Advances in Bioscience and Biotechnology
Vol.07 No.06(2016), Article ID:67762,12 pages
10.4236/abb.2016.76025
Mechanics of Centriole Microtubules
Ronald L. Huston
Department of Mechanical and Materials Engineering, University of Cincinnati, Cincinnati, OH, USA

Copyright © 2016 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 3June 2016; accepted 25 June 2016; published 28 June 2016
ABSTRACT
This is a review paper describing recent findings about the physical properties of centriolar microtubules. Microtubules are the principal structures making up the centrioles. The centrioles in turn are the principal agents in cell duplication and division (mitosis). The microtubules are seen to be long hollow cylinders: approximately 400 nm in length, with a 24 nm outside diameter, and a 5 nm wall thickness. Within the centrioles, the microtubules are arranged into nine parallel sets of triplets―thus numbering 27 parallel cylinders per centriole. Each normal eukaryotic (human and animal) cell, not in mitosis, has two perpendicular centrioles connected at their proximal (base) ends. During mitosis, these two become four, resulting in a total of 108 centriolar microtubules. The structure of the microtubules themselves is found to consist of 13 parallel filaments making up the cylinder walls. The filaments are composed of approximately 40 and β-tubulin connected end-to-end with their proximal (base) ends anchored in γ-tubulin. The longitudinal vibrations of the filaments are believed to create an electro-magnetic field within the cell which plays an important role in mitosis.
Keywords:
Microtubules, Centrioles, Tubulin, Cell Magnetism

1. Introduction
Adjacent to the nucleus in all human and animal (eukaryotic) cells is a cloud of electron dense proteins known as the “centrosome” [1] - [13] . Within the centrosome, and as a part of the centrosome, is a pair of centrioles―hollow cylinders, perpendicular to each other, and connected at their base (proximal) ends. The perpendicularity is due to one of the centrioles (the “daughter”) developing and growing on the side, at the base of the other centriole (the “mother”). The daughter is only about 80% as long as the mother [1] [14] - [17] . But with the daughter adjoining the mother at its base, the pair forms a perpendicular structure with nearly equal legs.
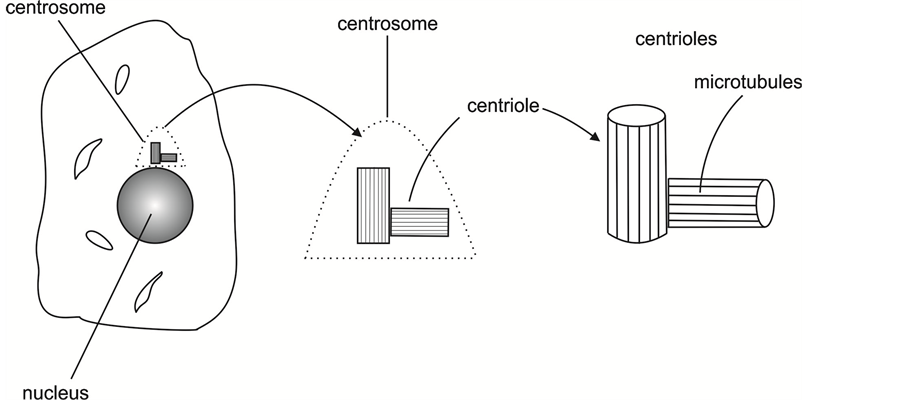
Figure 1 presents drawings of the nucleus, centrosome, and centrioles.
Each centriole cylinder consists of nine sets of triplets of microtubules, spaced evenly around the circumference,
Figure 1. Drawings of the nucleus, the centrosome, and the centrioles.
Figure 2. A centriole composed of microtubules (MTs).
and extending along the length of the centriole cylinder (see Figure 2). These microtubules (27 in total) form the subject matter of this paper. For efficiency in reading and writings, “microtubules” is abbreviated as “MT”.
Each MT is itself a hollow cylinder having the same length as the centriole but with only a small fraction of the centriole diameter. The sets of triplets of MTs have the appearance of “blades” around the centriole circumference, and the entire collection of MTs may be compared to heating pipes of a boiler. But unlike a boiler, and unlike other biological organs, the centriole set of MTs has no membrane cover.
The centrioles via their MTs are the principal agents of cell duplication and separation (mitosis): at the beginning of mitosis, the centrioles duplicate each other and at the same time the DNA of the nucleus is copied. The centriole pair becomes two pair, and then they separate with the newly formed pair migrating to the opposite side of the nucleus and establishing a new centrosome at that opposite site.
Once the nucleus has diametrically opposed centrosomes, the MTs of the embedded centrioles elongate and spread out about the nucleus and form the “mitotic spindle”. At the same time, the nuclear membrane, midway between the opposing centrioles, begins to soften and shrink away. The ends of the elongated microtubules then seek to connect to anchors, known as “kinetochores” [1] [2] [18] , within the dividing nucleus. Finally the kinatochore-connected MTs begin to shorten, pulling the nucleus apart.
Each half of the divided nucleus together with its centrosome then separate, divide the remainder of the cell interior (the “cytoplasm”) and thereby form two new cells.
While much remains to be discovered about each of these processes, as noted earlier, the focus herein is the mechanics of the MTs as structural elements of the centrioles.
