Agricultural Sciences
Vol.5 No.2(2014), Article ID:42549,5 pages DOI:10.4236/as.2014.52012
Preliminary evaluation of resistance genes in rice against bacterial leaf blight in Guilan Province—Iran
1Department of Plant Pathology, Rice Research Institute of Iran (RRII), Rasht, Iran
2Department of Plant Breeding, Rice Research Institute of Iran (RRII), Rasht, Iran; *Corresponding Author: Ebady_al@yahoo.com
Copyright © 2014 Maryam Khoshkdaman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Maryam Khoshkdaman et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received 8 October 2013; revised 11 January 2014; accepted 22 January 2014
KEYWORDS
Bacterial Leaf Blight; Iran; Isogenic Lines; Pyramiding Lines; Resistance Genes; RICE
ABSTRACT
The reactions of rice bacterial leaf blight races were identified in Guilan province—Iran on 12 near-isogenic lines and 14 pyramiding lines from International Network for Genetic Evaluation of rice (INGER) and 8 local and improved Iranian varieties were evaluated under natural photoperiod condition in the field. Inoculation was done at panicle initiation by clipping the sterilized scissors in the bacterial suspension to booting stage. Scoring of inoculated plants was made 21 days after inoculation. Infection levels of pyramiding lines containing two to five resistance genes, expect, IRBB53 and IRBB61 with respectively resistance gene combination, Xa5 + Xa13 and Xa4 + Xa5 + Xa7, were not so clear. Among near-isogenic lines IRBB1, IRBB2, IRBB4 and IRBB10 carrying resistance gene Xa1, Xa2, Xa4 and Xa10 were susceptible; IRBB8, IRBB11, IRBB3, IRBB5 and IRBB13 were moderately susceptible; (having resistance gene Xa8, Xa11, Xa3, Xa5 and Xa13) IRBB14, IRBB21 and IRBB7 with respectively resistance gene Xa14, Xa21 and Xa7 were moderately resistance to bacterial blight. Furthermore, most of the time gene combinations support the strategy of pyramiding appropriate resistance gene. Local varieties were more susceptible than improved varieties to leaf blight disease. Among local varieties, Tarom was the most susceptible. And also, there were no significant differences among improved varieties and all of them were moderately resistance.
1. INTRODUCTION
Bacterial leaf blight of rice (Oryzae sativa), caused by Xanthomonas oryzae pv. oryzae [1] is a very serious disease in rice growing regions of the world [2]. The disease has become a major rice disease in last three decades because of the introduction of modern cultivars, which is highly responsive to nitrogen fertilizer. Bacterial blight of rice is a widespread and destructive disease in irrigated and rain fed environments of Asia [3,4]. There are no effective ways of protecting rice from the disease other than by the development of resistant cultivars. Host resistance is an important component of an integrated disease management program for bacterial disease [5]. Bacterial leaf blight interacts with rice in a gene—for gene manner, which makes it on ideal model for studying plant—pathogen interactions [6,7]. So for, thirty resistance genes for Bacterial Blight have been identified and utilized in rice breeding programs. Five of them, Xa1 [8], Xa21 [9], Xa5 [10], Xa26 [11] and Xa7 [12] have been cloned as full length DNA sequences. The last decade has seen a dramatic advancement in our understanding of the molecular basis of the rice X. oryzae pv. oryzae interaction, with the cloning of the tow rice resistance (R) genes, Xa1 and Xa21 [8,9] the bacterial avirulence (avr) genes, avr-Xa5, avrXa7 and avr-Xa10 [13], the hypersensitive response and pathogenicity (hrp) genes [14] and many other virulence related genes [15-21]. Analyses of X. oryzae pv. oryzae genomic sequences showed that there are 15 alleles in the avr Bs3/pthA (avr/pth) family of effectors [22]. So far, the functions of avrXa7 [23], avr-Xa10 [24,25], avrXa5 [26] and avrXa27 have been investigated. However, only the interaction between avr-Xa27 and Xa27 has been fully characterized [12]. In Myanmar, on the basis of host-pathogen interactionMyanmar rice varieties were classified into four groups. Almost of Myanmar cultivars were belonged to Type II and III which were susceptible to Myanmar isolates of Xanthomonas oryzae pv. oryzae [27].
In Guilan province, previous results on BLB showed that there were four races of X. oryzae pv. oryzae [28]. The purpose of this study is to identify resistance sources for controlling the rice bacterial blight in Guilan—Iran.
