Advances in Bioscience and Biotechnology
Vol. 4 No. 2 (2013) , Article ID: 28404 , 6 pages DOI:10.4236/abb.2013.42024
The RAPD techniques used to assess the genetic diversity in Draba dorneri, a critically endangered plant species
![]()
Institute of Biology of the Romanian Academy, Bucharest, Romania
Email: catanarodica@yahoo.com
Received 12 December 2012; revised 13 January 2013; accepted 20 January 2013
Keywords: Draba dorneri; RAPD Techniques; Genetic Diversity; Endemic Plant Species; Retezat Mountains
ABSTRACT
A successful management and preservation of the natural populations is depending on accurate assessment of genetic diversity. Knowledge of genetic diversity within a population is important for the conservation of the species. Our aim was to assess the genetic diversity in Draba dorneri Heuff. population (Brassicaceae family)—an endemic plant species of conservative interest using Random Amplified Polymorphic DNA (RAPD) technique. The plant species is strictly protected at national level as well as at international level through “Convention on the Conservation of European Wildlife and Natural Habitats”, Bern, 1979 European Council. In this study, a total of 52 primers were scored initially. A total of 77 reproducible bands with an average of 6.41 bands per primer were obtained from the 12 primers selected. A cluster analysis (UPGAMA) was used to generate a dendrogram based on Dice coefficient. We found 67% similarity between the samples from the two analyzed slopes. Comparing with other rare plants species, our data revealed a higher level of genetic diversity in D. dorneri population in Retezat Mountains.
1. INTRODUCTION
Draba L., one of the largest genus of vascular plants in the Arctic, hosted about 350 species primarily in the N hemisphere, especially arctic, subarctic, alpine and subalpine regions [1]. Eleven species of genus Draba occur in Romania, nine of them being continental endemics. D. dorneri is an endemic plant restricted to Southern Carpathians (Romania). The distribution area is restricted to alpine and subalpine area of Retezat, Bucegi and Făgăraş Mountains [2]. It is a perennial caespitose plant, populations consists in small number of individuals found in the crevices of rock outcrops. The recognition characters are given by glabrous stem and leaf surface and ramified hairs on the leaf margins. The geological substrate is represented by both siliceous, granite or conglomerate rocks. Plant community includes species of Saxifraga, Thymus, Huperzia, Silene, Symphyandra, Phyteuma, Cetraria a.a.
According to 1997 IUCN Red List of Threatened Plants, the world status of D. dorneri Heuff is extinct and following the Red Book of Vascular Plant from Romania is considered a critically endangered plant. It is also mentioned on Anexe IIb, IVb of Habitats Directive, and the mountain ranges where Draba occurs are part of Natura 2000 network. There, the requirements for longterm conservation of D. dorneri are of prime importance. Currently, taking into account our monitoring in area of distribution during to last two years, the population is very poor being formed of few individuals distributed on a very small surface.
During the time, several studies concerning different aspects of Draba sp. were performed. In this way diverse aspects like systematics [3-8], cytology [4,7,9-11], breeding systems [12], molecular phylogeny [13-15], phylogeography [5], taxonomy [2,17-29], evolution of arctic Draba species using the microsatellites and QTL-mapping population [29,30] was covered.
Understanding the genetic variation within populations is essential to establish the proper conservation strategies for plant species, the preservation of genetic diversity of the populations being a fundamental goal of conservation biology [31]. The maintenance of genetic diversity has a considerable significance for the long-term survival of endangered plant species [32]. The ability of a species to adapt to environmental changes depends greatly on the genetic diversity in the species [33,34].
The analysis of the populations through RAPD showed the highly variable of the identified markers and the potential of them in population studies. Comparing with allozyme analysis, the RAPD method provides a much higher number of markers and it is now well established as a sensitive method for detecting genetic diversity [35]. The RAPD technique, a quick and relatively inexpensive method, is extensively used to analyze the genetic variability as bacteria, fungi and plants [36]. Despite of lower reproducibility, the RAPD method is generally more desirable because of cost-effectiveness studies involving a smaller number of samples. The RAPD technology was used to investigate the genetic diversity within and between populations and has been applied to many of rare or threatened plant species [37-46].
