Advances in Bioscience and Biotechnology
Vol.3 No.3(2012), Article ID:19881,5 pages DOI:10.4236/abb.2012.33026
Non-vesicle-bound free microRNAs could enter cells and affect gene expression
![]()
1Department of Medicine & Therapeutics, The Chinese University of Hong Kong, Hong Kong, China
2Division of Nephrology, The First Affiliated Hospital of Shenzhen University, Shenzhen, China
Email: ccszeto@cuhk.edu.hk
Received 3 February 2012; revised 23 March 2012; accepted 9 April 2012
Keywords: Cell Signaling; Fibrosis; Cytokine
ABSTRACT
Background: Cell-free microRNAs (miRNAs) exist in body fluid. Previous studies showed that cell-free miRNAs are partly bound in microvesicles, and could transfer between cells via fusion with cell membrane. Methods: We quantified the amount of a panel of miRNA targets in and outside microvesicles in human proximal tubular epithelial cell (HK2) medium by microarray and real-time quantitative polymerase chain reaction (RT-QPCR). Inter-cellular miRNA transfer was explored by medium transfer experiments. Results: We identified a portion of cell-free miRNAs that exists as non-vesicle bound, truly naked form. More importantly, these non-vesicle bound free miRNA could transfer between cells and exert biological effects. By miRNA microarray, we showed that the expression of many miRNA targets in HK-2 cells were altered, either upor down-regulated, after exposure to extrinsic free miRNAs. The miRNA-200 family was the most affected in our model, with a corresponding alteration in the messenger RNA expression of down-stream targets including ZEB1 and vimentin. Conclusion: Our results suggest that free miRNA may serve as an intercellular messenger, a phenomenon that needs further exploration.
1. INTRODUCTION
MicroRNAs (miRNAs) are small, non-coding, singlestranded, endogenous RNA molecules that regulate gene expression by promoting degradation of messenger RNA targets, inhibition of translation by binding to the 3’untranslated region (3’UTR), or altering mRNA metabolism by acting as molecular decoys for RNA-binding proteins [1-3]. MiRNAs has a profound influence on cellular gene expression; it regulates over 30% of all the genes in a cell, and participates in almost all major cellular activity [4].
While numerous studies focus on intra-cellular miRNAs, recent studies revealed the existence of cell-free miRNAs in blood, urine and cell culture medium [5-8]. Previous studies showed that these miRNAs are largely enclosed in microvesicles and could transfer between cells, possibly playing a role in intercellular communication ([8-11]). Besides within microvesicles, however, it is now realized that there is “naked” miRNAs freely present outside microvesicles because serum can be directly used as template for polymerase chain reaction without a formal miRNA extraction [5]. Nonetheless, the biological roles of these naked (non-vesicle bound) free miRNAs remain unclear.
2. MATERIALS AND METHODS
2.1. Cell Culture
The human kidney-2 (HK-2) cell line, which is a human proximal tubular cell line, was purchased from the American Type Culture Collection (Manassas, VA, USA). Keratinocyte serum-free media (K-SFM) (Invitrogen, Paisley, UK) supplemented with bovine pituitary extract (40 μg/mL), epidermal growth factor (5 ng/mL), 100 U/mL penicillin and 100 μg/mL streptomycin was used for cell culture. The cell lines were then maintained in a 37˚C, 5% CO2 humidified atmosphere. The media was changed every 2 to 3 days. HK-2 cells were subcultured in a 24-well plate and grown to 70% - 80% confluence.
2.2. RNA Extraction
MirVana™ miRNA isolation kit (Ambion, Inc. Austin, TX, USA) was used for the extraction of miRNA from HK-2 cells according to the manufacturer’s protocol. The mirVanaTM PARISTM Kit (Ambion, Inc. Austin, TX, USA) was used for the extraction of total RNA from the culture medium. In brief, the medium was first centrifuged at 15,000-g for 2 hours at 4˚C; 400 µl media were then mixed with 400 µl denaturing solution and 800 µl acid phenol chloroform and centrifuged for 10 minutes at 15,000-g. The upper aqueous phase (600 µl) was then mixed with 1.25 volumes of 100% ethanol and washed with washed 3 times on a filter cartridge. Total RNA was then eluted with 60 µl preheated (95˚C) water and stores at –80˚C till reverse transcription.
