Food and Nutrition Sciences
Vol. 2 No. 9 (2011) , Article ID: 8371 , 9 pages DOI:10.4236/fns.2011.29125
Modeling the Surface Cross-Contamination of Salmonella spp. on Ready-to-Eat Meat via Slicing Operation*
![]()
1Food Safety and Intervention Technologies Research Unit, Eastern Regional Research Center, Agricultural Research Service, United States Department of Agriculture, Wyndmoor, USA; 2Residue Chemistry and Predictive Microbiology Research Unit, Eastern Regional Research Center, Agricultural Research Service United States, Department of Agriculture, Wyndmoor, USA.
Email: {#Shiowshuh.sheen, Andy.hwang}@ars.usda.gov
Received July 20th, 2011; revised August 23rd, 2011; accepted August 30th, 2011.
Keywords: Salmonella, surface transfer, cross-contamination, modeling, slicing
ABSTRACT
Salmonella spp. surface transfer patterns/models between ready-to-eat (RTE) deli meat and a typical meat slicer are presented. A five-strain cocktail of Salmonella spp. was inoculated directly onto the rim of a round slicer blade at an initial level of ca. 4, 5, 6, 7 or 8 log CFU/blade (ca. 3, 4, 5, 6 or 7 log CFU/cm2 of the blade edge area). Thereafter, a RTE deli meat (ham) was sliced to a thickness of 1.5 - 2.0 mm. In another cross-contamination scenario, a clean blade was initially used to slice ham, which was pre-surface-inoculated with the Salmonella cocktail (ca. 2, 3, 4, 5, 6 or 7 log CFU/cm2 with a total area of 100 cm2), followed by slicing of an un-inoculated ham. The log CFU of Salmonella per ham slice was determined and empirical models were developed. The models, follow the decreasing Power law, predict the surface cross-contamination of Salmonella spp. (at any initial level) for sliced deli meat (ham) and will provide a useful tool in developing RTE meat risk assessments. Surface transfer patterns of three foodborne pathogens, i.e. Listeria monocytogenes, E. coli O157:H7 and Salmonella predicted by models are also presented.
1. Introduction
Salmonella is the commonly reported etiologic agent that causes bacterial foodborne disease outbreaks in the United States. Serotypes of Salmonella that have been implicated in causing salmonellosis were S. Typhimurium, S. Heidelberg, S. enteriditis, S. Montevideo and others. It is estimated that 1.4 million cases of salmonellosis occur in the US annually. The cost in the US associated with salmonellosis was estimated to be more than $2.9 billion [1]. Among the reported outbreaks of salmonellosis with known food vehicles, most were caused by the consumption of contaminated animal products in which poultry and poultry products are the frequent sources [2]. In the most recent report [3], Salmonella was found the leading cause of estimated hospitalizations and deaths, responsible for about 28 percent of deaths and 35 percent of hospitalizations due to known pathogens transmitted by food. In addition to poultry products, foods that have been implicated in salmonellosis out-breaks included ice cream [4], roast beef [5], ground beef [6], fermented sausage [7], peanut butter and spread [8,9], pistachios and pistachio products [9], vegetable sprouts [10], salad vegetables [11], almonds [12], coconut, chocolate, sauces and salad dressing, and cream-filled desserts and toppings [13], salami and ready-to-eat sausages [14]. The incidence of salmonellosis appears to be rising both in the US and in other industrialized nations [13]. Surveillance reports in European countries have indicated that 9% and 15% of human salmonellosis cases in Demark and the Netherlands, respectively, were associated with the consumption of contaminated pork [15]. In addition to pork, beef products have also be identified as food vehicles in outbreaks of salmonellosis [1]. Since there is a high possibility that Salmonella spp. can become established on environmental surfaces of beef and pork processing facilities, cross-contamination of Salmonella onto beef and pork products is highly possible. Further, added food ingredients may become a contamination source in RTE meat products which involved a recent salmonella outbreak and recalls of salami and RTE sausages [14].