The present paper deals with investigations related to genetic variability in D. dorneri population, an endemic species of conservative interest from Retezat Mountains, in Romania. According to the authors knowledge this is first description into field of genetic variability of D. dorneri from this area.
2. MATERIALS AND METHODS
Plant material: D. dorneri plant material was collected from 7 individuals of the natural population in Retezat Mountains. There, it occurs in a small area on the ridge that ascends from Glade Vǎlereasca to Retezat peak at an altitude of approximately 1850 m. Being a critically endangered plant species, the number of individuals from the natural population was reduced. The tissue was stored in silica gel at 25˚C until the DNA was extracted.
DNA extraction: Extraction of total genomic DNA was obtained by grinding small quantities of dried plant material (0.08 - 0.1 g) with quartz sand to obtain a powder that was re-suspended in 0.1 M TE buffer pH 8. For DNA isolation was used Genomic DNA purification kit, Fermentas, following the manufacturer protocol. The concentration of obtained DNA was between 16.7 - 973.45 ng/ml and 15.6 - 591.4 ng/ml after RNA-se treatment.
RAPD analysis: In order to evaluate the genetic diversity in D. dorneri population the RAPD techniques was used. The primers used for PCR amplification were purchased from Bio Basic Inc. The PCR reaction was perform using Fermentas kit (Dream TaqPCR master Mix and Dream Taq Green PCR Master Mix) in a BioRad C 1000 thermocycler. Each reaction was performed in a 12, 5 µl volume containing: 10 - 20 ng/µl DNA, 2.5 mM MgCl2, and 0.2 mM each dNTP, 1.5 µM primer, 0.312U Taq DNA polymerase or Dream Taq polymerase, PCR buffer and distilled water.
The amplification program consist in an initial denaturation step at 94˚C for 3 minutes, followed by 45 cycles of denaturation at 94˚C for 30 seconds, annealing at 37˚C for 30 seconds and elongation for 2 minutes at 72˚C and a final extension step at 72˚C.
The amplification products were separated by electrophoresis in 2% agarose gels in TBE buffer pH 8, added with 50 µg/ml ethidium bromide. The DNA fragments were visualized under UV light using a device DocuGel from BioRad. Molecular size of the amplification products were estimated by using a 100 bp Ladder from Bioscience with fragments between 100 - 2000 pb.
Data analysis: The DNA profiles were analyzed using Bio-Rad Quantity One software package for imaging and analyzing 1-D electrophoresis gels. The method for computing similarity in Quantity One is the Dice coefficient [47]. The formula is
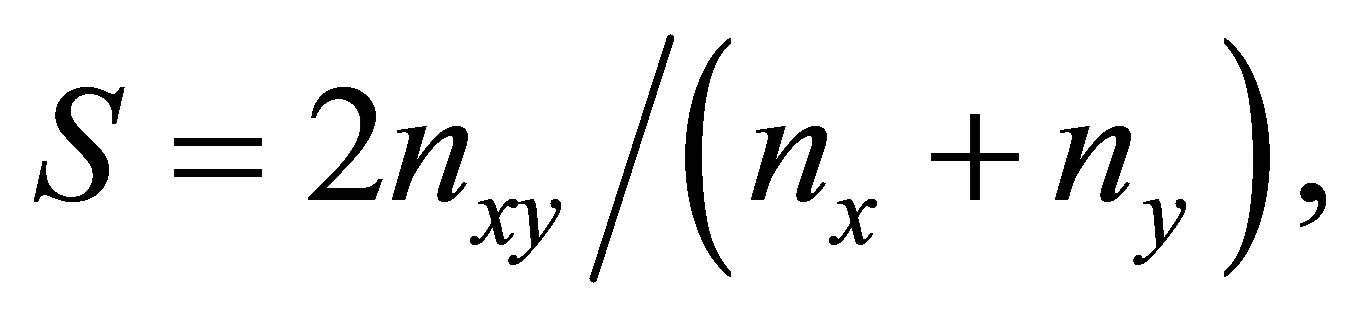
where nx and ny are the total number of fragments analyzed in individuals x and y, respectively, and nxy is the number of fragments shared by the two individuals. For the visualization of the similarity of samples, a phylogenetic tree was constructed based on unweighted pair group method using arithmetic averages (UPGAMA).
