Advances in Bioscience and Biotechnology
Vol.3 No.1(2012), Article ID:17184,7 pages DOI:10.4236/abb.2012.31002
Production, purification and characterization of D-aspartate oxidase from the fungus Trichoderma harzianum SKW-36
![]()
1Department of Biotechnology, Faculty of Life Sciences, Ritsumeikan University, Shiga, Japan
2Department of Applied Chemistry, Faculty of Life Sciences, Ritsumeikan University, Shiga, Japan
Email: *wakayama@sk.ritsumei.ac.jp
Received 5 October 2011; revised 14 November 2011; accepted 24 December 2011
Keywords: D-Aspartate Oxidase; Trichoderma harzianum; Purification; Characterization
ABSTRACT
Trichoderma harzianum Rifai SKW-36 produced two kinds of D-amino acid oxidizing enzymes. One enzyme was D-aspartate oxidase acting on acidic D-amino acids such as D-aspartate and D-glutamate and another one was D-amino acid oxidase acting on neutral D-amino acid such as D-phenylalanine and D-methionine. These enzymes in the cell-free extract were separated by DEAE-Toyopearl ion-exchange column chromatography. Casamino acids, peptone, and yeast extract as carbon and nitrogen sources were effective for the production of the enzymes. No D-amino acid tested induced the production of the enzymes. Casamino acid (0.33%) as carbon and nitrogen source gave a highest specific activity of D-aspartate oxidase among media tested. D-Aspartate oxidase, which was purified by four-step column chromatography in addition to ammonium sulfate precipitation, exhibited a subunit molecular mass of 40 kDa by SDS-PAGE analysis. D-Aspartate, D-glutamate and N-methyl-Daspartate were oxidized as substrates with the specific activities of 7.80 U/mg, 4.90 U/mg, and 4.22 U/mg, respectively. D-Asparagine, D-glutamine, D-alanine, and D-valine were slightly oxidized. No other Damino acids tested were inert. The enzyme exhibited relatively wide substrate specificity compared to Daspartate oxidases reported so far. The pH and temperature optima were 7.5 - 8.0 and 35˚C, respectively. The enzyme was stable at pH 6.0 - 9.0. About 75% of the enzyme activity was retained even after treating the enzyme at 50˚C for 10 min. The enzyme activity was inhibited not by benzoate and tartrate, but 60% and 24% by fumarate and malonate, respectively.
1. INTRODUCTION
D-Amino acid oxidase catalyzes the stereospecific oxidative deamination of D-amino acids to the corresponding α-keto acids, with producing ammonia and hydrogen peroxide in the course of the reaction. In general, the enzyme acting on D-amino acids except acidic D-amino acids, i.e. D-aspartate and D-glutamate, is so called D-amino acid oxidase. The enzyme acting on acidic Damino acids is called D-aspartate oxidase or D-glutamate oxidase according to substrate preference. D-Amino acid oxidases have been reported in microorganisms such as Cephalosporium acremonium [1], Fusarium solani [2], Fusarium oxysporum [3], Trigonopsis variabilis [4], Rhodotorula gracilis [5], and Candida boidinii [6] as well as in mammals such as pig [7], mouse [8], human [9]. D-Aspartate oxidase has been studied in human brain [10], Cryptococcus humicolus [11], and Fusarium sacchri var. elongatum [12]. D-Glutamate oxidase has been found in Octopus vulgaris [13] and Orconectes limosus [14]. The reactions catalyzed by these enzymes are expected to be exploited in biotechnological applications, for example, the production of α-keto acids from Damino acids, the resolution of the racemic mixtures of amino acids, and the analytical detection of D-amino acid.
We isolated Trichoderma harzianum Rifai SKW-36 as a producer of N-acyl-D-amino acid amidohydrolase, which catalyzes the hydrolysis of N-acyl-D-amino acid to D-amino acid and fatty acid [15]. Further study on D-amino acid metabolism of T. harzianum revealed that T. harzianum showed the activities of oxidative deamination towards neutral and acidic D-amino acids. Preliminary purification study showed that D-amino acid oxidizing activities detected in the cell-free extract of T. harzianum were separated into two different active portions by anionic ion-exchange column chromatography. One active portion showed D-amino acid oxidase activity and another did D-aspartate oxidase activity.
Some Trichoderma species are of industrial importance because they produce various enzymes useful in industry [16-18]. In particular, β-glucanases and chitinases from T. reesei have been well studies and widely used in pulp and food industries. However, no studies on D-amino acid metabolism and D-amino acid metabolizing enzyme of Trichoderma sp. have been studied. Moreover, there have been no reports describing the occurrence of two different kinds of D-amino acid oxidizing enzymes in fungi. The present paper describes the production condition and some properties of D-aspartate oxidase from T. harzianum Rifai SKW-36.