2.3. miRNA Labeling and Co-Culture
The whole extracted miRNAs were labeled with Label IT miRNA Labeling Kit, CyTM3 (Mirus, Inc. Madison, WI, USA) according to the manufacturer’s protocol. Labeled miRNAs were then added to the media of HK-2 cells. The media was changed after 24 hours and cells were observed with fluorescence microscope. In the co-culture experiment, the treated cells were further transferred to an insert and co-cultured with naïve HK-2 cells in another well for another 3 days.
2.4. Effect of Extrinsic miRNAs Exposure
The whole miRNAs extracted from HK-2 cells were added to the culture medium of fresh HK-2 cells. The latter cells were cultured for another 24 hours. The cells were then harvested for RNA extraction.
2.5. miRNA Microarray
The quantity of the extracted total RNA was determined using the Nanodrop (Thermo Fisher Scientific Inc. Wilmington, DE 19810, USA) and the quality was assayed using the Agilent 2100 Bioanalyzer Eukaryote Total RNA Nano assay (Agilent Technologies, Waldbronn, Germany). Briefly, 100 ng of total RNA was labeled and hybridized to the Agilent Human V3 miRNA Microarrays using the Agilent miRNA Complete Labeling and Hyb Kit (Agilent Technologies, Waldbronn, Germany) according to the manufacturer’s protocol. Scanning and image analysis were performed using the Agilent DNA Microarray Scanner (Agilent Technologies, Waldbronn, Germany) equipped with extended dynamic range (XDR) software. Data was then extracted from raw microarray image files using Feature Extraction Software (Version 10.5) (Agilent Technologies, Waldbronn, Germany). GeneSpring GX (Version 10.0) software (Agilent Technologies, Waldbronn, Germany) with default setting was used for data visualization and analysis.
2.6. Quantification of RNA Levels
Gene expression was quantified by real time quantitative polymerase chain reaction (RT-QPCR) using the ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). TaqMan® microRNA reverse transcription Kit (Applied Biosystems, Foster City, CA, USA) and High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) were used for reverse transcription. Commercially available Taqman primers and probes, including 2 unlabeled primers and 1 FAM™ dye-labeled TaqMan® MGB probe were used for all the targets (all from Applied Biosystems).
For miRNA, 1.67 µl total RNA was mixed with 1 µl specific primers, 0.05 µl 100 mM dNTPs (with dTTP), 0.5 µl 10× reverse transcription buffer, 0.33 µl (50 U) MultiScribe™ Reverse Transcriptase, 0.66 µl RNase inhibitor (20 U/µl) and made up to 5 µl with H2O. Reverse transcription was performed at 16˚C for 30 minutes, 42˚C for 30 minutes and 85˚C for 5 minutes. 1.67 µl media without RNA extraction was also used as template for reverse transcription. For mRNA, 10 µl total RNA was mixed with 2 µl specific primers, 0.8 µl 100 mM dNTPs (with dTTP), 2 µl 10× reverse transcription buffer, 1 µl (50 U) MultiScribe™ Reverse Transcriptase, 1 µl RNase inhibitor (20 U/µl) and made up to 20 µl with H2O. Reverse transcription was performed at 25˚C for 10 minutes, 37˚C for 120 minutes and 85˚C for 5 minutes. The resulting cDNA was stored in –80˚C until use.
