Advances in Bioscience and Biotechnology
Vol.4 No.6(2013), Article ID:33530,4 pages DOI:10.4236/abb.2013.46092
Occurrence of Alternaria alternata causing Alternaria blight in pigeonpea in India
![]()
International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, India
Email: #mamta.sharma@cgiar.org
Copyright © 2013 Mamta Sharma et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 21 March 2013; revised 10 May 2013; accepted 22 May 2013
Keywords: Climate Change; Internal Transcribed Spacer (ITS); PCR; Pigeonpea; Phylogenetic Tree
ABSTRACT
Blight symptoms on pigeonpea were observed in alarming proportion since 2009 onwards in Andhra Pradesh state of India. Alternaria blight infected pigeonpea plants were collected from Andhra Pradesh state of India to isolate and characterize the pathogen. The isolate proved pathogenic on pigeonpea cultivar ICPL 87119. Genetic characteristics were analyzed based on the sequence of the rDNA-internal transcribed spacer (ITS) region. The phylogenetic tree based on rDNAITS analysis showed that the Alternaria alternata causing Alternaria blight in pigeonpea is very distinct from the other Alternaria isolate reported from different host. This is the first report of molecular identification of Alternaria alternata causing Alternaria blight in pigeonpea.
1. INTRODUCTION
Pigeonpea [Cajanus cajan (L.) Millsp.] is the most versatile grain legume crop grown in the semi-arid tropical and subtropical regions in Asia, Africa, and America [1]. Globally, pigeonpea is cultivated in about 4.5 million ha, adding 3.48 million tonnes of grain to global food production [2]. India is a major pigeonpea producer in the world with a contribution of 75 - 80 percent. Pigeonpea is a major source of protein; enriches soil; provides fodder and fuel wood; and it is beneficial for arresting soil erosion [3,4]. Pigeonpea crop has a direct bearing on the economic and financial well-being and on the nutritional status of the subsistence farmers in the subcontinent as it is a low-input, rainfed crop and provides economic returns from each and every part of the plant. Howeveraverage yields of pigeonpea are low (450 - 670 kg per ha) and diseases are the major constraints to the high yield potential of pigeonpea cultivars. Pigeonpea is susceptible to many diseases [5]. Recently due to the changes in climate, blight symptoms on pigeonpea were observed in alarming proportion in major chickpea growing regions particularly in Andhra Pradesh and Karnataka from last 4 - 5 years. The disease was observed in 60% of the fields surveyed and incidence ranged from 20% - 80% during irrespective of cultivars sown. Infected plants in the field showed symptoms on all aerial parts of the plant (leaves, stems, buds, and pods). Symptoms on leaves were small, circular, necrotic spots that developed quickly forming typical concentric rings [6]. Later, these spots coalesced and caused blighting of leaves. Spots were initially light brown and later turned dark brown. On stems, spots were sunken with concentric rings. In severe infection, defoliation and drying of infected leaves, branches, and flower buds were observed (Figure 1).
Accurate identification of the pathogen is the preliminary step for further R&D on the biology of the pathogen and epidemiology of disease. Different molecular methods such as RAPD, RFLPs, DNA hybridization, AFLP, and DNA sequences [7-11] have been used to reveal the molecular identification as well as genetic polymorphism. No such reports regarding molecular identification of the causal organism of Alternaria blight of pigeonpea have been described yet.
Present study reports the frequent occurrence of Alternaria blight in pigeonpea and its emerging threat to pigeonpea production.
2. MATERIALS AND METHODS
2.1. Fungal Isolate Origin
Blight infected pigeonpea plants were collected from Andhra Pradesh state of India. Isolation was made by plating surface sterilized (0.8% sodium hypochlorite for 2 min) pieces from blight infected pigeonpea plant parts (leaves, stem, bud etc.) on potato dextrose agar (PDA) medium. Inoculated plates were incubated at 25˚C ± 2˚C for colony growth. The cultures were purified by single spore isolation. The culture was tentatively identified as Alternaria spp. based on standard mycological parameters and maintained on PDA slant at 4˚C for further study.