2. MATERIALS AND METHODS
2.1. Chemicals
2,4-Dinitrophenylhydrazine (DNPH), 2,2’-azino-bis(3- ethylbenzothiazoline-6-sulfonic acid (ABTS), catalase from bovine liver, and peroxidase from horse radish were purchased from Wako Pure Chemicals (Japan). D-Amino acids used were from Sigma (USA). The purity of all D-amino acid used in this study was more than 98%. DEAE-Toyopearl was purchased from Toso (Japan). EAH Sepharose 4B, HiTrap Q FF (5 cm × 1 cm) and HiTrap Butyl FF (5 cm × 1 cm) were purchased from GE Healthcare (UK). Immobilization of D-phenylalanine to Sepharose 4B was performed according to the manufacturer’s instruction. All other chemicals used were of analytical grade and commercially available.
2.2. Cultivation of T. harzianum Rifai SKW-36
T. harzianum SKW-36 was grown on a medium containing carbon source (~1.0%) and nitrogen source (~1.0%), 0.1 % K2HPO4, 0.1% KH2PO4, 0.05% MgSO4·7H2O, 0.1% yeast extract, and 0.075% streptomycin (pH 5.0). The culture was grown at 30˚C for 24 - 72 h with shaking in a 0.5 - l Sakaguchi flask containing 100 ml of medium or in 2 - l Sakaguchi flask containing 500 ml of medium.
2.3. Enzyme Assay
D-Amino acid oxidizing activities were assayed by measuring the formation of α-keto acids or hydrogen peroxide. Assay methods were as follows. α-Keto acid was determined by DNPH method. The standard reaction mixture for DNPH method contained 100 mM N-2-hydroxyethypiperazine-N’-2-ethane sufonic acid (HEPES) buffer (pH 8.0), 20 mM D-amino acid, 2 units of catalase, and the enzyme in a final volume of 0.5 ml. After 10-min incubation at 30˚C, 0.25 ml of 0.1% DNPH solution (in 2 M HCl) was added to the reaction mixture. After standing at room temperature for 3 min, 0.5 ml of 4 M NaOH solution was added to the mixture. The absorbance of dinitrophenylhydrazone formed was measured at 470 nm [19]. The amount of α-keto acid formed was calculated on the basis of calibration curve of each α-keto acid. Hydrogen peroxide was estimated by peroxidase-coupling method with ABTS. The standard reaction mixture for ABTS method contained 100 mM HEPES buffer (pH 8.0), 5 mM D-amino acid, 3 units of peroxidase, 1 mM ABTS, and the enzyme in a final volume of 1.0 ml. After 10-min incubation at 30˚C, the absorbance of ABTS oxidized form was measured at 470 nm [20].
2.4. Protein Determination
Protein concentration was determined by the method of Lowry et al. [21], using bovine serum albumin as a standard. It was also estimated by its absorbance at 280 nm in the fractionation of enzyme with column chromatography.
2.5. Purification of D-Aspartate Oxidase from T. harzianum
Cell growth and D-Asp oxidase activity of T. harzianum were examined at 24, 48, and 72 h in the casamino acid based medium. Maximum growth and enzyme activity were obtained after 48 h cultivation. T. harzianum SKW- 36 was cultivated aerobically at 30˚C for 48 h in a medium containing 0.33% casamino acids, 0.1% KH2PO4, 0.1% K2HPO4, 0.05% MgSO4·7H2O, 0.1% yeast extract, and 0.075% streptomycin (pH5.0). All subsequent purification steps were performed at 0˚C - 8˚C. The buffer, 10 mM potassium phosphate buffer, pH 7.0 was used throughout the purification unless otherwise stated. A 5 - l culture of T. harzianum SKW-36 was harvested by filtration with paper filter and washed twice in the buffer. After suspension containing about 50 g mycelium in 250 ml of the buffer, the cells were disrupted with ultrasonic oscillator (Nihonseiki Co., Japan) at 20 kHz for 40 min at 4˚C. The cell debris was removed by centrifugation (14,000× g) at 4˚C for 20 min. Crude extract was subjected to ammonium sulfate precipitation, and the active precipitate at a salt saturation between 30% and 60% was dissolved in the buffer and dialyzed against the buffer. The dialyzed enzyme solution was applied to an anionexchange column of DEAE-Toyopearl 650 M (f 3 × 50 cm), equilibrated with the buffer. Following an extensive wash with the buffer, the enzyme was eluted with 100 mM NaCl. The active fractions were applied to a Dphenylalanine-Sepharose affinity column (1.0 cm × 4.0 cm) equilibrated with the same buffer. Non-adsorbed protein was washed off thoroughly, and the enzyme was eluted with a liner gradient of NaCl from 0 to 130 mmol l–1 in the buffer. The active fractions were collected and dialyzed against the buffer. The enzyme solution was placed onto an ion-exchange column chromatography on HiTrap Q FF (5 cm × 1 cm), equilibrated with the buffer, and eluted with a linear gradient of NaCl from 0 to 220 mmol·l–1 in the buffer. The active fractions were collected and dialyzed against the buffer. Ammonium sulfate was added to the enzyme to 25% saturation. The enzyme solution was placed onto an hydrophobic interaction column chromatography on HiTrap Butyl FF (5 cm × 1 cm), equilibrated with the buffer containing 25% saturated ammonium sulfate. Following an extensive washing with 25% saturated ammonium sulfate, the elution of the enzyme was carried out with a linear gradient of ammonium sulfate from 25% to 0% saturation in the buffer. Subsequently, the active fractions were dialyzed against the buffer and concentrated with ultrafiltration system (Amicon, YM-10). The resulting enzyme preparation was stored at 4˚C.