Surface cross-contamination of foodborne pathogens on food products during processing or preparation has become a major concern to consumers and food manufacturers. Although risk assessment and analyses have been applied to monitor and to reduce the hazards, knowledge gaps related to surface cross-contamination at different processing stages need to be addressed for risk assessments [16]. Research that describe, simulate and model potential pathogen contamination may provide significant insight to enhance risk analyses, to ascertain the importance of equipment sanitation, and to improve equipment design. den Aantrekker et al. [17] reviewed the models for cross-contamination and recontamination of food products via three routes of contamination, i.e., equipment, air, and hands. They concluded that transfer rates for different recontamination scenarios and routes were needed to properly quantify the risk in a quantitative microbial risk assessment.
Mathematical models to predict the transfer of food pathogens between RTE meat and the slicer blade assist in assessing the risk of cross-contamination during slicing. The surface transfer models for Listeria monocytogenes during slicing were published by Aarnisalo et al. [18], Sheen [19], Sheen and Hwang [20]. Furthermore, E. coli O157:H7 transfer models were reported by PérezRodríguez et al. [21] and Sheen and Hwang [22]. Salmonella is another potential contaminant in the environment and in food ingredients. When the slicing equipment/operation involves multiple products, cross-contamination of Salmonella in RTE meats becomes highly possible if RTE products include other ingredients which may be Salmonella-contaminated [14]. It is important to understand the surface transfer patterns of different pathogens to improve food safety. The objectives of this study were to examine the transfer of Salmonella from a meat slicer to a ready-to-eat meat product and develop mathematical models to describe the transfer phenomenon.
2. Materials and Methods
2.1. Salmonella spp. Strains
A cocktail of five strains of Salmonella spp. including S. Copenhagen 8457 (pork isolate, FSIS), S. Typhimurium DT104, H3380 (clinical isolate, CDC), S. enteritidis H4386 (clinical isolate, CDC), S. Typhimurium FSIS 026 (beef isolate, FSIS), and S. Heidelberg 8456 (pork isolate, FSIS), was used in this study. A loopful of each strain was transferred from a stock culture stored at −80˚C into 10 ml of Brian Heart Infusion broth (BHI, Becton, Dickinson and Company, Sparks, MD) and incubated at 37˚C for 6 h. A loopful of each culture was then separately transferred into 10 ml of BHI broth and incubated at 37˚C for 24 h. Each strain was plated on Rappaport plates (Difco Lab, Sparks, Maryland) to determine the cell counts and adjusted to obtain equal cell concentration in the cocktail with 0.1% peptone water if needed. One ml of cell suspension from each strain was combined, and the mixture was further diluted with sterile 0.1% peptone water to the targeted level of Salmonella which was then applied to the blade or meat surface for transfer studies.
2.2. Delicatessen Slicer
A retail-scale, gravity-fed (45˚ angle) mechanical slicer (Model 3500, Globe Food Equipment Co., Dayton, OH) was used for ham slicing. The slicer was equipped with a 305 mm (12") diameter hollow ground knife (round blade) and operated at 300 revolutions per minute (rpm). The meat holding section was equipped with a 1.36 kg (3 lb) stainless steel end weight to deliver a consistent cut weight.
2.3. Ham Slicing and Microbial Inoculation
A RTE ham, from a local manufacturer, was used in the surface transfer experiment. The product contained 70% - 72% moisture, 18% - 20% protein, 1% - 2% carbohydrate, 2% salt and 2% - 3% fat. The average crosscut surface was about 150 mm × 90 mm (rectangular with round corners) and the whole ham weighed about 5 kg. The package-sealed ham was kept at 3˚C - 4˚C until used. The Salmonella spp. cocktail was evenly spread on each side of the blade’s sharp rim area. Ten drops, (total 10 × 10 μl/drop), were spread in 3 - 5 mm width and separated approximately 36˚ apart along the round blade edge. The same procedure was repeated for the other side of blade and the total area of blade edge inoculation was about 10 cm2 on each side. The inoculated blade was placed in a laminar airflow laboratory hood for approximately 20 - 30 min to allow for the drying of the inoculum and the slicer was then assembled. Ham was cut 1.5 - 2.0 mm in thickness and about 12 - 15 g in weight per slice. Each sliced sample was collected directly into a stomacher bag (Spiral Biotech, Norwood, MA) for Salmonella enumeration. Ham slices were individually weighed, mixed with an equal amount of 0.1% peptone water, and then stomached for 2 min (Bag Mixer, Model 400, St. Nom, France). Sample dilutions (50 ml to 1 ml) were spreadplated in duplicate onto Rappaport agar plates and incubated at 37˚C for 24 h.