2. MATERIALS AND METHODS
2.1. Rice Varieties and Preparation of Plants
Seeds of rice near-isogenic and pyramiding lines were obtained from the International Network for Genetic Evaluation of Rice (INGER) (Table 1). This Lines with local and improved Iranian varieties used to evaluation of resistance genes in rice plant against bacterial blight in Guilan province at Rice Research Institute of Iran. The 26 differential rice lines with a single gene or pyramid lines of two to five genes and 8 local and improved varieties were tested in this study. Rice Seeds were sown in nursery in natural condition in wet season. Individual seedlings transplanted in the field (twenty-one days after sowing). Spacing between and within rows is 20 cm × 20 cm. The length of rows is 3 m. Separate plots were used for different isolates. The fertilizer was applied according to the fertility status of the soil. The cultivation and the management of rice in the field proceeded as usual.
2.2. Inoculum Preparation and Inoculation
Bacterial isolates that were maintained at −80˚C were revived on NA (nutrient agar) slants at 27˚C for 48 h. Each isolates was transferred to NA slants and incubated an additional 24 h at 27˚C. Each bacterial colony on the slants were suspended with sterilized distilled water and adjusted to concentrations of approximately 109 cfu/ml prior to inoculation (29). Inoculation was don at panicle initiation to booting