The balance of the paper is divided into seven sections with the following section providing an overall description of the MT geometry. The next two sections describe the development of MTs and their resulting physical properties. The subsequent section (Section 5) then presents what appears to be currently known about MT dynamics. This is followed by a section indicating how MT development and behavior can go awry leading to cell death and disease―including cancer. The final section provides a brief discussion and concluding remarks.
2. Microtubule (MT) Geometry
The centriole cylinders are approximately 400 to 500 nm long and their diameters are approximately 200 nm [1] [2] . With the microtubules forming the length of the centrioles, the MTs are thus also 400 to 500 nm long, but their diameters are much smaller: The outside diameter is approximately 24 nm and the inside diameter is approximately 15 nm―thus creating a wall thickness of approximately 5 nm. (The use of the adverb “approximately” is due to the minute sizes of the structures. That is, precise measurements are elusive.)
The walls of the MT are composed of 13 filaments running lengthwise along the microtubule. These filaments in turn are composed of approximately 40 tubulin dimers connected end-to-end as in Figure 3.
The dimers are composed of alpha and beta-tubulin proteins with the alpha-tubulin toward the proximal end of the microtubule filament, and with a negative charge. Conversely the beta-tubulin is toward the distal end of the MT filament and with a positive charge.
Figure 4 shows a drawing of an alpha/beta-tubulin dimer.
With the α and β-tubulin being coiled proteins approximately 5 nm in thickness (the thickness of the microtubule wall), the dimer length is approximately 10 nm.
With the MT filament being approximately 400 to 500 nm long, there are approximately 40 to 50 dimers in a filament.
There are differing views on the longitudinal arrangement and positioning of adjacent filaments [19] - [22] : Some investigators suggest that the longitudinal position of the filaments is such that each alpha-tubulin is adjacent above and below, and to each side, by a beta-tubulin, and vice-versa. This arrangement presents a 45 degree spiral pattern on the wall of the microtubule as in Figure 5.
Figure 3. Microtubule (MT) model.
Figure 4. α/β dimer.
Figure 5. A spiral arrangement of α and β-tubulin.
Figure 6. Rings of α and β-tubulin.
Others have suggested a similar arrangement but with spiraling at a lesser angle. In the extreme case, with no spiral angle there are simply alternate rings of α and β-tubulin, as in Figure 6.
A difficulty with the spiral arrangements where each α-tubulin is adjacent on both sides with a β-tubulin (and vice-versa) is that with 13 filaments, the pattern becomes disrupted as it completes the MT circumference. That is, with an odd number of filaments (13), if say the α-tubulin is assigned an odd number (and the β-tubulin an even number) about the circumference, then the α-tubulin numbers: 1 and 13 come together so that two α-tubulin are adjacent (see Figure 7).
A difficulty with the ring arrangement of Figure 6 is that it is inconsistent with the dimer formation and the construction of the microtubule wall via γ-tubulin, as discussed in the following section.
Another, and apparently more plausible, view of the filament pattern is that with the filaments consisting of α/β-tubulin dimers, connected end-to-end, each filament can move longitudinally and independently of adjacent filaments. Also, the external surfaces of the filaments are smooth enabling the relative movement [23] .
Finally, the MTs making up the blades of the centrioles have differing lengths. The longest, labelled “A”, is closest to the axis; the shortest; labelled “C” is nearest to the outside wall; and then the middle one, known as “B”, has intermediate length (see Figure 8).
3. Microtubule (MT) Development
In the cell duplication cycle, before the separation begins, the DNA and the centrioles duplicate during a period known as the “S-phase” (this is during an intermediate phase of the cell cycle) [1] [2] [17] [24] . The stimuli for both the DNA separation and the cell duplication occur at the same time.
Much remains to be discovered. But in general terms, the centriole duplication occurs as follows: A collection of proteins within the centrosome act in a serial manner to form new microtubules. As a result of this process, the centrosome minus the centriole pair is known as the “microtubule organizing center” (MTOC). The proteins
Figure 7. End view of microtubule showing disrupted spiral pattern.
Figure 8. Varying MT filament lengths.
immediately surrounding the centrioles are known as “pericentriolar material”.
Of the more than 300 proteins in the MTOC only a few appear to play prominent roles in assembling the new centrioles with the new microtubules. These are: Plk1 and Plk4 (polo-like kinases one and four). Plk4 is also known as “SAK”; Asl (asterless); SAS4; SAS6; ZYG-1; STIL (SIL); p53; separase; Cdk2; cyclin E; and α, β, and γ-tubulin [24] .
The duplication of the centrioles, and thus also the creation of new MTs, begins with asterless making a small deposit on an outer MT near the base (the proximal end) of a centriole blade (on a “C-microtubule”). The asterless deposit then recruits Plk4 to form a patch atop and across the deposit. Next, this Plk4 patch, with the aid of SIL, recruits SAS6 in turn atop and across the Plk4 patch.
While this description of the development of the protein structure may not be precise, what is known is that identical base structures are built at the proximal ends of both the mother and daughter centrioles.