2.6. SDS-PAGE for Purity and Molecular Weight Determination
Purity and molecular weight of purified D-aspartate oxidase were analyzed by SDS-PAGE according to the method of Laemmli [22]. The proteins were visualized by staining with Coomassie Brilliant Blue (R-250).
3. RESULTS AND DISCUSSIONS
3.1. Occurrence of Two Different Types of D-Amino Acid Oxidizing Enzymes
The cell-free extract prepared from T. harzianum SKW- 36, which was cultured in the medium containing casamino acids as a sole carbon and nitrogen source, exhibited the oxidase activity toward the wide range of Damino acids including acidic D-amino acid (D-aspartate and D-glutamate) and basic D-amino acid (D-arginine and D-lysine) as well as neutral ones (D-phenylalanine and D-methionine). To date, no reports have been appeared concerning D-amino acid oxidases having such wide substrate specificity except for the mutant of Damino acid oxidase from R. gracilis [23]. These indicated that T. harzianum SKW-36 might produce at least two kinds of D-amino acid oxidases different from each other in substrate specificity. In ammonium sulfate fractionation, the activity toward acidic D-amino acids, D-aspartate and D-glutamate, was only found in the fraction precipitated at 30% - 60% saturation and the activity toward D-phenylalanine and D-arginine was found in precipitates at both 30% - 60% and 60% - 90% saturation (data not shown). Moreover, when the cell-free extract was subjected to DEAE-Toyopearl column chromatography, the activity toward acidic D-amino acids was detected in the fractions eluted by 100 mM NaCl and the activity toward neutral and basic D-amino acids was detected in the fractions eluted in the NaCl concentration between 100 and 150 mM (data not shown). These results suggested that T. harzianum SKW-36 produced at least two kinds of D-amino acid oxidizing enzymes, D-aspartate oxidase and D-amino acid oxidase, in the cells.
3.2. Effects of Carbon and Nitrogen Sources on Enzyme Activities
The effects of carbon and nitrogen sources on the activeties of these D-amino acid oxidases were tested (Table 1). The media containing casamino acids or peptone as carbon and nitrogen (C-N) sources provided relatively high activities of D-aspartate oxidase and D-amino acid oxidase. While glucose as a carbon source had a tendency to increase the growth, the addition of glucose resulted in the lower activities of both the enzymes. Inorganic nitrogen compounds such as ammonium sulfate and ammonium chloride were not good nitrogen source. For the production of D-aspartate oxidase, casamino acids was found to be an most effective C-N source, and the highest specific activity of the enzyme was obtained under the cultivation with 0.33% casamino acids. For the production of D-amino acid oxidase, peptone (0.5%) and yeast extract (0.5%) were determined as most effective C-N sources. Effect of D-amino acids on activities of the enzymes was examined, but no inducible effect of Damino acids and their acetyl derivatives on the production of both the enzymes was observed in T. harzianum (data not shown). The D-amino acid oxidases from F. oxysporum [3] and R. gracilis [5], and D-aspartate oxidase from C. humicolus [11] have been reported to be induced effectively by D-amino acids such as D-Ala and D-Asp, respectively. The result indicates that the production of D-aspartate oxidase and D-amino acid oxidase from T. harzianum could be regulated by the different type of expression mechanism from F. oxysporum, R. gracilis, and C. humicolus.