stage by clipping the sterilized scissors in bacterial suspension and by clipping off the leaves 2 - 3 cm from leaf tip [30]. To reduce the possible effects of high temperature on disease reactions, favors the entry of bacteria into infection courts in the presence of sufficient moisture on the leaf surface and maintain inoculation time consistency, the inoculation was conducted in the morning.
2.3. Disease Reaction
Disease severity was rated by measuring the lesion length from the leaf tips. Scoring was done at 21 days after inoculation (DAI). The scales to be used in scoring were those from the 4th edition of the Standard Evaluation System for Rice [31]. The data analyzed for the main effect and interaction effect between cultivars and bacterial isolates.
3. RESULTS
3.1. Reactions of Near-Isogenic Lines
Interactions between near-isogenic lines and races of X. oryzae pv. oryzae are shown in Table 2. The degree of disease reaction in this study showed different relationship between combination of rice varieties and bacterial races. Race 4 showed the most susceptible and Race 1
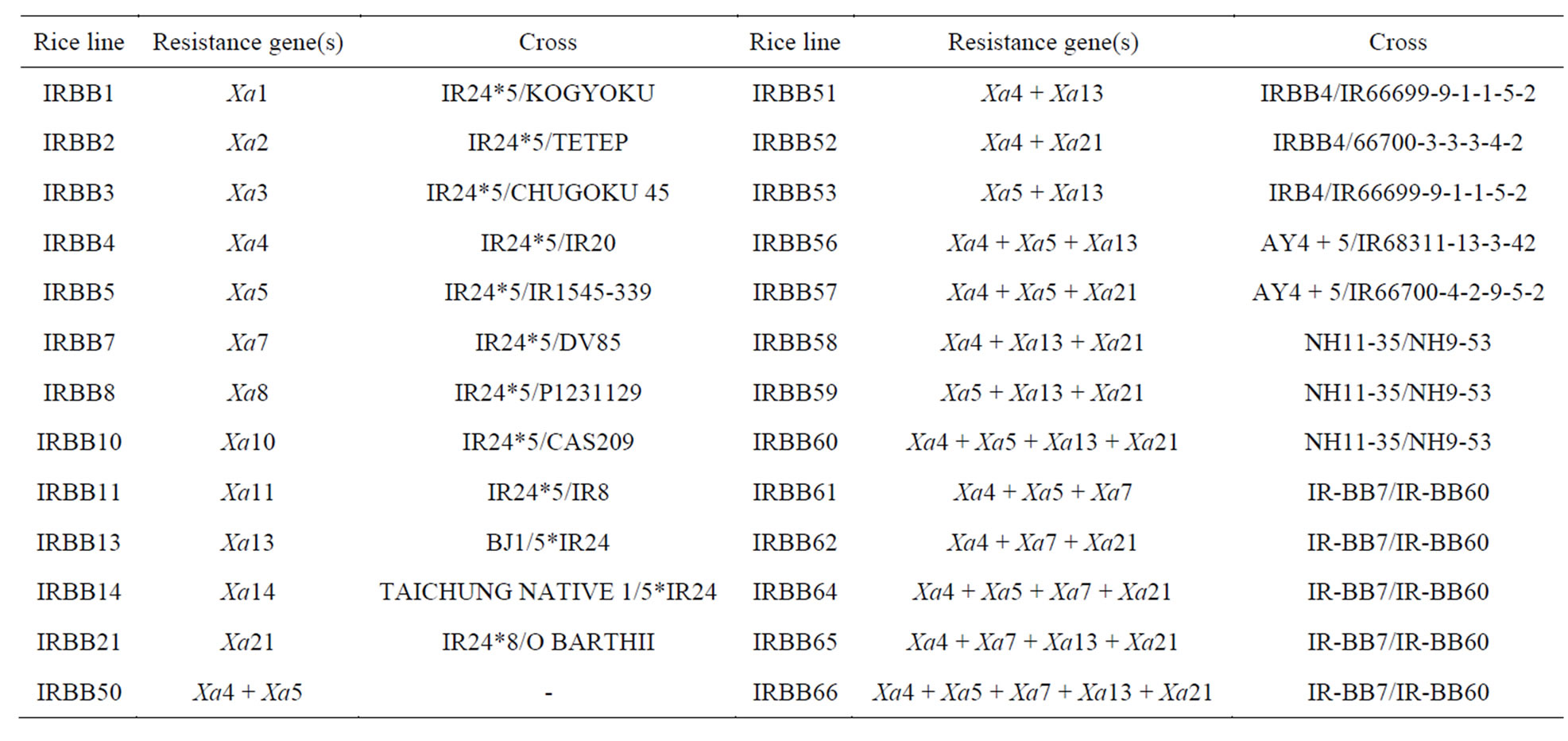
Table 1. Crosses and Resistance genes of differential rice lines used to characterize virulence of Xanthomonas oryzae pv. oryzae isolates in Iran.
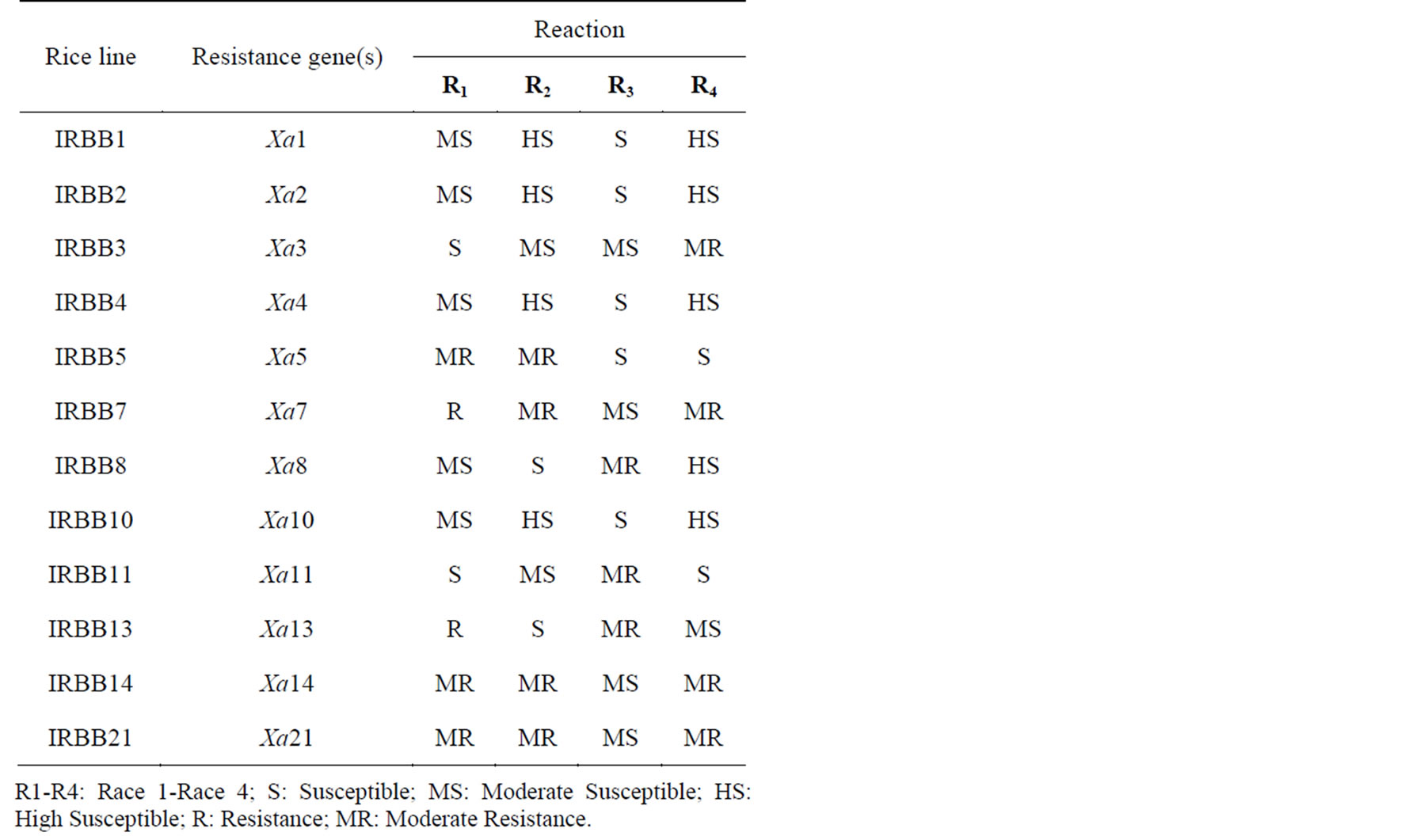
Table 2. Reaction of near-isogenic lines to races of X. oryzae pv. oryzae.