A 10 cm2 area on the blade (both sides), blade protecttion cover, ham holding device, and liquid/waste receiving cup were swabbed with a cotton-tip before inoculating the blade with Salmonella. The swab was placed in 9 ml of peptone water, vortexed for 10 sec, and then 1.0 ml of the sample was spread-plated onto duplicate Rappaport agar plates, and incubated for 24 h at 37˚C. This is to ensure the whole slicer was free of Salmonella before each experimental run. After each experiment, the slicer unit was disassembled and all of the removable parts were soaked in BacDown Detergent Disinfectant (Decon Labs, Inc., Bryn Mawr, PA) diluted 1:10 with warm tap water for at least 1.0 h at room temperature and then manually scrubbed and rinsed with hot water. The slicer surface were cleaned with 70% ethanol and air dried. The blade was further autoclaved (121˚C and 30 min) to destroy all Salmonella after every use.
To study the transfer of contaminated meat to a clean blade and then to an un-inoculated ham, a slab of ham (30 mm thick) was inoculated with Salmonella (20 × 10 µl, total count of 8, 7, 6, 5, or 4 log CFU per slab) on four sides, except top and bottom surfaces. The 20 × 10 µl was spread on the four sides of the ham slab using a bent plastic rod spreader in an area of about 100 cm2. This ensured that most of the Salmonella cells were transferred to the blade surface, compared to inoculation on all surface (six sides) including top and bottom, and maximized the transfer for the subsequent slicing of the un-contaminated ham [20].
2.4. Sampling Size of Sliced Ham
The sample size, i.e., number of slices collected, was determined by the initial Salmonella inoculation level. At 3 or 4 log CFU initial level the transfer result showed zero count at less than 70 - 80 slices. At higher inoculation levels, 6 log CFU and higher, about 200 - 250 slices were collected. Due to the large sample size, the ham slices collected from the first to the fortieth slice were all analyzed for Salmonella, then every 5th slice was analyzed, i.e., 45th, 50th, 55th, to the 100th, and then every 10th slice was tested. Each experiment was repeated three times.
2.5. Model Development and Statistical Analyses
TableCurve 2D version 5.01 (Systat Software Inc., Richmond, CA) software was used to derive the empirical models, where the Salmonella count per ham slice was the dependent variable vs. slice sequence number as the independent variable. Due to the lack of clear understanding and the complexity of microbial surface transfer mechanism, it was difficult to predict or select the model type, which may properly describe the transfer pattern. TableCurve 2D screened hundreds of equations for fitting the experimental data with regression analyses, and the results reported all selected models in an order of either F-statistic or coefficients of determination (R2) from high to low order. Model selection criteria in this study were F-statistic, t-test of each coefficient for parameter, simplicity and R2. A “best-fit” model was selected to further fulfill the following criteria: 1) decaying transfer counts and approaching zero for large slice number; 2) no singularity and divergence in prediction; 3) a simple model with fewer coefficients and parameters; 4) P > |t| (< 0.001) for all coefficients; 5) highly significant F-statistic results [i.e., P > F (<0.0001)]; 6) r2 higher than 0.6 (used as a reference in model selection).