2.2. Pathogenicity Test
To confirm pathogenicity, 8 - 10 days old seedlings of pigeonpea cultivar ICPL 87119 were grown in pots in three replications (5 plants/pots). Conidial suspension of Alternaria sp. was prepared from 7 days old culture grown on potato dextrose broth (PDB). Seedlings were spray inoculated with conidial (5 × 105 conidia/ml) suspension and covered with polythene covers and incubated at 28˚C ± 1˚C and 12 h photo period. Un-inoculated pots served as control. Polythene covers were removed after 48 hrs. Plants were regularly watered and monitored for disease development.
2.3. DNA Extraction
For DNA extraction, the pathogen was grown on PDB medium at 25˚C ± 2˚C for 8 days. Mycelia were harvested by filtering through Whatman filter Paper No. 1, washed repeatedly with distilled sterilized water to remove excess salts adhering to it. DNA extraction was based on the cetrimide tetradecyl trimethyl ammonium bromide (CTAB) method [12]. Mycelium (1 g) was crushed in liquid nitrogen and transferred into 7.5 ml pre-warmed (65˚C) DNA extraction buffer [1 M TrisHCl (pH 8.0), 5 M NaCl, 0.5 M EDTA (pH 8.0) and 2% CTAB], mixed well and incubated in a water bath at 65˚C with gentle shaking for 45 min. Equal volume of chloroform: isoamyl alcohol (24:1 v/v) was added and mixed gently to denature proteins and centrifuged at 12,857 g for 10 min. DNA was precipitated with 0.6 volume of chilled ethanol and 0.1 volume of 3 M sodium acetate (pH 5.2) and centrifuged at 18,514 g for 15 min. The pellets were washed twice with chilled 70% ethanol, dried at room temperature, re-suspended in 100 μl sterile TE (10 mM Tris-HCl buffer and 1 mM EDTA—pH 8) and stored at −20˚C deep freezer. Isolated DNA was electrophorased in 1.0% agarose gels to check the quality and concentration.
2.4. Sequencing of rDNA-ITS Region
The pathogen Alternaria sp. identified tentatively on morphological level was characterized at molecular level using rDNA-ITS region sequencing. ITS 1 and ITS 4 primer pair were used for PCR amplification of the pathogen [13]. The PCR reaction contained 50 ng genomic DNA, 1X PCR buffer (Thermo scientific), 1.5 mM MgCl2, 0.2 mM dNTPs, 0.25 μM of each primer and 1.0 unit Dream Taq DNA Polymerase (Thermo scientific) in a 50-μL reaction volume. The PCR program was: one cycle at 94˚C for 2 min, 35 cycles of 94˚C for 30 s, 55˚C for 30 s, and 72˚C for 30 s, and one cycle of 72˚C for 5 min, and then held at 4˚C. PCR amplicon was purified with a Wizard® SV Gel and PCR Clean up system (Promega, USA) according to the manufacturer protocol and then sequenced from commercial service (Xcelris Labs Limited, Ahmedabad).
2.5. Genetic Data Analyses
Sequence identity matrix of Alternaria sp. was generated using Bioedit Sequence Alignment Editor (version 5.0.9) [14]. After multiple alignment, phylogenetic analysis was done in MEGA 4.0 software [15] using the default parameters of one character-based algorithm. The bootstrapped consensus phylogenetic tree was generated for each of these algorithms with 500 replication.
3. RESULTS AND DISCUSSION
3.1. Fungal Isolate
The fungus produced abundant, branched, septate, brownish mycelia. Conidiophores were simple, olive-brown, septate, variable in length with terminal conidia, which were solitary or in short chains. Mature conidia measures from 10 - 30 × 5 - 12 µm, short conical beak or beakless, narrowly ellipsoid to ovoid and elongated on branching chains. Juveniles were narrowly elliptical, 3 - 7 transepta, 1 - 5 longisepta and have individual chains of 5 - 15 conidia. Conidial characteristics from culture were similar to the conidia isolated from infected plants. Based on the morphological characters (Figures 1 and 2), the organism was identified as Alternaria alternata.