For mRNA expression, the primer and probe set was deliberately designed across the intron-exon boundary so as not to detect probable genomic DNA. For RT-QPCR, 10 µl universal master mix, 1 µl primer and probe set, 1.33 µl cDNA and 7.67 µl H2O were mixed to make a 20 µl reaction volume. Each sample was run in triplicate. RT-QPCR were performed at 50˚C for 2 minutes, 95˚C for 10 minutes, followed by 40 cycles at 95˚C for 15 seconds and 60˚C for 1 minute. β-Glucuronidase (GUSB, Applied Biosystems) and RNU48 (Applied Biosystems) were used as house-keeping genes to normalize the mRNA and miRNA expression respectively. Results were analyzed with Sequence Detection Software version 2.0 (Applied Biosystems). In order to calculate the differences of expression level for each target among samples, the DDCT method for relative quantitation was used.
2.7. Statistical Analysis
Statistical analysis was performed by SPSS for Windows software version 15.0 (SPSS Inc., Chicago, IL). All the results were presented in mean ± SD. Since the gene expression data are highly skewed, Mann-Whitney U test was used to compare gene expression levels between groups. A P value of below 0.05 was considered statistically significant. All probabilities were two-tailed.
3. RESULTS
3.1. Free miR-200a in Culture Medium
The miR-200a level in culture medium, with and without RNA extraction, which represent vesicle-bound and truly free miRNAs, is summarized in Figure 1. Although the level of miR-200a in culture medium without RNA extraction was significantly lower than that with RNA extraction (15.24 ± 0.65 vs 19.48 ± 0.82, P < 0.001) (Figure 1), the former was easily detectable, indicating that there is a substantial quantify of non-vesicle bound free miRNAs in the culture medium.
3.2. Free miRNAs Could Enter Cells
We then examine whether free miRNA in culture medium could gain entrance into the cytosol. After CyTM3- abeled miRNA was added into the culture medium, fluorescence could be detected in HK-2 cells as early as 5 minutes, and it continued to accumulate in the following 24 hours (Figure 2). When the cells pre-treated with CyTM3 labeled miRNA were then co-cultured with ordinary HK2 cells, fluorescence could also be detected in the latter after 72 hours.
3.3. Change in Micro-RNA Expression in Response to Extrinsic miRNA
We further explored the change in miRNA expression profiles of HK-2 cells in response to treatment of extrinc miRNA for 24 hours by microRNA expression microbarray. The results are summarized in Tables 1 and 2. Of the 866 miRNA species in the microarray, 308 were detected in HK-2 cells. The expression of 206 species remained unchanged, 86 were down-regulated, while 16 were up-regulated. The change in miRNA expression was confirmed by RT-QPCR (details not shown).

Figure 1. Levels of total and non-vesicle bound free miR-200a in the culture medium of HK-2 cell.
Figure 2. Fluorescence in HK-2 cells after exposure to CyTM3 labeled miRNAs for 24 hours.

Table 1. Micro-RNA species that were significantly down-regulated in HK-2 cells after treated by extrinsic miRNAs for 24 hours.

Table 2. Micro-RNA species that were significantly up-regulated in HK-2 cells after treated by extrinsic miRNAs for 24 hours.
3.4. Change in Messenger RNA Expression in Response to Extrinsic miRNA
We note that the down-regulation miRNA species were mostly related to epithelial-mesenchymal transition. As a result, we further explored the mRNA expression of ZEB1, ZEB2 and vimentin in HK-2 cells following exposure to extrinsic free miRNA for 24 hours (Figure 3). Briefly, the expression of ZEB2 remained unchange, while that of vimentin and ZEB1 increased following exposure to extrinsic free miRNA, although the change in ZEB1 expression did not reach statistical significance (details not shown).
4. DISCUSSION
In line with a previous study [5], which showed that serum and plasma can be directly used as template for RT-QPCR because of the presence of free miRNA in body fluid, we found that there was a substantial expression of miRNAs in HK-2 cell medium. Our results suggest the existence of “naked” miRNA in cell culture medium, presumably released by the cells under culture.
It is often believed that miRNAs in microvesicles could be transferred between cells via fusion of microvesicles to cell membrane. In contrast, our study showed that non-vesicle bound free miRNAs could also enter cells and transfer between them. The mechanism of how non-vesicle bound free miRNAs gain entrance to living cells is not clear. In fact, miRNAs are big, negatively charged molecules that should theoretically be repelled by cell membrane. Nonetheless, living cells has been found to be able to take up other nucleic acids in various in vitro models [12,13].