After this protein base structure is established, the SAS6 expands symmetrically growing nine outward spokes tangent to the centriole surface. While these spokes are growing they recruit γ-tubulin along their lengths. Finally, the γ-tubulin deposits form the basis for the development of three new MTs along each of the nine spokes.
The complexity of this structure requires regulation to keep it from going awry. This regulation, and to some extent the protein recruitment, is aided by still other proteins: SAS4; Bld10/Cep135; ZYG-1; p53; and probably others as well.
The γ-tubulin deposits on the SAS6 spokes are believed to be suppliers of the α/β-tubulin dimers from the MTOC for construction of the MT [25] - [27] . Although, as before, the details are still somewhat sketchy, the γ-tubulin deposit appears to take the shape of a ring with overlapping ends―having the appearance of a lock-washer [19] . There are 13 ports along this ring through which the α/β dimers are inserted from the MTOC to form the microtubule filaments. This process is known as the “protofilament method” [19] [22] [28] [29] .
The γ-tubulin ring is known as the “γ-tubulin ring complex” (γ-TuRC) and/or as “gammasome” [20] [21] . The 13 ports are composed of another tubulin known as the “γ-tubulin small complex” (γ-TuSC). The γ-TuSC is like cylindrical channels perpendicular to the γ-TuRC.
In another process of microtubule nucleation, known as the “template method,” the α/β tubulin dimers are inserted through the γ-Tu-SC pores, but then pushed along the γ-TuRC forming a spiral pattern for the microtubule.
There are unresolved issues with each of these methods: 1) In the protofilament method, if each protofilament is independent of its neighbor, an intact microtubule filament 400 nm long would be like a cantilever beam with an 80 to 1 aspect ratio. 2) In the template method, the spiraling of the α/β-tubulin dimers does not conform to the 13 filament structure of the microtubule without a seam of duplicate tubulin, and thus disrupting the symmetry. Also, the spiraling does not allow for independent longitudinal movement of the filaments―a phenomenow believed to be necessary for the microtubule dynamics and the creation of electro-magnetic fields.
Finally, there are two other tubulins contributing to the development of the microtubules: δ and ε-tubulin. δ-tubulin is instrumental in the development of the three microtubule blades in each of the nine blades of the centriole [30] [31] , while ε-tubulin is instrumental in the development and maintenance of the paricentriolar material [23] .
4. Physical Properties of Microtubules (MTs)
In spite of measurement difficulties, there is still considerable data about the structural and electrical properties of the alpha/beta-tubulin dimers and the complete microtubule structure.
As noted earlier an MT has the shape of a hollow cylinder, approximately 400 nm long, with inside and outside diameters of 15 and 25 nm respectively. The wall thickness is thus approximately 5 nm―the approximate thickness of an α/β-tubulin dimer.
The outside wall of an MT cylinder is smooth―presumably to facilitate MT filament movement within the centrosome. The inside wall, however, does not possess the same degree of smoothness [23] .
From a structural perspective MTs are relatively stiff in tension and compression but weak in shear (torsion) [32] . The elastic modulus E is estimated to be approximately 109 Pa (N/m2) [33] . From the known geometric dimensions the second moment of area I of the cross-section is calculated to be: 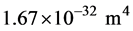 [34] . Then the flexural rigidity EI becomes approximately:
[34] . Then the flexural rigidity EI becomes approximately: 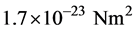 [35] .
[35] .
Equation (1) summarizes these results:
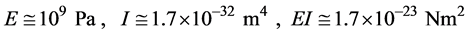 . (1)
. (1)
For structural analysis, there are two other parameters which may be of interest: 1) Poisson’s ratio ν; and 2) the shear modulus G. Poisson’s ratio is the ratio of transverse (lateral) strain to longitudinal strains, and is also known as the “transverse-contraction ratio”. For stiff materials ν is approximately: 0.3. The shear modulus, also known as the “modulus of rigidity”, may then be evaluated using the expression [34] :
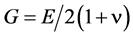 . (2)
. (2)
Then G becomes:
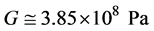 . (3)
. (3)
Experiments also reveal electrical properties of the MTs: The MTOC is thought to be composed of electron-dense material [4] [36] - [38] . Therefore the MT base is given a negative charge. Consequently, the α/β dimer has a negative charge at the base (or proximal) end of the α-tubulin and a positive charge at the distal end of the β-tubulin [39] [40] .
It is believed that the electro-magnetic properties of a cell occur due to independent longitudinal vibrations of the MT filaments, thus oscillating the positive and negative charges of the α/β tubulin dimers [23] [41] - [44] .
For a single dimer, the dipole moment is of the order of 10−26 Cm (Coulomb meters) [43] - [46] .
Finally, the masses of the α-tubulin and the β-tubulin proteins are essentially the same at approximately 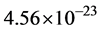 kg, so that the dimer mass is approximately
kg, so that the dimer mass is approximately 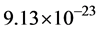 kg.
kg.
5. Microtubule (MT) Dynamics
MTs are the most dynamic of all the cell organelles. They have continuous movement―analogous to respiration and blood flow. MT dynamics occur by the supply of the α/β dimers being recruited from the MTOC and then inserted at the base of the MT by the γ-TuRC. Each dimer insertion by the γ-TuRC tends to lengthen the MT. But then, at the distal end, the dimers break away and go into the cytoplasm. This breakup tends to shorten the MT.