3. RESULTS
From a total of 52 primers scored initially, 25 RAPD primers were selected for analysis based on their amplification products. A total of 77 reproducible bands with an average of 6.41 bands per primer were obtained from the 12 primers selected (Table 1). The size of amplified fragments varied between 200 and 1500 bp. The percentage of polymorphic loci in the analyzed samples was 78.94%. The highest number of polymorphic bands (10) was obtained by primers S69 and S235 and lowest (1) by primer S90 and S197 (Table 1, Figure 1).
Nowadays, many rare plant species are currently threatened by extinction due to the destruction, the fragmentation of habitats and the isolation of predominantly small plant populations [48], the ecological parameters having influences on the genetic differentiation of populations [49].
In this study, 12 primers were analyzed for assessing the genetic diversity in D. dorneri samples collected from the eastern and western slope of the Retezat Mountains. The presence of the species on the eastern slope summarizes a small number of individuals where it grows in association with less sun-loving plants. On the western slope, where solar radiation is high are installed clonal individuals more developed, that include even over 100 floral stems.
A number of 77 amplified products were produced, ranging in size from 200 to 1500 bp. From 12 primers tested, only one primer (S6) produced amplification products that were monomorphic across all the plants. Most of the primers generated high percent of polymerphic loci, which means that genetic polymorphism in the population, is generally high.