Previous studies have shown that the miRNAs transferred by the fusion of microvesicles to cell membrane would increase the specific miRNA expression in target cells [8]. In this study, however, we found that the change in target cell miRNA expression was diverse after exposure to naked miRNAs, and the levels of many
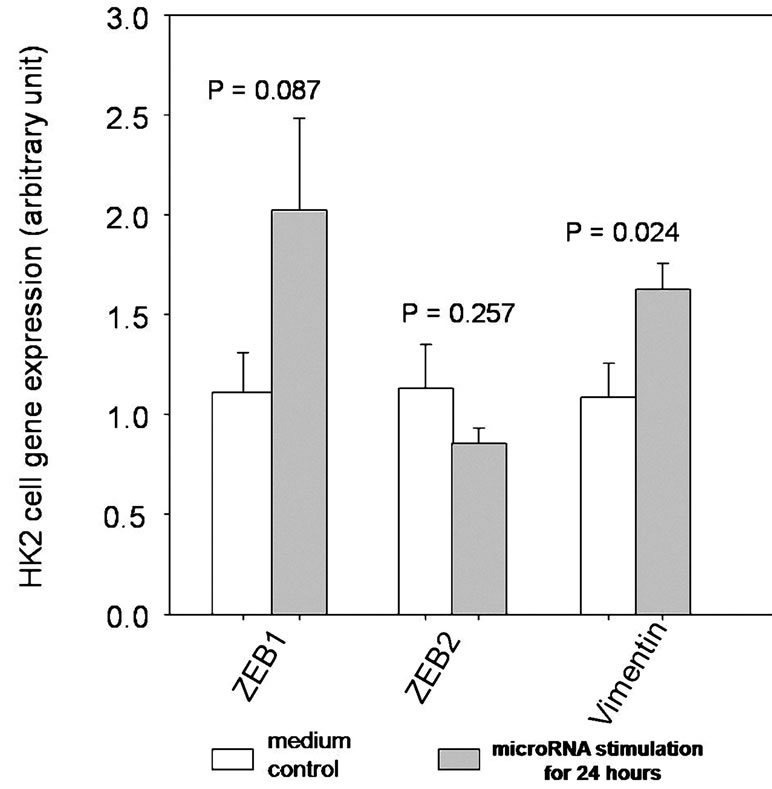
Figure 3. Expression levels of ZEB1, ZEB2 and vimentin in HK-2 cells after exposure to extrinsic free miRNAs for 24 hours.
miRNA species actually decreased. It was possible that free miRNAs gaining entrance into the cell might trigger a negative feedback, resulting in a down-regulation of miRNA production. It should be noted that we did not use specific miRNA species in this study; the physiological effect of individual miRNA species needs to be further investigated.
We found that exposure to extrinsic free miRNA have biological effect. Specifically, the messenger RNA expression of ZEB1 and vimentin in HK-2 cells, which are known to be regulated by miRNA 200 family [14-16], were altered. It should be noted that intracellular expression of miRNA 200 family was down-regulated upon exposure to extrinsic free miRNAs, which may lead to repression of ZEB2 translation to nucleus, resulting in a change in vimentin expression.
There are a few shortcomings of our study. Firstly, we used the whole miRNA in the culture medium for our study. The free miRNAs extracted from microvesiclefree medium could be used because the species and quantity of non-vesicle bound free miRNAs may be quite different from the total miRNAs present in the culture medium. Secondly, a chemically synthesized, specific miRNA species might be needed for the study to exclude the interaction within a group of miRNAs. Further, two types of cells might be used to study intercellular miRNA transfer.
In conclusion, we found that there exists non-vesicle bound free miRNA in the culture medium of HK-2 cells. We also showed that these non-vesicle bound free miRNAs could gain entrance to other HK-2 cells and exert biological effects (i.e. affecting the expression of miRNA and mRNA expression). The role of free miRNA as intercellular messenger needs further exploration.