3.2. Pathogenicity Test
The pathogen proved pathogenic on pigeonpea cultivar ICPL 87119 and identical disease symptoms as observed

Figure 1. (A)Alternaria blight symptom on leaves and (B) aerial plant parts (leaves, stem, buds and pods).
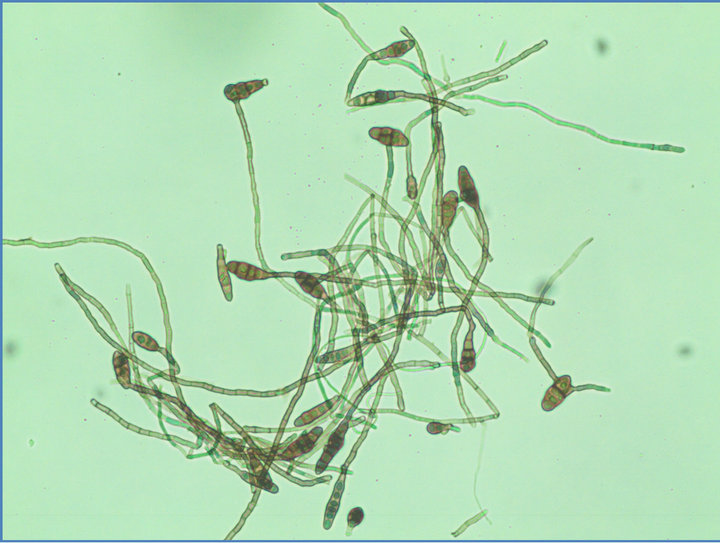
Figure 2. Conidial and mycelial morphology of Alternaria alternata.
in the field symptoms on leaves were small, circular, necrotic spots that developed quickly forming typical concentric rings. Later, these spots coalesced and caused blighting of leaves. Spots were initially light brown and later turned dark brown were observed 10 days after inoculation.No symptoms were observed on control plants.
3.3. Sequencing of rDNA-ITS Region
To confirm the molecular identity of A. alternata, the rDNA-ITS region of the present isolate was amplified and sequenced. The rDNA sequence was deposited in the GenBank database under the accession no JQ074093. The size of the PCR product was 490 bp. BLAST analysis of the 490-bp amplicon showed 100% similarity with A. alternata sequences (JN542524) reported from other host from India. To assess the relationships with other Alternaria spp. the corresponding genomic regions of 17 Alternaria sp. showing 99% - 100% sequence identity with present isolate were obtained from GenBank and a phylogenetic tree was constructed. Phylogenetic analysis revealed that the present Alternaria alternata infecting pigeonpea grouped separately from other Alternaria spp. reported from other host spp. (Figure 3).
The rDNA-ITS analyses performed on genomic DNA of A. alternata isolate revealed the presence of high level of genetic diversity with other Alternaria isolates infecting other host. The rDNA-ITS has the unique potential for providing information across an entire genome. The isolates showed strong genetic similarity within a range of 99% to 100%, indicating high level of identity among them irrespective of host. In phylogenetic tree, the present isolate grouped separately from the other isolates from different host indicating A. alternata infecting pigeonpea is distinct from other Alternaria spp. reported from different host.
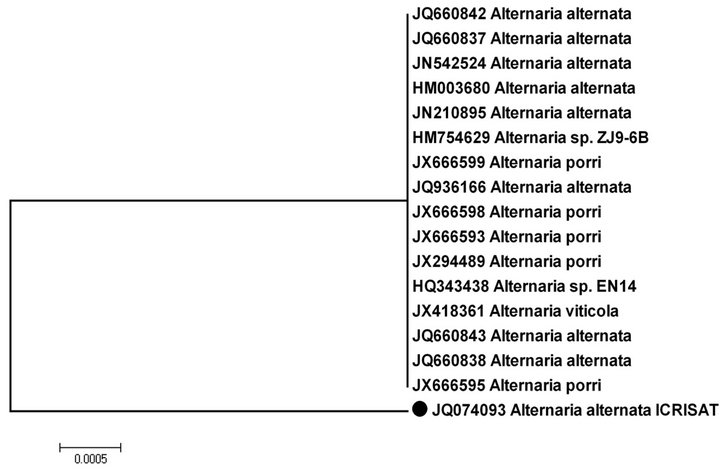
Figure 3. Phylogeny tree showing relationships among the related Alternaria spp. with Alternaria alternata (●) based on their ITS sequences.