3.3. Purification of D-Aspartate Oxidase
Enzyme purification started with about 2.4 g of soluble proteins, exhibiting a specific activity of 1.61 × 10–2 U·mg–1. D-Asp oxidase was enriched 2.35-fold by ammonium sulfate precipitation. After consecutive purification steps using anion exchange chromatography, Dphenylalanine-Sepharose affinity chromatography, and hydrophobic interaction chromatography, the purified enzyme was obtained with a specific activity of 4.61 U·mg–1, with a final recovery of 0.78 mg protein and a total yield of 9.2% (Table 2). SDS-PAGE revealed it
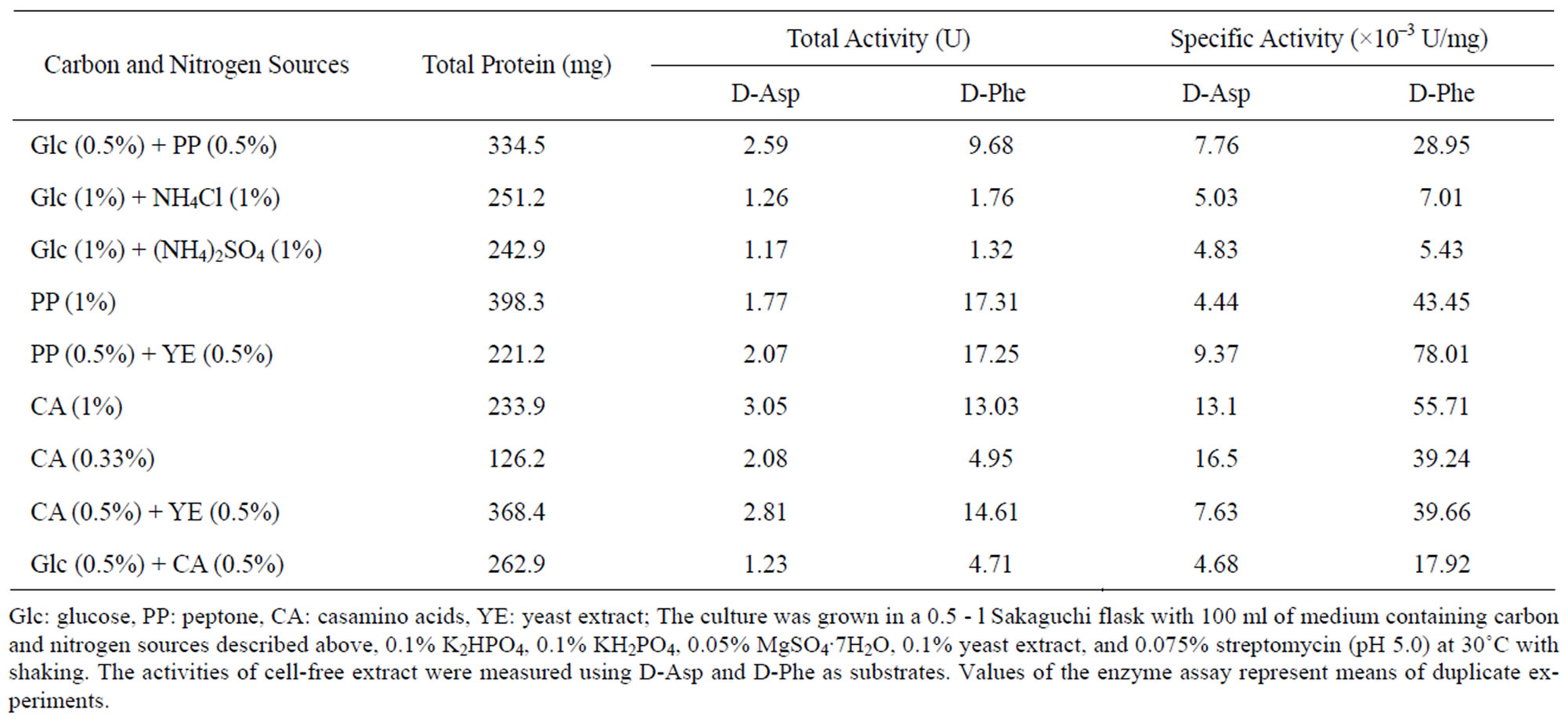
Table 1. Effect of carbon and nitrogen sources on the activity of D-aspartate oxidase and D-amino acid oxidase from T. harzianum.
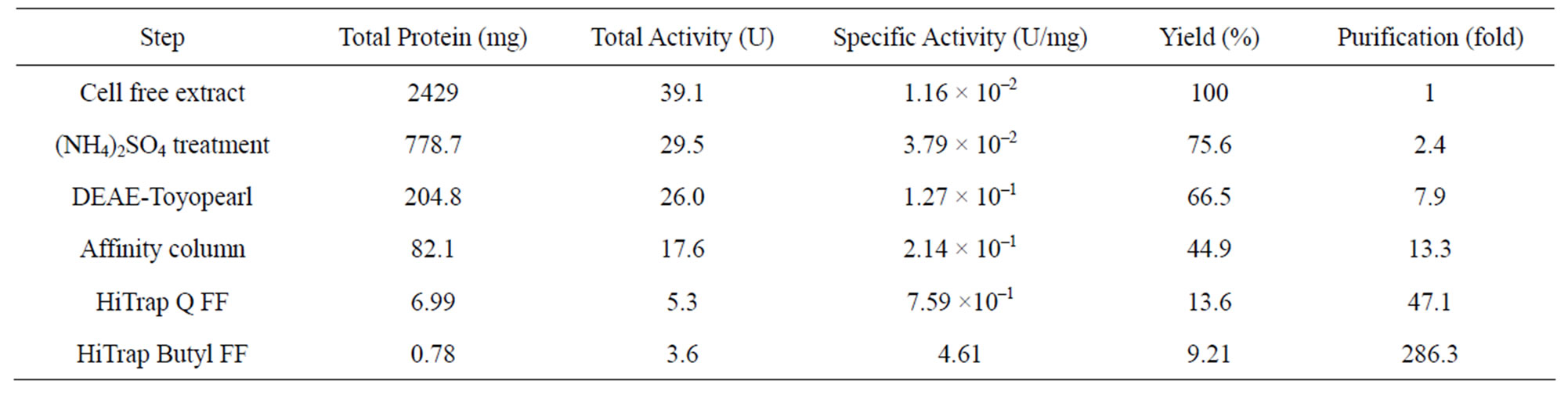
Table 2. Purification of D-aspartate oxidase from T. harzianum.
had a subunit molecular mass of 40 kDa (Figure 1). The monomer subunits of both human brain D-aspartate oxidase and C. humicolus D-aspartate oxidase have been reported to be the molecular masses of 37 and 40 kDa, respectively [10,11]. These are almost the same values with that of T. harzianum D-aspartate oxidase.