The continual lengthening and shortening of the MTs is known as “dynamic instability”, and it is characterized in four phases: 1) growth, 2) shrinkage, 3) catastrophe, and 4) rescue [47] - [50] .
Growth is simply MT elongation and shrinkage is MT shortening. Catastrophe is the transition from growth to shrinkage; and correspondingly rescue is the transition from shrinkage to growth.
Growth and shrinkage may occur alone or simultaneously. Catastrophe occurs when the growth either stops or the growth rate is less than the shrinkage rate. Similarly, rescue occurs when the shrinkage either stops or the shrinkage rate is less than the growth rate.
Superpose upon the ongoing lengthening and shortening of the MTs as a whole, the individual filaments oscillate longitudinally. Due to the small tubulin mass the oscillation frequency is high: Estimates range from 107 to 1010 Hz [51] - [53] .
The 13 filaments making up the MT wall are believed to keep interactive vibration modes between the filaments to a minimum: 13 is a prime number immediately following 12. With 12 being the product of 4 and 3, if there were only 12 filaments, there would tend to be close coupling of the vibrations across the diameter and at 50˚, 60˚, and 90˚.
Finally it is believed that the γ-tubulin in some way controls the MT dynamics in general and the vibration in particular. The mechanism of this control, however, is unknown.
6. Microtubules (MTs) Going Awry
In view of all that is involved in MT geometries and their development, microtubule properties, and microtubule function, it is easy to envision something (or many things) going awry. When this happens, the centriole is negatively affected, leading possibly to cell death, or worse, to ongoing disease such as tumorigenesis or cancer.
Among the most common defects is a disruption in centriole geometry due to an overgrowth of MTs. Perhaps the most dangerous is the case of “flowering” [6] [54] where there is an excess in the number of centrioles. Figure 9 shows a conceptual imaging of flowering where a mother centriole may have two or more daughters simultaneously. When this happens the cell has extra or supernumerary centrioles.
Figure 9. A mother centriole with two daughters (“flowering”).
Virtually all cancer cells contain supernumerary centrioles [3] [10] [38] [55] - [86] . These extra centrioles tend to cluster together―presumably due to electromagnetic attraction. In an earlier paper [6] , it is suggested that these centriole clusters create an enhanced electromagnetic field and that this enhanced field can then serve as a biomarker for cancer cell identification and therapy.
Much additional work is needed to validate these assertions and to explore their application.
7. Conclusions
This brief review, like many reviews, raises questions with virtually all of the assertions. Admittedly, many of the assertions are based upon limited experimental data. The experimental limitations are primarily due to the minute size of the microtubules, and even more so for the smaller tubulin dimers. Improvements in experimental techniques, however, are likely to clarify many of the unresolved issues.
If, however, the findings and assertions presented herein are generally correct and indicative of microtubule physics, these findings and assertions could be useful in pointing the way to additional research.
Perhaps the most interesting finding is the apparent development of electromagnetic fields via MT oscillations. This needs to be explored in greater detail. If this further exploration leads to finding an enhanced field about centriole clusters in cancer cells, the enhanced electromagnetic field could serve as a biomarker for cancer cells, leading in terms to better imaging and therapies.
Acknowledgements
Support for this research was provided by the Schafer Foundation for Centriolar Research and is sincerely appreciated. Also, the consultation and encouragement of Bardyl Tirana and Roger Adelman is acknowledged.
Cite this paper
Ronald L. Huston, (2016) Mechanics of Centriole Microtubules. Advances in Bioscience and Biotechnology,07,266-277. doi: 10.4236/abb.2016.76025
References
- 1. Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K. and Watson, J.D. (1994) Molecular Biology of the Cell. 3rd Edition, Garland Publishing, New York.
- 2. Hardin, J., Bertoni, G. and Kleinsmith, L.J. (2015) Becker’s World of the Cell. 8th Edition, Benjamin Cummings, Boston.
- 3. Kramer, A., Neben, K. and Ho, A.D. (2002) Centrosome Replication, Genomic Instability and Cancer. Leukemia, 16, 767-775.
http://dx.doi.org/10.1038/sj.leu.2402454 - 4. Lange, B.M. and Gull, K. (1996) Structure and Function of the Centriole in Animal Cells: Progress and Questions. Trends in Cell Biology, 6, 348-352.
http://dx.doi.org/10.1016/0962-8924(96)10033-7 - 5. Tassin, A.-M. and Bornens, M. (1999) Centrosome Structure and Microtubule Nucleation in Animal Cells. Biology of the Cell, 91, 343-354.
http://dx.doi.org/10.1111/j.1768-322X.1999.tb01092.x - 6. Huston, R.L. (2015) Using the Electromagnetics of Cancer’s Centrosome Clusters to Attract Therapeutic Nanoparticles. Advances in Bioscience and Biotechnology, 6, 172-181.
http://dx.doi.org/10.4236/abb.2015.63017 - 7. Mennella, V., Agard, D.A., Huang, B. and Pelletier, L. (2014) Amorphous No More: Subdiffraction View of the Pericentriolar Material Architecture. Trends in Cell Biology, 24, 188-197.