5. ACKNOWLEDGEMENTS
GW, BCHK and CCS designed the experiments; GW and KBL performed the laboratory experiments; GW, KMC and CCS performed data analysis; GW and CCS wrote the manuscript; PKTL took care of the administrative and regulatory aspect of the study. This work was supported in part by the CUHK research accounts 6901031 and 7101215, and the National Science Foundation of China (NSFC) grant number 81000287. All authors declare we have no conflict of interest.
REFERENCES
- Bartel, D.P. (2004) MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116, 281-297. doi:10.1016/S0092-8674(04)00045-5
- Vasudevan, S., Tong, Y. and Steitz, J.A. (2007) Switching from repression to activation: MicroRNAs can upregulate translation. Science, 318, 1931-1934. doi:10.1126/science.1149460
- Eiring, A.M., Harb, J.G., Neviani, P., Garton, C., Oaks, J.J., Spizzo, R., et al. (2010) miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell, 140, 652-665. doi:10.1016/j.cell.2010.01.007
- Lewis, B.P., Burge, C.B. and Bartel, D.P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15-20. doi:10.1016/j.cell.2004.12.035
- Chen, X., Ba, Y., Ma, L., Cai, X., Yin, Y., Wang, K., et al. (2008) Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research, 18, 997-1006. doi:10.1038/cr.2008.282
- Mitchell, P.S., Parkin, R.K., Kroh, E.M., Fritz, B.R., Wyman, S.K., Pogosova-Agadjanyan, E.L., et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy Sciences USA, 105, 10513-10518.
- Gilad, S., Meiri, E., Yogev, Y., Benjamin, S., Lebanony, D., Yerushalmi, N., et al. (2008) Serum microRNAs are promising novel biomarkers. PLoS One, 3, e3148. doi:10.1371/journal.pone.0003148
- Yuan, A., Farber, E.L., Rapoport, A.L., Tejada, D., Deniskin, R., Akhmedov, N.B. and Farber, D.B. (2009) Transfer of microRNAs by embryonic stem cell microve sicles. PLoS One, 4, e4722. doi:10.1371/journal.pone.0004722
- Hunter, M.P., Ismail, N., Zhang, X., Aguda, B.D., Lee, E.J., Yu, L., et al. (2008) Detection of microRNA expression in human peripheral blood microvesicles. PLoS One, 3, e3694. doi:10.1371/journal.pone.0003694
- Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J.J. and Lotvall, J.O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654- 659. doi:10.1038/ncb1596
- Chen, T.S., Lai, R.C., Lee, M.M., Choo, A.B., Lee, C.N. and Lim, S.K. (2010) Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Research, 38, 215-224. doi:10.1093/nar/gkp857
- Probst, J., Weide, B., Scheel, B., Pichler, B.J., Hoerr, I., Rammensee, H.G. and Pascolo, S. (2007) Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Therapy, 14, 1175-1180. doi:10.1038/sj.gt.3302964
- Wolff, J.A. and Budker, V. (2005) The mechanism of naked DNA uptake and expression. Advances in Genetics, 54, 3-20. doi:10.1016/S0065-2660(05)54001-X
- Paterson, E.L., Kolesnikoff, N., Gregory, P.A., Bert, A.G., Khew-Goodall, Y. and Goodall, G.J. (2008) The microRNA-200 family regulates epithelial to mesenchymal transition. Scientific World Journal, 8, 901-904. doi:10.1100/tsw.2008.115
- Miska, E.A. (2008) MicroRNAs—Keeping cells in formation. Nature Cell Biology, 10, 501-502. doi:10.1038/ncb0508-501
- Kato, M., Zhang, J., Wang, M., Lanting, L., Yuan, H., Rossi, J.J. and Natarajan, R. (2007) MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-betainduced collagen expression via inhibition of E-box repressors. Proceedings of the National Academy Sciences USA, 104, 3432-3437. doi:10.1073/pnas.0611192104