The percentage of polymorphic bands (78.94%) obtained in D. dorneri samples was higher than in other endangered plants like Paeonia suffruticosa 22.5%, P.
Table 1. Total number of amplified fragments and number of polymorphic fragments generated by 12 random primers in D. dorneri samples.

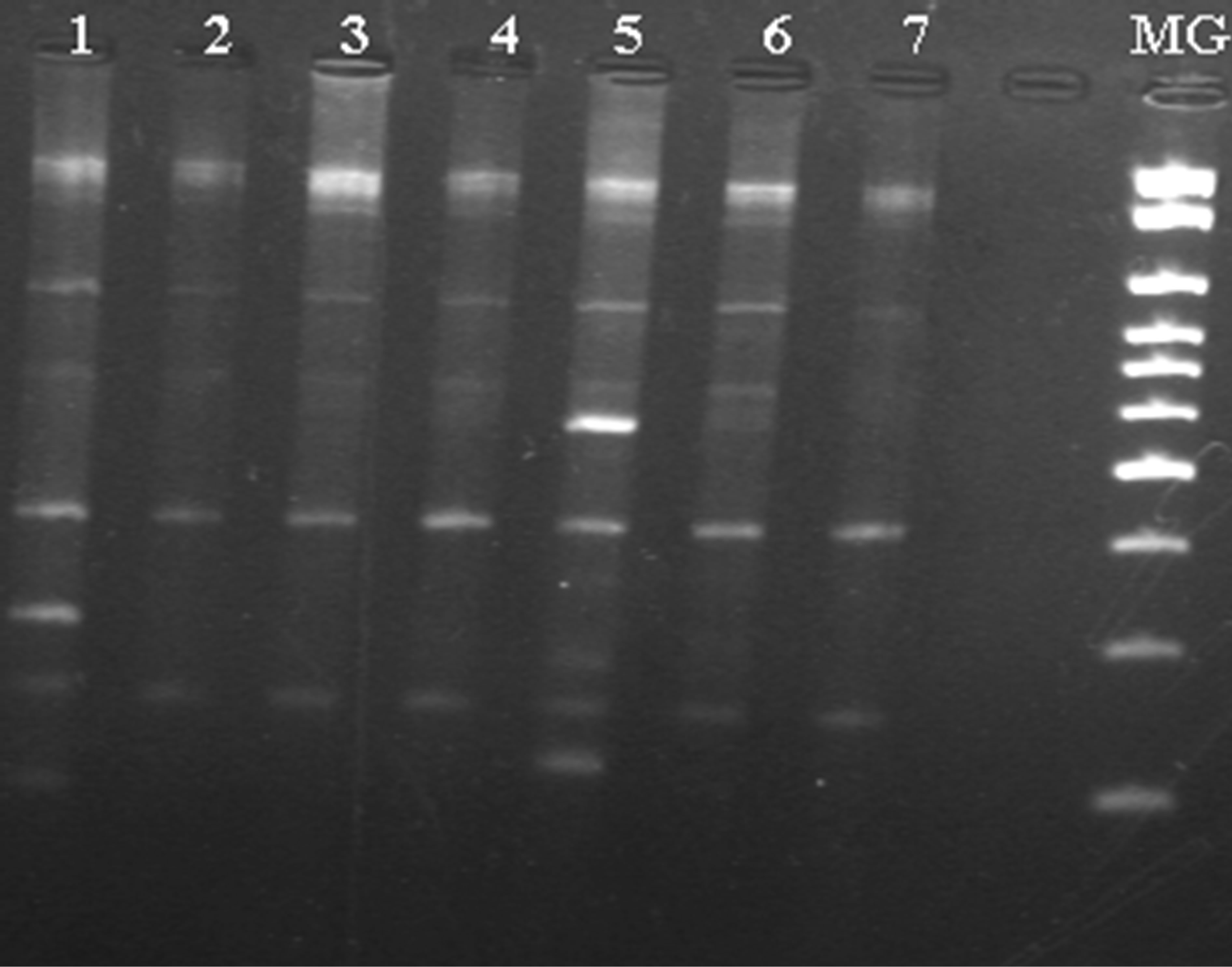
Figure 1. The RAPD profiles of Draba dorneri Heuff. generated by S105 primer. Samples 1 - 7 D. dorneri individuals. MG- 100 bpDNA ladder.
rockii 27.6% [37], Lactoris fernandeziana 24.5% [50], Allium aseae 40% - 63% [47], Cathaya argyrophylla 32% [51], Dacydium pierrei 33.3% [48]. Comparing with these percentages our data showed that the genetic variability of this species is not low. The maintenance of existing levels of genetic diversity has a considerable significance for the long-term survival of endangered plants.
In order to represent the relationships among samples, a cluster analysis (UPGAMA) was used to generate a dendrogram based on Dice coefficient (Figure 2).
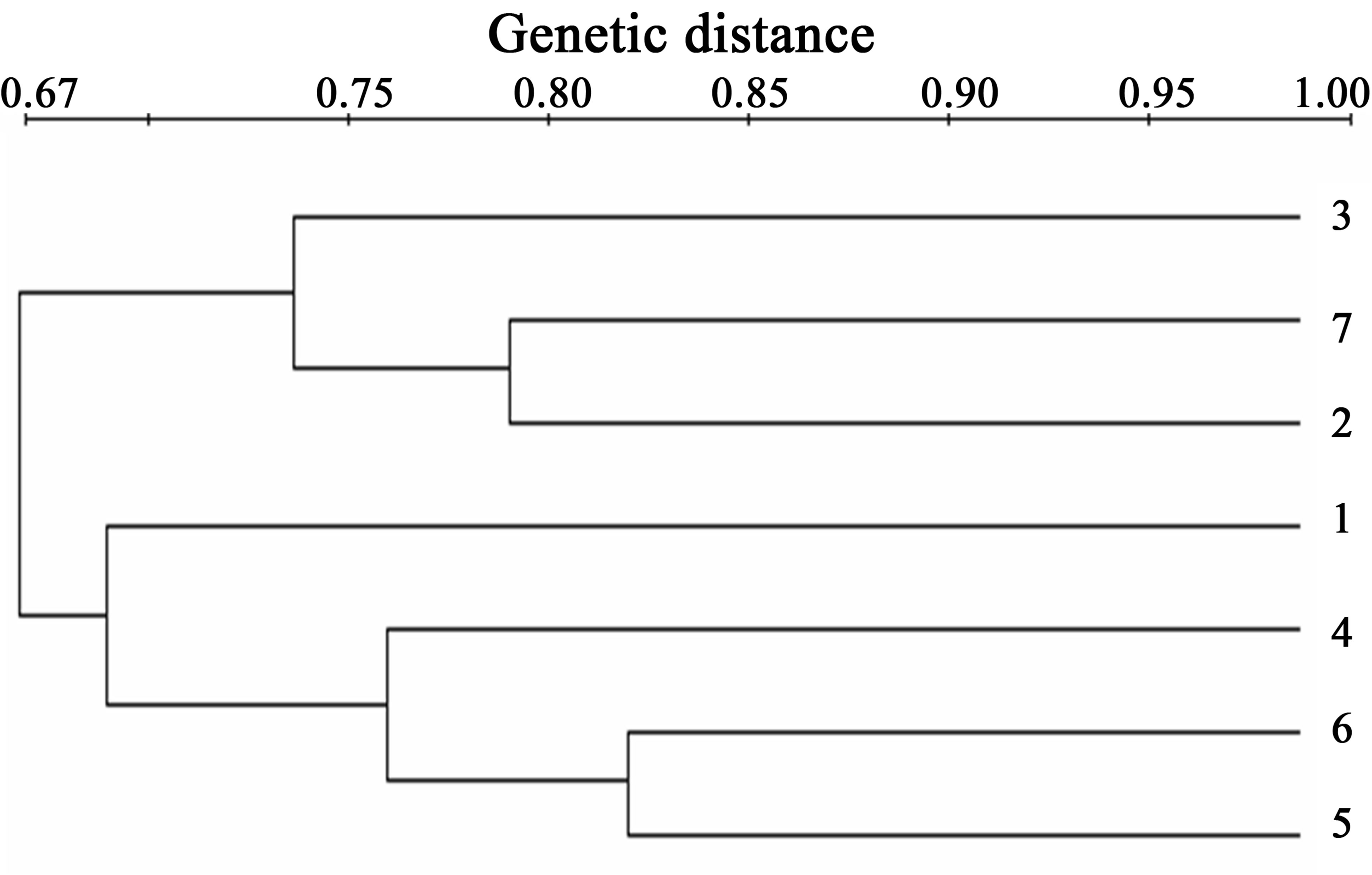
Figure 2. Dendrogram of the seven individuals from natural population of D. dorneri from Retezat Mounatins, constructed using UPGAMA based on Dice coefficient. 1, 2, 3—samples from Western slope; 4, 5, 6—samples from Eastern Slope, 7— sample from the second site. The scale indicates the genetic distance between individuals.
The similarity between the samples from Eastern Slope (samples 4, 5, 6) is 76% and 74% for the samples from Western slope (samples 1, 2, 3). The sample 7, collected from a few meters from the sample 2 present a high similarity (80%) with this. The similarity between the samples from the slopes is 67%. This percent of polymorphism coincide with the small distance between the 2 slopes. We may conclude that the samples are part from the same population. Despite of the high percentage of polymorphic bands identified by RAPD, D. dorneri have no spreading power.
Our results showed that the similarity represented by the polymorphism of RAPD bands between the individuals from the two slopes of the Retezat Mountains is 67%. The cluster analysis (UPGAMA) used to distinguish between individuals showed that samples can not be clustered in different populations.
Despite of the small size of the population, which generally exhibit lower levels of genetic diversity, our results based on the RAPD techniques showed that the genetic diversity of this critically endangered plant species is not low.
A good strategy to protect this critically endangered plant species is to protect more of their habitat.
4. FUTURE PERSPECTIVES
We intend to increase the number of samples from the same and different locations. Also, we intend to check our results concerning the genetic diversity founded using RAPD through other molecular tools.
5. ACKNOWLEDGEMENTS
The study in Retezat Mountains was supported financially by Conservation of bioand geodiversity, as support of sustainable development and economic and social growth in Hateg County—Retezat area, Project RO 0023 (RO-00056 MF SEE). The authors thanks to Dr. Madalin Enache for helpful discussion during to manuscript preparation
REFERENCES
- Science Press (Beijing) and Missouri Botanical Garden Press (2001) Draba. Flora of China, 8, 66. EFloras.org
- Dragulescu, C. (2008) Draba dorneri. Natura 2000 in Romania Species Fact-Sheet, 238-239.