http://dx.doi.org/10.1016/j.tcb.2013.10.001 - 8. Loncarek, J. and Khodjakov, A. (2009) Ab ovo or de nous? Mechanisms of Centriole Duplication. Molecules and Cells, 27, 135-142.
http://dx.doi.org/10.1007/s10059-009-0017-z - 9. Bettencourt-Dias, M. and Glover, D.M. (2007) Centrosome Biogenesis and Function: Centrosomics Brings New Understanding. Nature Reviews Molecular Cell Biology, 8, 451-463.
http://dx.doi.org/10.1038/nrm2180 - 10. Lingle, W.L. and Salisbury, J.L. (1999) Altered Centrosome Structure Is Associated with Abnormal Mitoses in Human Breast Tumors. American Journal of Pathology, 155, 1941-1951.
http://dx.doi.org/10.1016/S0002-9440(10)65513-7 - 11. Barenz, F., Mayilo, D. and Gruss, O.J. (2011) Centriolar Satellites: Busy Orbits around the Centrosome. European Journal of Cell Biology, 90, 983-989.
http://dx.doi.org/10.1016/j.ejcb.2011.07.007 - 12. Cunha-Ferreira, I., Bento, I. and Bettencourt-Dias, M. (2009) From Zero to Many: Control of Centriole Number in Development and Disease. Traffic, 10, 482-498.
http://dx.doi.org/10.1111/j.1600-0854.2009.00905.x - 13. Delattre, M. and Gonczy, P. (2004) The Arithmetic of Centrosome Biogenesis. Journal of Cell Science, 117, 1619-1630.
http://dx.doi.org/10.1242/jcs.01128 - 14. Bornens, M. (2012) The Centrosome in Cells and Organisms. Science, 335, 422-426.
http://dx.doi.org/10.1126/science.1209037 - 15. Azimzadeh, J. and Bornens, M. (2007) Structure and Duplication of the Centrosome. Journal of Cell Science, 120, 2139-2142.
http://dx.doi.org/10.1242/jcs.005231 - 16. Marieb, E.N. (1991) Human Anatomy and Physiology. 3rd Edition, Chapter 3, Benjamin/Cummings Publishing, Redwood City, 60-101.
- 17. Guest, D. (1996) Biology Smart, the Princeton Review. Random House, New York, 133-135.
- 18. DeLuca, J.G. (2010) Kinetochore-Microtubule Dynamics and Attachment Stability. Methods in Cell Biology, 97, 53-79.
http://dx.doi.org/10.1016/S0091-679X(10)97004-0 - 19. Moritz, M., Braunfeld, M.B. Guénebaut, V., Heuser, J. and Agard, D.A. (2000) Structure of the Gamma-Tubulin Ring Complex: A Template for Microtubule Nucleation. Nature Cell Biology, 2, 365-370.
http://dx.doi.org/10.1038/35014058 - 20. Kollman, J.M., Polka, J.K., Zelter, A., Davis, T.N. and Agard, D.A. (2010) Microtubule Nucleating γ-TuSC Assembles Structures with 13-Fold Microtubule-Like Symmetry. Nature, 466, 879-882.
http://dx.doi.org/10.1038/nature09207 - 21. Wiese, C. and Zheng, Y. (2000) A New Function for the γ-Tubulin Ring Complex as a Microtubule Minus-End Cap. Nature Cell Biology, 2, 358-364.
http://dx.doi.org/10.1038/35014051 - 22. Teixidó-Travesa, N., Roig, J. and Lüders, J. (2012) The Where, When and How of Microtubule Nucleation—One Ring to Rule Them All. Journal of Cell Science, 125, 4445-4456.
http://dx.doi.org/10.1242/jcs.106971 - 23. Pokorny, J., Hasek, J. and Jelínek, F. (2005) Endogenous Electric Field and Organization of Living Matter. Electromagnetic Biology and Medicine, 24, 185-197.
http://dx.doi.org/10.1080/15368370500379566 - 24. Huston, R.L. (2016) A Review of Centriole Activity, and Wrongful Activity, during Cell Division. Advances in Bioscience and Biotechnology, 7, 169-182.
http://dx.doi.org/10.4236/abb.2016.73015 - 25. Strnad, P. and Gonczy, P. (2008) Mechanisms of Procentriole Formation. Trends in Cell Biology, 18, 389-396.
http://dx.doi.org/10.1016/j.tcb.2008.06.004 - 26. Schiebel, E. (2000) γ-Tubulin Complexes: Binding to the Centrosome, Regulation and Microtubule Nucleation. Current Opinion in Cell Biology, 12, 113-118.
http://dx.doi.org/10.1016/S0955-0674(99)00064-2 - 27. Lin, T.-C., Neuner, A. and Schiebel, E. (2015) Targeting of γ-Tubulin Complexes to Microtubule Organizing Centers: Conservation and Divergence. Trends in Cell Biology, 25, 296-307.
http://dx.doi.org/10.1016/j.tcb.2014.12.002 - 28. Oakley, B.R., Paolillo, V. and Zheng, Y. (2015) γ-Tubulin Complexes in Microtubule Nucleation and Beyond. Molecular Biology of the Cell, 26, 2957-2962.