3.4. Characterization of D-Aspartate Oxidase
3.4.1. Effects of pH and Temperature on Enzyme Activity
The optimal pH and temperature for D-aspartate oxidase from T. harzianum were determined to be pH 7.5 - 8.0 and 35˚C, respectively (Figures 2(a) and (c)). The optimal pH and temperature of D-aspartate oxidase from T. harziaum were similar with those of D-aspartate oxidase from C. humicola [11]. At relatively wide range of pH between 6.0 and 9.0, the enzyme was stable, but by treating at 50˚C for 10 min at pH 5.5, 80% of the activity was lost, but at pH 10, 70% of the optimal enzyme activeity was still retained after 10 min. The enzyme was stable up to 30˚C, and by treatment at 50˚C for 10 min, only 25%
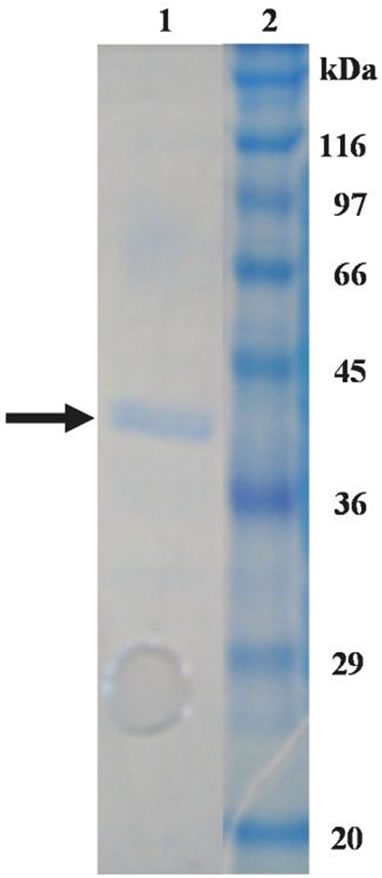
Figure 1. SDS-PAGE analysis of Daspartate oxidase from T. harzianum. lane 1: purified enzyme; lane 2: protein markers.
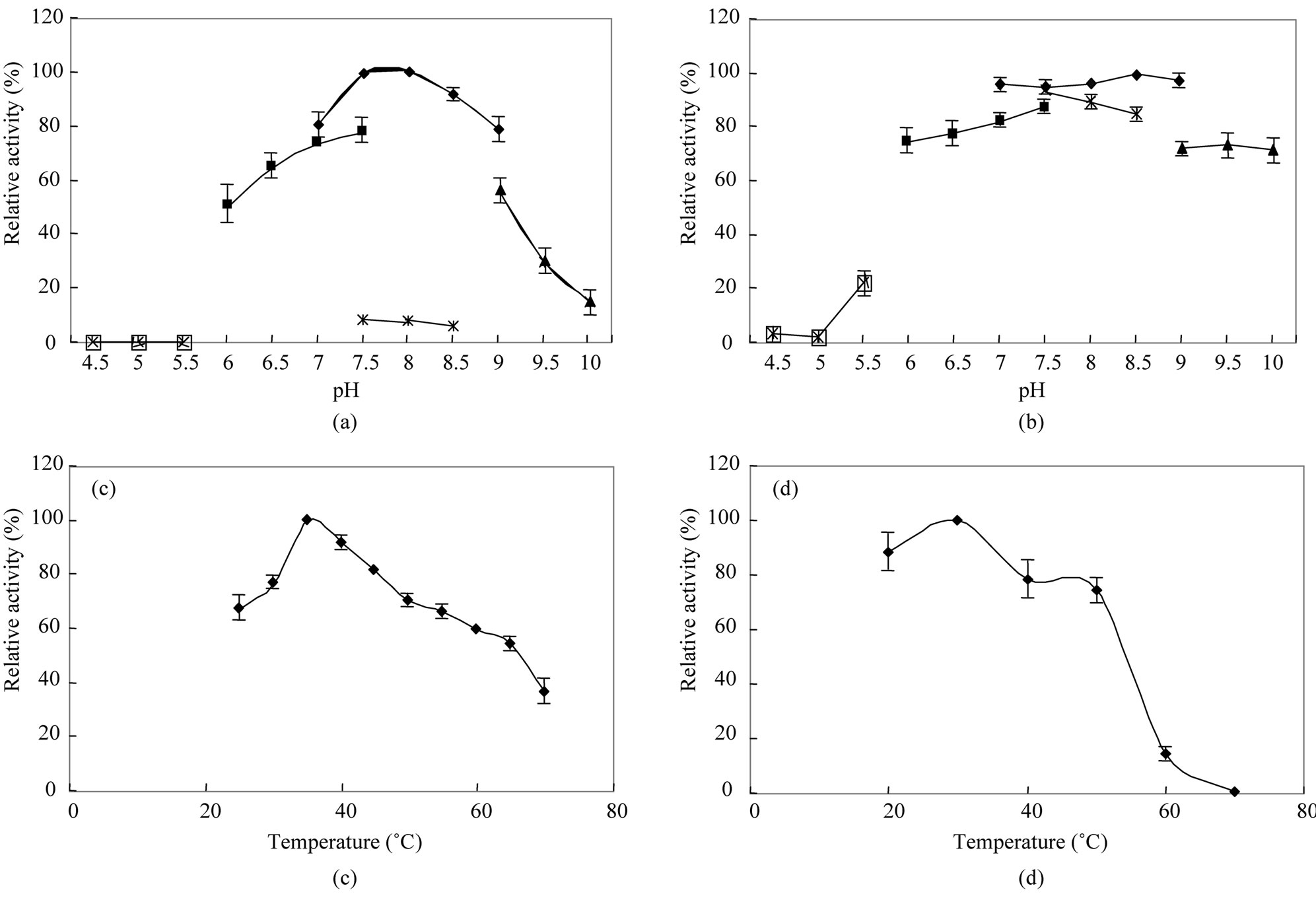
Figure 2. Effects of pH and temperature on enzyme activity and stability. (a) Enzyme activity was measured at various pH values with the following buffers: acetate buffer (pH 4.5 - 5.5; -□-), potassium phosphate buffer (pH 6 - 7.5; -■-), Tris·HCl (pH 7.5 - 8.5; -×-), HEPES buffer (pH 7.0 - 9.0; -♦-), and CHES buffer (pH 9 - 10; -▲-) and (b) Enzyme stability was evaluated after its incubation at various pHs at 50˚C for 10 min; (c) Enzyme activity was assayed at various temperatures under the standard reaction mixture and (d) Enzyme stability was evaluated after its incubation at various temperatures for 10 min. Error bars indicate standard deviations (n = 3).
of the optimal activity was lost (Figures 2(b) and (d)).