- Al-Shehbaz, I.A. (1990) A note on the Chilean endemic Draba thlaspiformis (Brassicaceae). Journal of the Arnold Arboretum, 71, 385-387.
- Al-Shehbaz, I.A. (2002) Six new species of Draba (Brassicaceae) from the Himalayas. Novon, 12, 314-318. doi:10.2307/3393072
- Al-Shehbaz, I.A. and Koch, M.A. (2003) Drabopsis is united with Draba (Brassicaceae). Novon, 13, 172-173. doi:10.2307/3393511
- Al-Shehbaz, I.A. (2004) Novelties and notes on miscellaneous Asian Brassicaceae. Novon, 14, 153-157.
- Al-Shehbaz, I.A. and Windham, M.D. (2007) New or noteworthy North American Draba (Brassicaceae). Harvard Papers in Botany, 12, 409-419. doi:10.3100/1043-4534(2007)12[409:NONNAD]2.0.CO;2
- Al-Shehbaz, I.A. (2007) Two new species of Draba and Eutrema (Brassicaceae) from Sichuan, China. Harvard Papers in Botany, 11, 277-279. doi:10.3100/1043-4534(2007)11[277:TNSODA]2.0.CO;2
- Anand, A., Rao, C.S., Eganathan, P., et al. (2004) Saving an endangered Taxon: Syzygium travancoricum Gamble: A case study focusing on its genetic diversity and reintroduction. Physiology and Molecular Biology of Plants, 10, 233-242.
- Ayres, D.R. and Ryan, F.J. (1999) Genetic diversity and structure of the narrow endemic Wyethia reticulata and its congener W. bolanderi (Asteraceae) using RAPD and allozyme techniques. American Journal of Botany, 86, 344-353. doi:10.2307/2656756
- Barrett, S.C.H. and Kohn, J.R. (1991) Genetic and evolutionary consequences of small population sizes in plants: Implications for conservation. In: Falk, D.A. and Holsinger, K.A., Eds., Genetics and Conservation of Rare Plants, Oxford University Press, New York, 3-30.
- Beilstein, M.A. and Windham, M.D. (2003) A phylogenetic analysis of Western North American Draba (Brassicaceae) based on nuclear ribosomal DNA sequences from the ITS region. Systematic Botany, 28, 584-592.
- Brauner, S., Crawford, D.J. and Stuessy, T.F. (1992) Ribosomal DNA and RAPD variation in the rare plant family Lactoridaceae. American Journal of Botany, 79, 1436- 1439. doi:10.2307/2445144
- Brochmann, C. (1992) Polyploid evolution in artic-alpine Draba (Brassicaceae). Sommerfeltia, 4, 1-37.
- Brochmann, C., Borgen, L. and Stedje, B. (1993) Crossing relationships and chromosome numbers of Nordic populations of Draba (Brassicaceae), with emphasis on the D. alpina complex. Nordic Journal of Botany, 3, 121-147. doi:10.1111/j.1756-1051.1993.tb00023.x
- Dannemann, A. (2000) Der einfluss von fragmentierung und populationsgrösse auf die genetische variation und fitness von seltenen pflanzenarten am beispiel von Biscutella Laevigata (Brassicaceae). Cramer, Berlin.
- Dice, L.R. (1945) Measures of the amount of ecologic association between species. Ecology, 26, 297-302. doi:10.2307/1932409
- Dihoru, Ghe. and Negrean, G. (2009) Cartea rosie a plantelor vasculare din Romania. Editura Academiei Romane, Bucuresti.
- Elven, R. and Al-Shehbaz, I.A. (2008) Draba simmonsii (Brassicaceae), a new species of the D. micropetala complex from the Canadian Arctic Archipelago. Novon, 18, 325-329. doi:10.3417/2007178
- Fu, C., Qiu, Y. and Kong, H. (2003) RAPD analysis for genetic diversity in Changium smyrnioides (Apiaceae), an endangered plant. Botanical Bulletin of Academia Sinica, 44, 13-18.