http://dx.doi.org/10.1091/mbc.E14-11-1514 - 29. Bouissou, A., et al. (2009) γ-Tubulin Ring Complexes Regulate Microtubule plus End Dynamics. The Journal of Cell Biology, 187, 327-334.
http://dx.doi.org/10.1083/jcb.200905060 - 30. Chang, P., Giddings Jr., T.H., Winey, M. and Stearns, T. (2003) ε-Tubulin Is Required for Centriole Duplication and Microtubule Organization. Nature Cell Biology, 5, 71-76.
http://dx.doi.org/10.1038/ncb900 - 31. Inclán, Y.F. and Nogales, E. (2000) Structural Models for the Self-Assembly and Microtubule Interactions of γ-, δ- and ε-Tubulin. Journal of Cell Science, 114, 413-422.
- 32. Kis, A., Kasas, S., Babic, B., Kulik, A.J., Benoit, W., Briggs, G.A., Schonenberger, C., Catsicas, S. and Forró, L. (2002) Nanomechanics of Microtubules. Physical Review Letters, 89, Article ID: 248101.
http://dx.doi.org/10.1103/PhysRevLett.89.248101 - 33. Bicek, A.D., Tuzel, E., Kroll, D.M. and Odde, D.J. (2007) Analysis of Microtubule Curvature. Methods in Cell Biology, 83, 237-268.
http://dx.doi.org/10.1016/S0091-679X(07)83010-X - 34. Beer, F.P. and Johnston Jr., E.R. (1994) Mechanics of Materials. 2nd Edition, McGraw-Hill, New York.
- 35. Cassimeris, L., Gard, D., Tran, P.T. and Erickson, H.P. (2001) XMAP215 Is a Long Thin Molecule that Does Not Increase Microtubule Stiffness. Journal of Cell Science, 114, 3025-3033.
- 36. Dutcher, S.K. (2001) The Tubulin Fraternity: Alpha to Eta. Current Opinion in Cell Biology, 13, 49-54.
http://dx.doi.org/10.1016/S0955-0674(00)00173-3 - 37. Mahen, R. and Venkitaraman, A.R. (2012) Pattern Formation in Centrosome Assembly. Current Opinion in Cell Biology, 24, 14-23.
http://dx.doi.org/10.1016/j.ceb.2011.12.012 - 38. Lingle, W.L., Barrett, S.L., Negron, V.C., D’Assoro, A.B., Boeneman, K., Liu, W., Whitehead, C.M., Reynolds, C. and Salisbury, J.L. (2002) Centrosome Amplification Drives Chromosomal Instability in Breast Tumor Development. Proceedings of the National Academy of Sciences of the United States of America, 99, 1978-1983.
http://dx.doi.org/10.1073/pnas.032479999 - 39. Pokorny, J. (2003) Viscous Effects on Polar Vibrations in Microtubules. Electromagnetic Biology and Medicine, 22, 15-29.
http://dx.doi.org/10.1081/JBC-120020349 - 40. Raynaud-Messina, B. and Merdes, A. (2007) γ-Tubulin Complexes and Microtubule Organization. Current Opinion in Cell Biology, 19, 24-30.
http://dx.doi.org/10.1016/j.ceb.2006.12.008 - 41. Cifra, M., Havelka, D. and Kucera, O. (2010) Electric Oscillations Generated by Collective Vibration Modes of Microtubules. Proceedings of SPIE, 7376, 73760N.
- 42. Pokorny, J., Hasek, J. and Jelínek, F. (2005) Electromagnetic Field of Microtubules: Effects on Transfer of Mass Particles and Electrons. Journal of Biological Physics, 31, 501-514.
http://dx.doi.org/10.1007/s10867-005-1286-1 - 43. Cifra, M., Pokorny, J., Havelka, D. and Kucera, O. (2010) Electric Field Generated by Axial Longitudinal Vibration Modes of Microtuble. Biosystems, 100, 122-131.
http://dx.doi.org/10.1016/j.biosystems.2010.02.007 - 44. Havelka, D., Cifra, M., Kucera, O., Pokorny, J. and Vrba, J. (2011) High-Frequency Electric Field and Radiation Characteristics of Cellular Microtubule Network. Journal of Theoretical Biology, 286, 31-40.
http://dx.doi.org/10.1016/j.jtbi.2011.07.007 - 45. Pokorny, J. (2012) Physical Aspects of Biological Activity and Cancer. AIP Advances, 2, Article ID: 011207.
http://dx.doi.org/10.1063/1.3699057 - 46. Havelka, D. and Cifra, M. (2009) Calculation of the Electromagnetic Field around a Microtubule. Acta Polytechnica, 49, 58-63.
