gene in Infiltrating Macrophages of Human Renal Cell Carcinomas,” Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, Vol. 7, No. 7, 2001, pp. 2145-2151.
- J. Chen, S. Li, Z. Yang, G. Lu and H. Hu, “Correlation between NDRG1 and PTEN Expression in Endometrial Carcinoma,” Cancer Science, Vol. 99, No. 4, 2008, pp. 706-710. doi:10.1111/j.1349-7006.2008.00749.x
- P. Lachat, P. Shaw, S. Gebhard, N. van Belzen, P. Chaubert and F. T. Bosman, “Expression of NDRG1, a Differentiation-Related Gene, in Human Tissues,” Histochemistry and Cell Biology, Vol. 118, No. 5, 2002, pp. 399-408. doi:10.1007/s00418-002-0460-9
- N. Barboule, V. Baldin, S. Jozan, S. Vidal and A. Valette, “Increased Level of p21 in Human Ovarian Tumors Is Associated with Increased Expression of CDK2, Cyclin A and PCNA,” International Journal of Cancer, Vol. 76, No. 6, 1998, pp. 891-896.
- F. Terauchi, A. Okamoto, T. Nagashima, Y. Kobayashi, T. Moritake, Y. Yamamoto, et al., “Clinical Significance of p21(WAF1/CIP1) and p53 Expression in Serous Cystadenocarcinoma of the Ovary,” Oncology Reports, Vol. 14, No. 2, 2005, pp. 363-368.
- L. G. Buchynska, I. P. Nesina, N. P. Yurchenko, O. O. Bilyk, V. N. Grinkevych and V. S. Svintitsky, “Expression of p53, p21WAF1/CIP1, p16INK4A and Ki-67 Proteins in Serous Ovarian Tumors,” Experimental Oncology, Vol. 29, No. 1, 2007, pp. 49-53.
- A. Marchetti, F. Buttitta, S. Pellegrini, G. Bertacca, A. Lori and G. Bevilacqua, “Absence of Somatic Mutations in the Coding Region of the waf1/cip1 Gene in Human Breast, Lung and Ovarian Carcinomas—A Polymorphism at Codon-31,” International Journal of Oncology, Vol. 6, No. 1, 1995, pp. 187-189.
- M. Esteller, “Epigenetics in Cancer,” The New England Journal of Medicine, Vol. 358, No. 11, 2008, pp. 1148- 1159. doi:10.1056/NEJMra072067
- I. Ibanez de Caceres, C. Battagli, M. Esteller, J. G. Herman, E. Dulaimi, M. I. Edelson, et al., “Tumor Cell-Specific BRCA1 and RASSF1A Hypermethylation in Serum, Plasma, and Peritoneal Fluid from Ovarian Cancer Patients,” Cancer Research, Vol. 64, No. 18, 2004, pp. 6476-6481. doi:10.1158/0008-5472.CAN-04-1529
- D. H. Shen, K. Y. Chan, U. S. Khoo, H. Y. Ngan, W. C. Xue, P. M. Chiu, et al., “Epigenetic and Genetic Alterations of p33ING1b in Ovarian Cancer,” Carcinogenesis, Vol. 26, No. 4, 2005, pp. 855-863. doi:10.1093/carcin/bgi011
- W. Feng, R. T. Marquez, Z. Lu, J. Liu, K. H. Lu, J. P. Issa, et al., “Imprinted Tumor Suppressor Genes ARHI and PEG3 Are the Most Frequently Down-Regulated in Human Ovarian Cancers by Loss of Heterozygosity and Promoter Methylation,” Cancer, Vol. 112, No. 7, 2008, pp. 1489-1502. doi:10.1002/cncr.23323
- K. Selvendiran, L. Tong, S. Vishwanath, A. Bratasz, N. J. Trigg, V. K. Kutala, et al., “EF24 Induces G2/M Arrest and Apoptosis in Cisplatin-Resistant Human Ovarian Cancer Cells by Increasing PTEN Expression,” The Journal of Biological Chemistry, Vol. 282, No. 39, 2007, pp. 28609-28618. doi:10.1074/jbc.M703796200
- H. Wu, S. Wang, D. Weng, H. Xing, X. Song, T. Zhu, et al., “Reversal of the Malignant Phenotype of Ovarian Cancer A2780 Cells through Transfection with WildType PTEN Gene,” Cancer Letters, Vol. 271, No. 2, 2008, pp. 205-214. doi:10.1016/j.canlet.2008.06.018
- Y. Takei, Y. Saga, H. Mizukami, T. Takayama, M. Ohwada, K. Ozawa, et al., “Over Expression of PTEN in Ovarian Cancer Cells Suppresses I.P. Dissemination and Extends Survival in Mice,” Molecular Cancer Therapeutics, Vol. 7, No. 3, 2008, pp. 704-711. doi:10.1158/1535-7163.MCT-06-0724
- Z. Ju, A. R. Choudhury and K. L. Rudolph, “A Dual Role of p21 in Stem Cell Aging,” Annals of the New York Academy of Sciences, Vol. 1100, 2007, pp. 333-344. doi:10.1196/annals.1395.036
- A. Viale, F. De Franco, A. Orleth, V. Cambiaghi, V. Giuliani, D. Bossi, et al., “Cell-Cycle Restriction Limits DNA Damage and Maintains Self-Renewal of Leukaemia Stem Cells,” Nature, Vol. 457, No. 7225, 2009, pp. 51-56. doi:10.1038/nature07618
NOTES
*Conflict of interest statement: The authors declare that there are no conflicts of interest.
#Corresponding author.