- Grundt, H.H., Popp, M., Brochmann, C., et al. (2004) Polyploid origins in a circumpolar complex in Draba (Brassicaceae) inferred from cloned nuclear DNA sequences and fingerprints. Molecular Phylogenetics and Evolution, 32, 695-710. doi:10.1016/j.ympev.2004.04.006
- Grundt, H.H., Obermayer, R. and Borgen, L. (2005) Ploidal levels in the arctic alpine polyploidy Draba lactea (Brassicaceae) and its low-ploid relatives. Botanical Journal of the Linnean Society, 147, 333-347. doi:10.1111/j.1095-8339.2005.00377.x
- Jordon-Thaden, I.E. and Koch, M.A. (2008) Diversity patterns in the genus Draba: A first global perspective. Plant Ecology and Diversity, 1, 255-263. doi:10.1080/17550870802349112
- Jordan-Thaden, I.E., Hase, I., Al-Shehbaz, I., et al. (2010) Molecular phylogeny and systematics of the genus Draba (Brassicaceae) and identification of its most closely related genera. Molecular Phylogenetics and Evolution, 55, 524-540. doi:10.1016/j.ympev.2010.02.012
- Jover, M.A., del Castillo-Agudo, L., Garcia-Carrascosa, M., et al. (2003) Random amplified polymorphic DNA assessment of diversity in western Mediterranean populations of the seagrass Posidonia oceanica. American Journal of Botany, 9, 364-369. doi:10.3732/ajb.90.3.364
- Keiper, F.J. and McConchie, R. (2000) An analysis of genetic variation in natural populations of Sticherus flabellatus [R. Br. (St John)] using amplified fragment length polymorphism (AFLP) markers. Molecular Ecology, 9, 571-581. doi:10.1046/j.1365-294x.2000.00901.x
- Koch, M.A. and Al-Shehbaz, I.A. (2002) Molecular data indicates complex intraand intercontinental differentiation of American Draba (Brassicaceae). Annals of the Missouri Botanical Garden, 89, 88-109. doi:10.2307/3298659
- Kölliker, R., Stadelmann, F.J., Reidy, B., et al. (1998) Fertilization and defoliation frequency affect genetic diversity of Festuca pratensis Huds. in permanent grasslands. Molecular Ecology, 7, 1557-1567. doi:10.1046/j.1365-294x.1998.00486.x
- Maki, M. and Hories, S. (1999) Random amplified polymorphic DNA (RAPD) markers reveal less genetic variation in the endangered plant Cerastium fisherianum var. mollethan in the widespread conspecific C. fisherianum var. fisherianum (Caryophyllaceae). Molecular Ecology, 8, 145-150. doi:10.1046/j.1365-294X.1999.00517.x
- Martín, C., González-Benito, M.E. and Iriondo, J.M. (1997) Genetic diversity within and among populations of a threatened species: Erodium paularense Fern. Gonz. & Izco. Molecular Ecology, 6, 813-820. doi:10.1111/j.1365-294X.1997.tb00135.x
- Mulligan, G.A. and Findlay, J.N. (1970) Sexual reproduction and agamospermy in the genus Draba. Canadian Journal of Botany, 48, 269-270. doi:10.1139/b70-040
- Mulligan, G.A. (1974) Cytotaxonomic studies of Draba nivalis and its close allies in Canada and Alaska. Canadian Journal of Botany, 52, 1793-1801. doi:10.1139/b74-232
- Nebauer S.G., del Castillo-Agudo, L. and Segura, J. (1999) RAPD variation within and among natural populations of outcrossing willow-leaved foxglove (Digitalis obscura L.). Theoretical and Applied Genetics, 98, 985-994. doi:10.1007/s001220051159
- Nebauer, S.G., del Castillo-Agudo, L. and Segura, J. (2000) An assessment of genetic relationships within the genus Digitalis based on PCR-generated RAPD markers. Theoretical and Applied Genetics, 100, 1209-1216. doi:10.1007/s001220051426
- Neel, M.C. and Ellstrand, N.C. (2003) Conservation of genetic diversity in the endangered plant Eriogonum ovalifolium var. vineum. Conservation Genetics, 4, 337- 352. doi:10.1023/A:1024017029933
- Nybon, N. and Bartish, I. (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspectives in Plant Ecology, Evolution and Systematics, 3, 93- 114. doi:10.1078/1433-8319-00006
- Pei, Y.L., Zou, Y.P., Yin, Z., et al. (1995) Preliminary report of RAPD analysis in Paeonia suffruticosa subsp. spontanea and P. rockii. Acta Phytotaxonomica Sinica, 33, 350-356.