- 47. Janson, M.E., de Dood, M.E. and Dogterom, M. (2003) Dynamic Instability of Microtubules Is Regulated by Force. The Journal of Cell Biology, 161, 1029-1034.
http://dx.doi.org/10.1083/jcb.200301147 - 48. Kline-Smith, S.L. and Wolczak, C.E. (2002) The Microtubule-Destabilizing Kinesin XKCM1 Regulates Microtubule Dynamic Instability in Cells. Molecular Biology of the Cell, 13, 2718-2731.
http://dx.doi.org/10.1091/mbc.E01-12-0143 - 49. Walczak, C.E. (2000) Microtubule Dynamics and Tubulin Interacting Proteins. Current Opinion in Cell Biology, 12, 52-56.
http://dx.doi.org/10.1016/S0955-0674(99)00056-3 - 50. Dogterom, M, Kerssemakers, J.W.J., Romet-Lemonne, G. and Janson, M.E. (2005) Force Generation by Dynamic Microtubules. Current Opinion in Cell Biology, 17, 67-74.
http://dx.doi.org/10.1016/j.ceb.2004.12.011 - 51. Portet, S., Tuszyński, J.A., Hogue, C.W.V. and Dixon, J.M. (2005) Elastic Vibrations in Seamless Microtubules. European Biophysics Journal, 34, 912-920.
http://dx.doi.org/10.1007/s00249-005-0461-4 - 52. Shawki, M.M. and Farid, A. (2014) Low Electric Field Parameters Required to Induce Death of Cancer Cells. Electromagnetic Biology & Medicine, 33, 159-163.
http://dx.doi.org/10.3109/15368378.2013.800105 - 53. Pokorny, J., Pokorny, J. and Kobilková, J. (2013) Postulates on Electromagnetic Activity in Biological Systems and Cancer. Integrative Biology, 5, 1439-1446.
http://dx.doi.org/10.1039/c3ib40166a - 54. Huston, R.L. (2014) On Centrioles, Microtubules, and Cellular Electromagnitism. Journal of Nanotechnology in Engineering and Medicine, 5, Article ID: 021003.
- 55. Duensing, A., Liu, Y., Perdreau, S.A., Kleylein-Sohn, J., Nigg, E.A. and Duensing, S. (2007) Centriole Overduplication through the Concurrent Formation of Multiple Daughter Centrioles at Single Material Templates. Oncogene, 26, 6280-6288.
http://dx.doi.org/10.1038/sj.onc.1210456 - 56. Schockel, L., Mockel, M., Mayer, B., Boos, D. and Stemmann, O. (2011) Cleavage of Cohesion Rings Coordinates the Separation of Centrioles and Chromatids. Nature Cell Biology, 13, 966-972.
http://dx.doi.org/10.1038/ncb2280 - 57. Nigg, E.A. and Raft, J.W. (2009) Centrioles, Centrosomes, and Cilia in Health and Disease. Cell, 139, 663-678.
http://dx.doi.org/10.1016/j.cell.2009.10.036 - 58. Kobayashi, T. and Dynlacht, B.D. (2011) Regulating the Transition from Centriole to Basal Body. Journal of Cell Biology, 193, 435-444.
http://dx.doi.org/10.1083/jcb.201101005 - 59. Ganem, N.J., Godinho, S.A. and Pellman, D. (2009) A Mechanism Linking Extra Centrosomes to Chromosomal Instability. Nature, 460, 278-282.
http://dx.doi.org/10.1038/nature08136 - 60. Nigg, E.A. (2002) Centrosome Aberrations: Cause or Consequence of Cancer Progression? Nature Reviews Cancer, 2, 815-825.
http://dx.doi.org/10.1038/nrc924 - 61. Korzeniewski, N., Hohenfellner, M. and Duensing, S. (2012) CANDI Promotes PLK4-Mediated Centriole Overduplication and Is Frequently Disrupted in Prostate Cancer. NEOPLASIA, 14, 799-806.
http://dx.doi.org/10.1593/neo.12580 - 62. Nigg, E.A. (2006) Origins and Consequences of Centrosome Aberrations in Human Cancers. International Journal of Cancer, 119, 2717-2723.
http://dx.doi.org/10.1002/ijc.22245 - 63. Godinho, S.A., Kwon, M. and Pellman, D. (2009) Centrosomes and Cancer: How Cancer Cells Divide with Too Many Centrosomes. Cancer and Metastasis Reviews, 28, 85-98.
http://dx.doi.org/10.1007/s10555-008-9163-6 - 64. Sluder, G. and Nordberg, J.J. (2004) The Good, the Bad and the Ugly: The Practical Consequences of Centrosome Amplification. Current Opinion in Cell Biology, 16, 49-54.
http://dx.doi.org/10.1016/j.ceb.2003.11.006 - 65. Brinkley, B.R. (2001) Managing the Centrosome Numbers Game: From Chaos to Stability in Cancer Cell Division. Trends in Cell Biology, 11, 18-21.
http://dx.doi.org/10.1016/S0962-8924(00)01872-9 - 66. D’Assoro, A.B., Lingle, W.L. and Salisbury, J.L. (2002) Centrosome Amplification and the Development of Cancer. Oncogene, 21, 6146-6153.
http://dx.doi.org/10.1038/sj.onc.1205772 - 67. Tsou, M.F. and Stearns, T. (2006) Mechanism Limiting Centrosome Duplication to Once per Cell Cycle. Nature, 442, 947-951.
http://dx.doi.org/10.1038/nature04985 - 68. Bettencourt-Dias, M. and Glover, D.M. (2009) SnapShot: Centriole Biogenesis. Cell, 136, 188.e1-188.e2.