3.4.2. Kinetic Properties and Inhibitors
Table 3 shows the kinetic properties of D-aspartate oxidase from T. harzianum. D-Aspartate, D-glutamate, and N-methyl-D-aspartate (NMDA) were the preferred substrates and oxidized at the Vmax values of 7.80, 4.90, and 4.22 U/mg with Km values of 2.29, 3.48, and 0.62 mM, respectively. The Km value of D-aspartate oxidase from T. harzianum was similar with those of human brain and C. humicolus D-aspartate oxidases, 2.70 and 3.65 mM, respectively [10,11]. But the Km value for NMDA was rather different from those of human brain and C. humicolus D-aspartate oxidases, 6.80 and 28.0 mM, respec tively. In addition to these acidic D-amino acids, D-asparagine, D-glutamine, D-alanine, and D-valine were also slightly oxidized as low-reactive substrates at 12%, 8.8%, 9.3%, and 7.6% of the activity toward D-aspartate, respectively. It has been reported that the mutant M213R of D-amino acid oxidase from R. gracilis acted on D-aspartate and D-glutamate in addition to its original
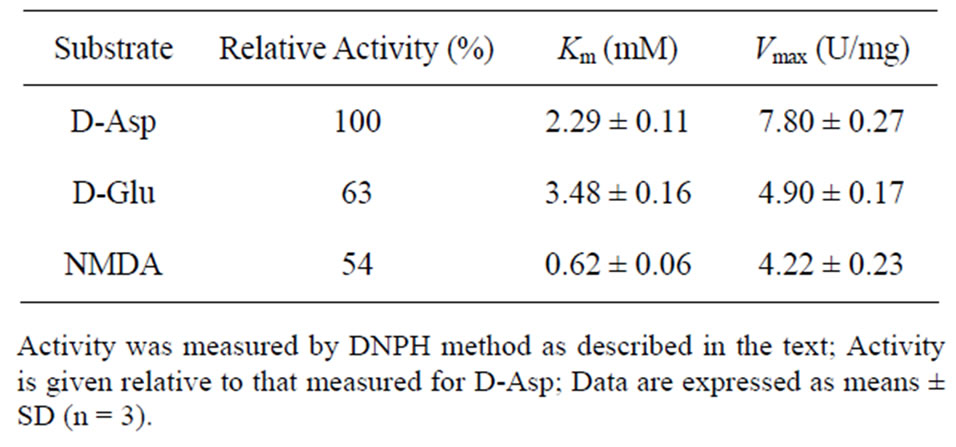
Table 3. Kinetic properties of D-aspartate oxidase from T. harzianum.
substrates [23], but no acidic D-amino acid oxidase catalyzing oxidative deamination of D-asparagine, D-glutamine, and D-alanine has been reported. D-Aspartate oxidase from T. harzianum showed no catalytic activity towards both L-aspartate and L-glutamate. It might be a new type of enzyme in substrate specificity, which acts on more wide rage of substrates than the other D-Asp oxidases reported previously. Inhibitory effects of carboxylic acids and metals were investigated (Table 4).