- Prasad, M.N.V., Padmalatha, K., Jayaram, K., et al. (2007) Medicinal plants from Deccan ecoregion, India: Traditional knowledge, ethnopharmacology, cultivation, utilization, biotechnology and conservation—Opportunities and impediments. Medicinal and Aromatic Plant Science and Biotechnology, 1, 155-208.
- Price, R.A. and Rollins, R.C. (1991) New taxa of Draba (Cruciferae) from California, Nevada and Colorado. Harvard Papers in Botany, 3, 71-77.
- Reisch, C. and Gmünd, S. (2001) Climatic oscillations and the fragmentation of plant populations—Genetic diversity within and among populations of the glacial relict plants Saxifraga paniculata (Saxifragaceae) and Sesleria albicans (Poaceae). Biologie und Vorklinische Medizin der Universität Regensburg, Berlin.
- Rollins, R.C. (1984) Draba (Cruciferae) in Mexico and Guatemala. Contributions from the Gray Herbarium, 213, 1-10.
- Rollins, R.C. and Price, R.A. (1988) High-elevation Draba (Cruciferae) of the White Mountains of California and Nevada. Aliso, 12, 17-27.
- Rossetto, M., Weaver, P.K. and Dixon, K.W. (1995) Use of RAPD analysis in devising conservation strategies for the rare and endangered Grevillea scapigera (Proteaceae). Molecular Ecology, 4, 321-329. doi:10.1111/j.1365-294X.1995.tb00225.x
- Scheen, A.C., Elven, R. and Brochmann, C. (2002) A molecular-morphological approach solves taxonomic controversy in arctic Draba (Brassicaceae). Canadian Journal of Botany, 80, 59-71. doi:10.1139/b01-132
- Skrede, I., Brochmann, C., Borgen, L., et al. (2008) Genetics of intrinsic postzygotic isolation in a circumpolar plant species Draba nivalis (Brassicaceae). Evolution, 62, 1840-1851. doi:10.1111/j.1558-5646.2008.00418.x
- Skrede, I., Carlsen, T., Rieseberg, L.H., et al. (2008) Microsatellites for three distantly related genera in the Brassicaceae. Conservation Genetics, 10, 643-648. doi:10.1007/s10592-008-9598-x
- Smith, J.F. and Pham, T.V. (1996) Genetic diversity of the narrow endemic Allium aaseae (Alliaceae). American Journal of Botany, 83, 717-726. doi:10.2307/2445848
- Su, Y.J., Wang, T. and Huang, C. (1999) RAPD analysis of different population of Dacydium pierrei. Acta Scientiarum. Naturalium Universitatis Sunyatseni, 38, 99-101.
- Tansley, S.A. and Brown, C.R. (2003) RAPD variation in the rare and endangered Leuca dendronelimense (Proteaceae): Implications for their conservation. Biological Conservation, 95, 39-48. doi:10.1016/S0006-3207(00)00015-X
- Widmer, A. and Baltisberrger, M. (1999) Molecular evidence for allopolyploid speciation and a single origin of the narrow endemic Draba ladina (Brassicaceae). American Journal of Botany, 86, 1282-1291. doi:10.2307/2656776
- Wang, X.Q., Zou, Y.P., Zhang, D.M., et al. (1996) RAPD analysis for genetic polymorphism in Cathaya argyrophylla. Science in China, 26, 437-441.