http://dx.doi.org/10.1016/j.cell.2008.12.035 - 69. Loffler, H., Fechter, A., Liu, F.Y., Poppelreuther, S. and Kramer, A. (2013) DNA Damage-Induced Centrosome Amplification Occurs via Excessive Formation of Centriolar Satellites. Oncogene, 32, 2963-2972.
http://dx.doi.org/10.1038/onc.2012.310 - 70. Nigg, E.A. and Stearns, T. (2011) The Centrosome Cycle: Centriole Biogenesis, Duplication and Inherent Asymmetries. Nature Cell Biology, 13, 1154-1160.
http://dx.doi.org/10.1038/ncb2345 - 71. Rodrigues-Martins, A., Riparbelli, M., Callaini, G., Glover, D.M. and Bettencourt-Dias, M. (2008) From Centriole Biogenesis to Cellular Function: Centrioles Are Essential for Cell Division at Critical Developmental Stages. Cell Cycle, 7, 11-16.
http://dx.doi.org/10.4161/cc.7.1.5226 - 72. Vulprecht, J., David, A., Tibelius, A., Castiel, A., Konotop, G., Liu, F., Bestvater, F., Raab, M.S., Zentgraf, H., Izraeli, S. and Kramer, A. (2012) STIL Is Required for Centriole Duplication in Human Cells. Journal of Cell Science, 125, 1353-1362.
http://dx.doi.org/10.1242/jcs.104109 - 73. Kwon, M., Godinho, S.A., Chandhok, N.S., Ganem, N.J., Azioune, A., Thery, M. and Pellman, D. (2008) Mechanisms to Suppress Multipolar Divisions in Cancer Cells with Extra Centrosomes. Genes & Development, 22, 2189-2203.
- 74. Pihan, G.A., Wallace, J., Zhou, Y. and Doxsey, S.J. (2003) Centrosome Abnormalities and Chromosome Instability Occur Together in Pre-Invasive Carcinomas. Cancer Research, 63, 1398-1404.
- 75. Holland, A.J., Lan, W., Niessen, S., Hoover, H. and Cleveland, D.W. (2010) Polo-Like Kinase 4 Kinase Activity Limits Centrosome Overduplication by Autoregulating Its Own Stability. The Journal of Cell Biology, 188, 191-198.
http://dx.doi.org/10.1083/jcb.200911102 - 76. Lingle, W.L. and Salisbury, J.L. (2000) The Role of the Centrosome in the Development of Malignant Tumors. Current Topics in Developmental Biology, 49, 313-339.
http://dx.doi.org/10.1016/S0070-2153(99)49015-5 - 77. Goepfert, T.M., Adigun, Y.E., Zhong, L., Gay, J., Medina, D. and Brinkley, W.R. (2002) Centrosome Amplification and Overexpression of Aurora A Are Early Events in Rat Mammary Carcinogenesis. Cancer Research, 62, 4115-4122.
- 78. Marx, J. (2001) Do Centrosome Abnormalities Lead to Cancer? Science, 292, 426-429.
http://dx.doi.org/10.1126/science.292.5516.426 - 79. Marthiens, V., Rujano, M.A., Pennetier, C., Tessier, S., Paul-Gilloteaux, P. and Basto, R. (2013) Centrosome Amplification Causes Microcephaly. Nature Cell Biology, 15, 731-740.
http://dx.doi.org/10.1038/ncb2746 - 80. Maiato, H. and Logarinho, E. (2014) Mitotic Spindle Multipolarity without Centrosome Amplifications. Nature Cell Biology, 16, 386-394.
http://dx.doi.org/10.1038/ncb2958 - 81. Yang, Z., Loncarek, J., Khodjakov, A. and Rieder, C.I. (2008) Extra Centrosomes and/or Chromosomes Prolong Mitosis in Human Cells. Nature Cell Biology, 10, 748-751.
- 82. Leber, B., Maier, B., Fuchs, et al. (2010) Proteins Required for Centrosome Clustering in Cancer Cells. Science Translational Medicine, 2, 33-38.
http://dx.doi.org/10.1126/scitranslmed.3000915 - 83. Pihan, G.A., Purohit, A., Wallace, J., Knecht, H., Woda, B., Quesenberry, P. and Doxsey, S.J. (1998) Centrosome Defects and Genetic Instability in Malignant Tumors. Cancer Research, 58, 3974-3985.
- 84. Lingle, W.L., Lutz, W.H., Ingle, J.N., Maihle, N.J. and Salisbury, J.L. (1998) Centrosome Hypertrophy in Human Breast Tumors: Implications for Genomic Stability and Cell Polarity. Proceedings of the National Academy of Sciences of the United States of America, 95, 2950-2955.
http://dx.doi.org/10.1073/pnas.95.6.2950 - 85. Bornens, M. (2008) Organelle Positioning and Cell Polarity. Nature Reviews Molecular Cell Biology, 9, 874-886.
http://dx.doi.org/10.1038/nrm2524 - 86. Fukasawa, K. (2007) Oncogenes and Tumour Suppressors Take on Centrosomes. Nature Reviews Cancer, 7, 911-924.
http://dx.doi.org/10.1038/nrc2249