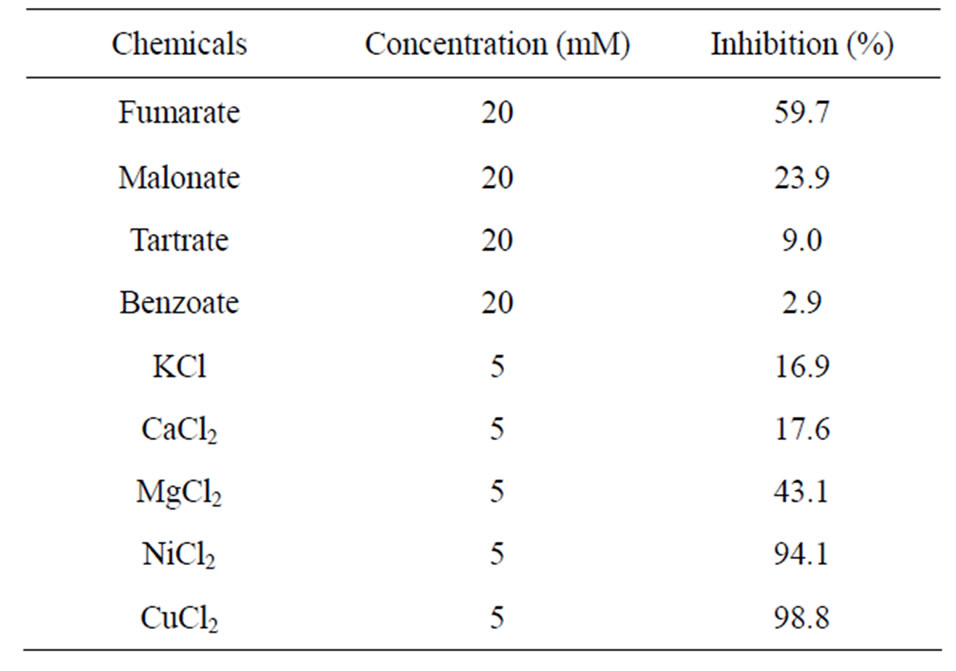
Table 4. Inhibition of D-aspartate oxidase from T. harzianum by carboxylic acids and metals.
NiCl2 and MgCl2 produced 94% and 43% inhibition at 5 mM, respectively, whereas CuCl2 inhibited the enzyme activity completely at 5 mM. Benzoate which is known as an effective inhibitor for mammalian D-amino acid oxidase was not effective for inhibition to D-aspartate oxidase from T. harzianum. Fumarate was the effective inhibitor followed by malonate. On the other hand, Daspartate oxidase from C. humicolus was inhibited by malonate but not fumarate [11].
4. CONCLUSIONS
This is the first paper describing the occurrence of the two different types of D-amino acid oxidizing enzymes in fungi. D-Aspartate oxidase, one of them, was purified from T. harzianum Rifai SKW-36 and characterized. In fungi, a little study on D-amino acid metabolism-related enzymes including D-amino acid oxidizing enzyme have been reported. T. harzianum produces N-acyl-D-amino acid amidohydrolase which acts on N-acyl derivatives of neutral D-amino acids [15]. This suggests that D-amino acid oxidase might be involved in the degradation of D-amino acids produced from N-acyl-D-amino acids by the action of N-acyl-D-amino acid amidohydrolase in T. harzianum. But N-acyl-D-amino acid amidohydrolase acting on N-acyl derivatives of acidic D-amino acid has not been reported in T. harzianum. Physiological role of D-aspartate oxidase in T. harzianum is interesting. To construct high expression system of D-aspartate oxidase in E. coli and yeast and D-aspartate oxidase defective mutant of T. harzianum, molecular cloning of the gene is under planning. Further studies will provide us more information and insight on fungal D-amino acid oxidizing enzymes and D-amino acid metabolism.
REFERENCES
- Benz, F., Liersch, M., Nuesch, J. and Treichler, H.J. (1971) Methionine metabolism and Cephalosporin C synthesis in Cephalosporium acremonium D-amino acid oxidase. European Journal of Biochemistry, 20, 81-88. doi:10.1111/j.1432-1033.1971.tb01365.x
- Isogai, T., Ono, H., Ishitani, Y., Kojo, H., Ueda, Y. and Kohsaka, M. (1990) Structure and expression of cDNA for D-amino acid oxidase active against Cephalorporin C from Fusarium solani. Journal of Biochemistry, 108, 1063-1069.
- Gabler, M. and Fischer, L. (1999) Production of a new D-amino acid oxidase from the fungus Fusarium oxysporum. Applied and Environmental Microbiology, 65, 3750-3753.