expressed the lowest susceptible among the races. Hashemi Cultivar (maybe has not major functional gene for resistance to Iranian races) was used as susceptible check. However, none of NILs was highly resistant or resistant to races of X. oryzae pv. oryzae in Guilan province.
3.2. Reaction of Pyramiding Lines
Interactions of pyramiding lines expressed in Table 3. Pyramiding lines containing two to five resistance genes except, IRBB53 and IRBB61 were significantly more resistant to the most races than their respective individual Xa gene, IRBB53 and IRBB61 with respectively resistance gene combination Xa5 + Xa13 and Xa4 + Xa5 + Xa7 were moderately resistance to bacterial races. Khazar Cultivar (maybe has major functional gene for resistance to Iranian races) was used as resistant check. High level of resistance was observed in combination of four and five dominant resistance genes Xa4 + Xa5 + Xa13 + Xa21, Xa4 + Xa5 + Xa7 + Xa21 and Xa4 + Xa5 + Xa7 + Xa13 + Xa21 (IRBB60, IRBB64 and IRBB66 respectively).
3.3. Reaction of Local and Improved Varieties
Iranian varieties were tested for their resistance to the four races (Table 4). Local varieties were more susceptible than improved varieties to leaf blight disease. Among local varieties, Tarom was the most susceptible. Between
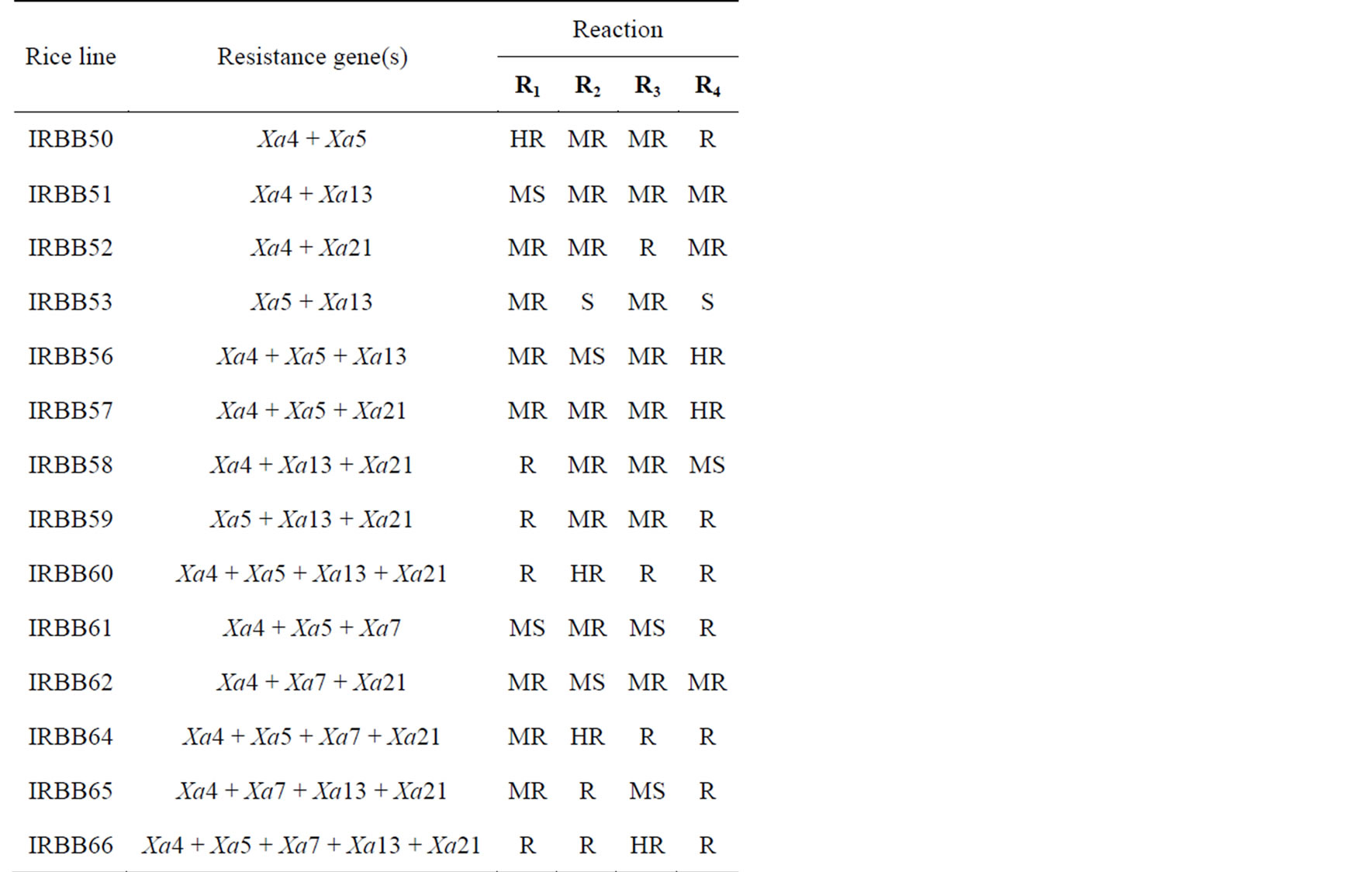
Table 3. Reaction of pyramiding lines to races of X. oryzae pv. oryzae.
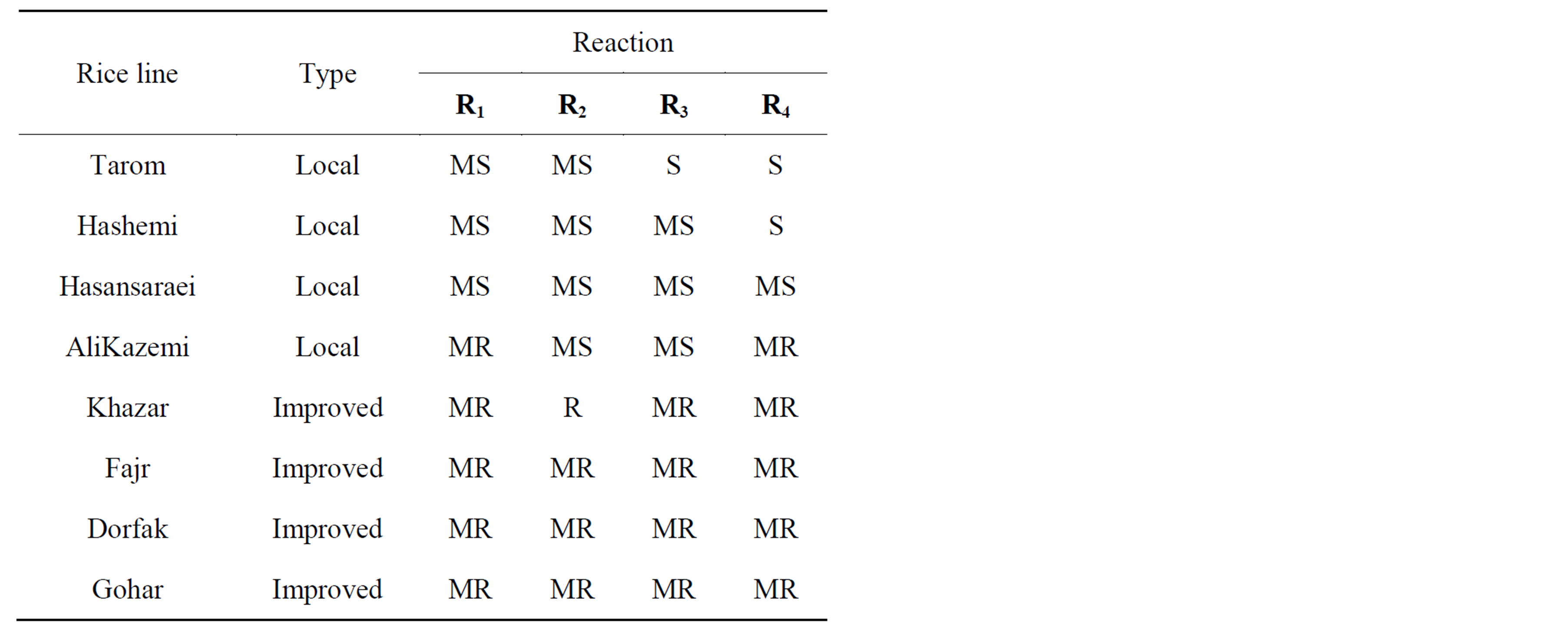