4. CONCLUSSION
Alternaria alternata causing Alternaria blight in pigeonpea was characterized first time at molecular level using rDNA-ITS region. rDNA-ITS analysis indicated that Alternaria alternata causing Alternaria blight in pigeonpea is a distinct species from other reported Alternaria spp. infecting different host. In our earlier findings, we [16] have reported Alternaria tenuissima causing Alternaria blight in pigeonpea.
REFERENCES
- Van der Maesen, L.J.G. (1990) Pigeonpea: Origin, history, evolution and taxonomy. In: Nene, Y.L., Hall, S.D. and Sheila, V.K., Eds., The Pigeonpea, CAB International, Wallingford, 15-46.
- FAOSTAT (2009) Food and Agriculture Organization of the United Nations, Rome. http://faostat.fao.org/
- Ae, N., Arihara, J., Okada, K., Yoshihara, T. and Johansen, C. (1990) Phosphorus uptake by pigeonpea and its role in cropping system of the Indian subcontinent. Current Science, 248, 477-480.
- Saxena, K.B., Kumar, R.V. and Rao, P.V. (2002) Pigeonpea nutrition and its improvement. Journal of Crop Production, 5, 227-260. doi:10.1300/J144v05n01_10
- Nene, Y.L., Shiela, V.K. and Sharma, S.B. (1984) A world list of chickpea (Cicer arietinum) and pigeonpea (Cajanus cajan) pathogen. ICRISAT Pulse Pathology Progress Report, 32, 19.
- Kannaiyan, J. and Nene. Y.L. (1977) Alternaria leaf spot of pigeonpea. Tropical Grain Legume Bulletin, 9, 34.
- Tanabe, K., Tsuge, T. and Nishimura, S. (1990) Potential application of DNA restriction fragment length polymorphisms to the ecological studies of Alternaria alternata Japanese pear pathotype. Annals of Phytopathological Society of Japan, 55, 361-365. doi:10.3186/jjphytopath.55.361
- Adachi, Y., Watanabe, H., Tababe, K., Doke, N., Nishimura, S. and Tsuge, T. (1993) Nuclear ribosomal DNA as a probe for genetic variability in the Japanese pear pathotype of Alternaria alternata. Applied and Environmental Microbiology, 59, 3197-3205.
- Morris, P.F., Connolly, M.S. and St Clair, D.A. (2000) Genetic diversity of Alternaria alternata isolated from tomato in California assessed using RAPDs. Mycological Research, 104, 286-292. doi:10.1017/S0953756299008758
- Aradhya, M.K., Chan, H.M. and Parfitt, D.E. (2001) Genetic variability in the pistachio late blight fungus, Alternaria alternata. Mycological Research, 105, 300-306. doi:10.1017/S0953756201003677
- Peever, T.L., Ibañez, A., Akimitsu, K. and Timmer, L.W. (2002) Worldwide phylogeography of the citrus brown spot pathogen, Alternaria alternata. Phytopathology, 92, 794-802. doi:10.1094/PHYTO.2002.92.7.794
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight DNA. Nucleic Acids Research, 8, 4321-4325. doi:10.1093/nar/8.19.4321
- White, T.J., Bruns, T., Lee, S. and Talyor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninisky, J.J. and White, T.J. Eds., PCR protocols: a guide to methods and applications, Academic Press, San Diego, 315-322.
- Hall, T.A. (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows95/98/NT. Nucleic Acids Symposium Series, 41, 95- 98.
- Tamura, K., Dudley, J., Nei, M. and Kumar, S. (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599. doi:10.1093/molbev/msm092
- Sharma, M., Ghosh, R., Mangala, U.N., Saxena, K.B. and Pande, S. (2012) Alternaria tenuissima Causing Alternaria Blight on Pigeonpea [Cajanus cajan (L.) Millsp.] in India. Plant Disease, 96, 907.
NOTES
*Mamta Sharma and Raju Ghosh were the co-first authors.
#Corresponding author.