- Schroder, T. and Anderson, J. R. (1996) Studies on the inactivation of the flavoprotein D-amino acid oxidase from Trigonopsis variabilis. Applied Microbiology and Biotechnology, 45, 458-464. doi:10.1007/BF00578456
- Molla, G., Motteran, L., Piubelli, L., Pilone, M.S. and Pollegioni, L. (2003) Regulation of D-amino acid oxidase expression in the yeast Rhodotorula gracilis. Yeast, 20, 1061-1069. doi:10.1002/yea.1023
- Yurimoto, H., Hasegawa, T., Sakai, Y. and Kato, N. (2001) Characterization and high-level production of D-amino acid oxidase in Candida boidinii. Bioscience, Biotechnology, and Biochemistry, 65, 627-633. doi:10.1002/yea.1023
- Fukui, K., Watanabe, F., Shibata, T. and Miyake, Y. (1987) Molecular cloning and sequence of cDNAs encoding porcine kidney D-amino acid oxidase. Biochemistry, 26, 3612-3618. doi:10.1021/bi00386a054
- Tada, M., Fukui, K., Momoi, K. and Miyake, Y. (1990) Cloning and expression of a cDNA encoding mouse kidney D-amino acid oxidase. Gene, 90, 293-297. doi:10.1016/0378-1119(90)90193-U
- Fukui, K. and Miyake, K. (1992) Molecular cloning and chromosomal localization of human gene encoding Damino acid oxidase. Journal of Biological Chemistry, 267, 18631-18638.
- Setoyama, C. and Miura, R. (1997) Structural and functional characterization of the human brain D-aspartate oxidase. Journal of Biochemistry, 121, 798-803.
- Yamada, R., et al., (1996) Purification and properties of D-aspartate oxidase from Cryptococcus humicolus UJ1. Biocimica et Biophysica Acta, 1294, 153-158. doi:10.1016/0167-4838(96)00012-X
- Wakayama, M., Nakashima, S., Sakai, K. and Moriguchi, M. (1994) Isolation, enzyme production and characterization of D-aspartate oxidase from Fusarium sacchari var. elongatum Y-105. Journal of Fermentation and Bioengineering, 78, 377-379. doi:10.1016/0922-338X(94)90284-4
- Rocca, E. and Ghiretti, F. (1958) Purification and properties of D-glutamic acid oxidase from Octopus vulgaris. Lam. Archives of Biochemistry and Biophysics, 77, 336- 349. doi:10.1016/0003-9861(58)90081-X
- Urich, K. (1968) D-Glutamat oxidase aus Antennendruse des Flusskrebses Orconectes limosus: Reinigung und Charakterisierung. Zeitschrift fur Naturforschung, 23b, 1508- 1511.
- Wakayama, M., Kitahata, S., Manoch, L., Tachiki, T., Yoshimune, K. and Moriguchi, M. (2004) Production, purification and properties of D-aminoacylase from a newly isolated Trichoderma sp. SKW-36. Process Biochemistry, 39, 1119-1124. doi:10.1016/S0032-9592(03)00227-9
- Iqbal, H. M. N., Ahmed, I., Zia, M.A. and Irfan, M. (2011) Purification and characterization of the kinetic parameters of cellulose produced from wheat straw by Trichoderma viride under SSF and its detergent compatibility. Advances in Bioscience and Biotechnology, 2, 149-156. doi:10.4236/abb.2011.23024
- Juhasz, T., Szengyel, Z., Reczey, K., Siika-Aho, M. and Viikari, L. (2005) Characterization of cellulases and hemicellulases by Trichoderma reesei on various carbon sources. Process Biochemistry, 40, 3519-3525. doi:10.1016/j.procbio.2005.03.057
- Nampoothiri, M.K., Baiju, T.V., Sandhya, C., Sabu, A., Szakacs, G. and Pandey, A. (2004) Process optimization for antifungal chitinase production by Trichoderma harzianum. Process Biochemistry, 39, 1583-1590. doi:10.1016/S0032-9592(03)00282-6
- Kawano, C., Katsuki, H., Yoshida, T. and Tanaka, S. (1962) A method for extraction and determination of 2, 4-dinitrophenylhydrazones of keto acids. Analytical Biochemistry, 3, 361-368. doi:10.1016/0003-2697(62)90064-7
- Floch, C., Alarcon-Gutierrez, E. and Criquet, S. (2007) ABTS assay of phenol oxidase activity in soil. Journal of Microbiological Methods, 71, 319-324. doi:10.1016/j.mimet.2007.09.020
- Lowry, O.H., Rosebrough, N.J., Farr, A.L. and Randall, R.J. (1951) Protein measurement with the folin phenol reagent. Journal of Biological Chemistry, 193, 265-275.
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685. doi:10.1038/227680a0
- Sacchi, S., Lorenzi, S., Molla, G., Pilone, M.S., Rossetti, C. and Pollegioni, L. (2002) Engineering the substrate specificity of D-amino-acid oxidase. Journal of Biological Chemistry, 277, 27510-27516. doi:10.1074/jbc.M203946200
NOTES
*Corresponding author.