Table 4. Reaction of pyramiding lines to races of X. oryzae pv. oryzae.
improved varieties weren’t very important differential and all of them were moderately resistance.
4. DISCUSSION
The major goals of this project was to develop rice cultivars with effective resistance genes to Xanthomonas oryzae pv. oryzae. In this study, we establish the level of resistance to the bacterial blight pathogen conferred by a single gene individually and multiple gene combinations. Previous results on the basis of host-pathogen interaction showed that there were four bacterial leaf blight races in Iran [28]. These races were tested on 12 international differentials of near-isogenic lines and 14 varieties of pyramided lines and 8 local and improved varieties to support a gene deployment approach to managing the disease using resistant cultivars. The phenotype reactions between the single-gene lines and isolates were clear and easily classified into virulence or avirulence patterns. Among 12 near-isogenic lines with single gene, IRBB1 (Xa1) was low resistance and IRBB7 (Xa7) was high resistance on the basis of percentages of the compatible strains, also IRBB7 (Xa7) was not resistance to all tested isolates. These results indicate that IRBB21 (Xa21), IRBB14 (Xa14) and especially IRBB7 (Xa7) was resistant to the most isolates. Among Iranian varieties were tested in this project. Local varieties were more susceptible than improved varieties to leaf blight disease. Among local varieties, Tarom was the most susceptible. And also, there were no significant differences among improved varieties and all of them were moderately resistance.
Generally, in comparison between near-isogenic and pyramiding lines, the percent lesion area on pyramiding lines was significantly fewer than near-isogenic lines with the single resistant gene. But the Xa5 and Xa13 combination in IRBB53 was less resistant than gene Xa13 in IRBB13. As a result, none of the near-isogenic lines was resistant to all the races tested. IRBB7 (Xa7), IRBB14 (Xa14) and IRBB21 (Xa21) had a good resistant to the most of the races, but pyramiding lines, expect, IRBB53 and IRBB61 were significantly more resistant to the most races. Therefore a breeding program should be initiated to transfer these resistance genes from the differential varieties to high quality Iranian rice varieties to control the disease effectively.
There are obvious differences in the field-testing of the rice with the bacterial blight from different countries due to different agroecosystems [32]. Except for cultivar types, different terrains, climates, periods of rice planting and other factors may be associated with the population diversity and virulent variation of Xanthomonas oryzae pv. oryzae.
ACKNOWLEDGEMENTS
We thank Rice Research Institute of Iran (RRII) for preparation of field, plant material and all finance to doing this project.
REFERENCES
- Swings, J., Vander Mooter, M., Vauterin, L., Hoste, B., Gillis, M., Mew, T.W. and Kersteres, K. (1990) Reclassification of the causal agents of bacterial blight (Xanthomonas campestris pv. oryzicola) of rice as pathovars of Xanthomonas oryzae (ex Ishiyama, 1922) sp. nov. nom, rev. International Journal of Systematic and Evolutionary Microbiology, 40, 309-311. http://dx.doi.org/10.1099/00207713-40-3-309
- Niño-Liu, D.O., Ronald, P.C. and Bogdanove, A.J. (2006) Xanthomonas oryzae pathovars: Model pathogens of a model crop. Molecular Plant Pathology, 7, 303-324. http://dx.doi.org/10.1111/j.1364-3703.2006.00344.x
- Mew, T.W. (1987) Current status and future prospects of research on bacterial blight of rice. Annual Review of Phytopathology, 25, 359-382. http://dx.doi.org/10.1146/annurev.py.25.090187.002043
- Ou, S.H. (1985) Rice diseases. Common Wealth Mycological Institute, Kew.
- Mew, T.W. and Khush, G.S. (1981) Breeding for bacterial blight resistance in rice at IRRI. Proceedings of the 5th International Conference of Plant Pathologic Bacteria, Centro International de Agricultura Tropical (CIAT), Cali (Colombia), 16-23 August 1981, 504-510.
- Ronald, P.C. and Leung, H. (2002) The rice genome: The most precious things are not jade and pearls. Science, 296, 58-59. http://dx.doi.org/10.1126/science.1071337
- Chen, G.Y., Zou, L.F., Wang, X.P., Xiang, Y. and Wang, J.S. (2004) Pathogenicity determinants of Xanthomonas oryzae pv. Oryzae. Chinese Agricultural Sciences, 37, 1301-1307.
- Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z.X., Kono, I., Kurata, N., Yano, M., Iwata, N. and Sasaki, T. (1998) Expression of Xa1, a bacterial blightresistance gene in rice is induced by bacterial inoculation. Proceedings of the National Academy of Sciences USA, 95, 1663-1668.
- Song, W.Y., Wang, G. and Chen, L.L. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science, 270, 1804-1806. http://dx.doi.org/10.1126/science.270.5243.1804
- Anjali, S.I. and Susan, R.M. (2004) The rice bacterial blight resistance gene Xa5 encodes a novel form of disease resistance. Molecular Plant-Microbe Interactions, 17, 1348-1354. http://dx.doi.org/10.1094/MPMI.2004.17.12.1348
- Sun, X.L., Cao, Y.L., Yang, Z.F., Xu, C.G., Li, X.H., Wang, S.P. and Zhang, Q.F. (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. The Plant Journal, 37, 517-527. http://dx.doi.org/10.1046/j.1365-313X.2003.01976.x
- Gu, K.Y., Yang, B., Tian, D.S., Wu, L.F., Wang, D.J., Sreekala, C., Yang, F., Chu, Z.Q., Wang, G.L., White, F.F. and Yin, Z.C. (2005) R-gene expression induced by a type-III effector triggers disease resistance in rice. Nature, 435, 1122-1125. http://dx.doi.org/10.1038/nature03630
- Hopkins, C.M., White, F.F., Choi, S.H., Guo, A. and Leach, J.E. (1992) Identification of a family of avirulence gene from Xanthomonas oryzae pv. oryzae. Molecular Plant-Microbe Interactions, 5, 451-459. http://dx.doi.org/10.1094/MPMI-5-451
- Zhu, W., Magbanua, M.M. and White, F.F. (2000) Identification of tow novel hrp-associated genes in the hrp gene cluster of Xanthomonas oryzae pv. Oryzae. Journal of Bacteriology, 182, 1844-1853. http://dx.doi.org/10.1128/JB.182.7.1844-1853.2000
- Shen, Y., Sharma, P., da Silva, F. and Ronald, P. (2002) The Xanthomonas oryzae pv. oryzae rax\p and raxQ genes encode an ATP sulfurylase and adenosine 5—phosphosulphate kinase that are required for AvrXa21 avirulence activity. Molecular Microbiology, 44, 37-38. http://dx.doi.org/10.1046/j.1365-2958.2002.02862.x
- Xu, G.W. and Gonzales, C.F. (1989) Evaluation of TN4431- induced protease mutants of Xanthomonas campestris pv. oryzae for growth in plants and pathogenicity. Phytopathology, 79, 1210-1215.
- Ray, S.K., Rajeshwari, R. and Sonti, R.V. (2000) Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Molecular Plant-Microbe Interactions, 13, 394-401. http://dx.doi.org/10.1094/MPMI.2000.13.4.394
- Rajeshwari, R. and Sonti, R.V. (2000) Stationary-phase variation due to transposition of novel insertion elements in Xanthomonas oryzae pv. Oryzae. Journal of Bacteriology, 182, 4797-4802. http://dx.doi.org/10.1128/JB.182.17.4797-4802.2000
- Dharmapuri, S. and Sonti, R.V. (1999) A transposon insertion in the gumG homologue of Xanthomonas oryzae pv. oryzae causes loss of extracellular polysaccharide production and virulence. FEMS Microbiology Letters, 179, 53-59. http://dx.doi.org/10.1111/j.1574-6968.1999.tb08707.x
- Dharmapuri, S., Yashitola, J., Vishnupriya, M.R. and Sonti, R.V. (2001) Novel genomic locus with atypical G+C content that is required for extracellular polysaccharide production and virulence in Xanthomonas oryzae pv. oryzae. Molecular Plant-Microbe Interactions, 14, 1335- 1339. http://dx.doi.org/10.1094/MPMI.2001.14.11.1335
- Goel, A.K., Rajagopal, L. and Sonti, R.V. (2001) Pigment and virulence deficiencies associated with mutations in the aroE gene of Xanthomonas oryzae pv. oryzae. Applied and Environmental Microbiology, 67, 245-250. http://dx.doi.org/10.1128/AEM.67.1.245-250.2001
- Lee, B.M., Park, Y.J., Park, D.S., et al., (2005) The genome sequence of Xanthomonas oryzae pathovar KACC- 10331, the bacterial blight pathogen of rice. Nucleic Acids Research, 33, 577-586. http://dx.doi.org/10.1093/nar/gki206
- Yang, B., Zhu, W., Johnson, L.B. and White, F.F. (2000) The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent, nuclearlocalized, double-stranded DNA binding protein. Proceeding of the National Academy of Science, USA, 97, 9807-9812. http://dx.doi.org/10.1073/pnas.170286897
- Zhu, W., Yang, B., Chittoor, J.M., Johnson, L.B. and White, F.F. (1998) AvrXA10 contains an acidic transcriptional activation domain in the functionally conserved C terminus. Molecular Plant—Microbe Interactions, 11, 824-832. http://dx.doi.org/10.1094/MPMI.1998.11.8.824
- Zhu, W., Yang, B., Wills, N., Johnson, L.B. and White, F.F. (1999) The C terminus of AvrXA10 can be replaced by the transcriptional activation domain of VP16 from the herpes simplex virus. The Plant cell, 11, 1665-1674.
- Bai, J., Choi, S.H., Ponciano, G., Leung, H. and Leach, J.E. (2000) Xanthomonas oryzae pv. oryzae avirulence genes contribute differently and specifically to pathogen aggressiveness. Molecular Plant-Microbe Interactions, 13, 1322-1329. http://dx.doi.org/10.1094/MPMI.2000.13.12.1322
- Aye, S.S., Matsumoto, M., Kaku, H., Goto, T., Furuya, N. and Yoshimura, A. (2006) Evaluation of resistance in rice plants to Myanmar isolates of Xanthomonas oryzae pv. oryzae. Journal of faculty Agriculture Kyushu University, 52, 17-21.
- Khoshkdaman, M., Ebady, A.A. and Kahrizi, D. (2012) Evaluation of pathogenicity and race classification of Xanthomonas oryzae pv. oryzae in Guilan Province—Iran. Agricultural Science, 4, 557-561. http://dx.doi.org/10.4236/as.2012.34066
- Fang, Z.D., Xu, Z.G., Guo, C.J., Chen, Y.L., Deng, X.R., Li, H.S. and Zhu, L.M. (1981) Variability of pathogenicity of rice bacterial leaf blight organism. Journal of Nanjing Agricultural Technology College, 1, 1-11.
- Kauffman, H.E., Reddy, A.P.K., Hsieh, S.P.Y. and Merca, S.D. (1973) An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Disease Reporter, 57, 537-541.
- IRRI (1996) Standard evaluation system for rice. 4th Edition, IRRI, The Philippines.
- Ardales, E.Y., Leung, H., Vera Cruz, C.M., Leach, J.E., Mew, T.W. and Nelson, R.J. (1996) Hierarchical analysis of spatial variation of the rice bacterial blight pathogen across agroecosystems in the Philippines. Phytopathology, 86, 241-252. http://dx.doi.org/10.1094/Phyto-86-241