3. Results and Discussion
3.1. Surface Transfer of Salmonella from Inoculated Blade to Ham
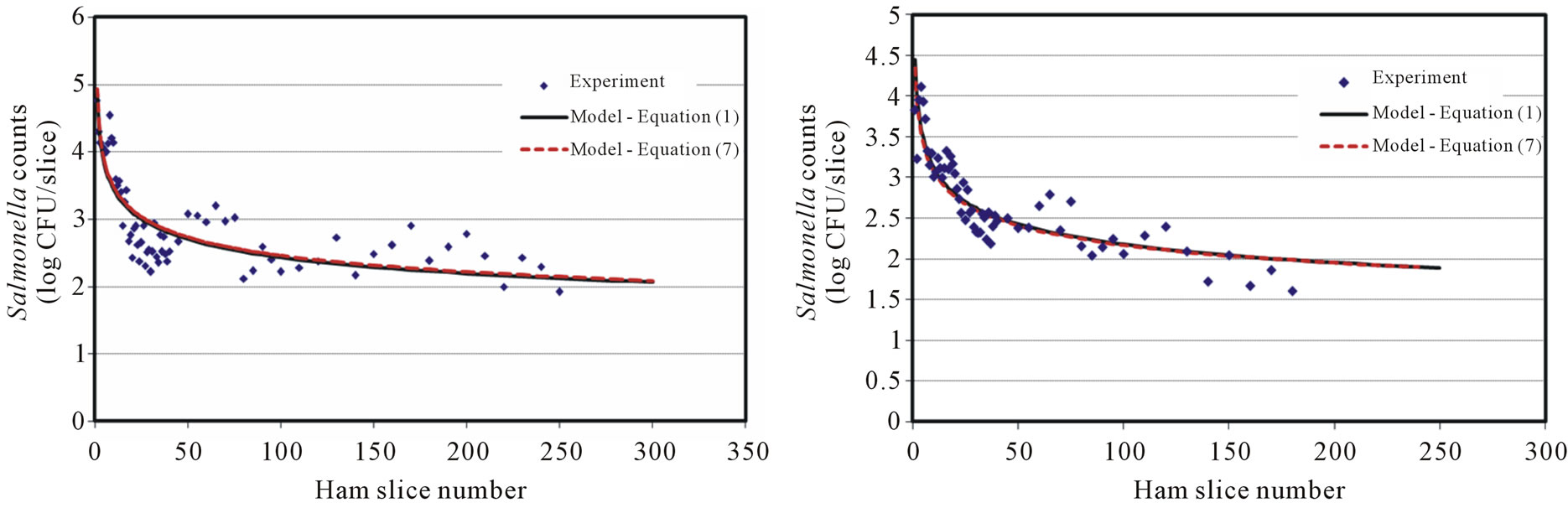
The Salmonella counts recovered from the first few slices were 1 - 2 logs higher than the immediately following data points (Figures 1(a)-1(c)), but approximately 2 - 3 logs less than the level of inoculum on the blade. The number of cells recovered from samples showed a continuously decreasing pattern (with some fluctuations) as the slicing progressed. The 8-log CFU (7.8 ± 0.2) contamination level may require testing of several hundred slices to visibly demonstrate the whole decreasing trend. Figures 1(b) and 1(c) show the 7 log CFU (6.8 ± 0.2) and 5 log CFU (4.8 ± 0.3) per blade contamination transfer results, respectively, which indicated a transfer pattern of an initial high Salmonella transfer followed by a sharp decrease, then, a slight increase (Figure 1(c)), and decrease again as the slicing proceeded. When a 4-log CFU level per blade inoculation was tested, the transfer counts showed a decreasing trend but with a random pattern in which a few slices with zero count (not detected) were observed before the 25th slice. There was no model suitable to describe this kind of experimental surface transfer trend, so only few surface transfer reports (with experimental data) for low level foodborne pathogen cross-contamination are available in the literature. Lin and others [23] investigated the cross-contamination of L. monocytogenes between processing equipment and deli meats (turkey, salami and bologna) with 1, 2, and 3 log CFU per blade inoculation and observed a similar low and random transfer result. They also confirmed positive and negative results by enrichments, and a 3-log CFU per blade inoculation showed that L. monocytogenes was positive in 12/200 (6%), 7/200 (3.5%), and 1/200 (0.5%) for turkey, salami and bologna samples, respectively.
A typical 2 - 3 log cell count reduction was observed at the initial transfer of sliced ham at high level inocula (≥5 log CFU) cases. When this pattern applied to low
 (a)
(a)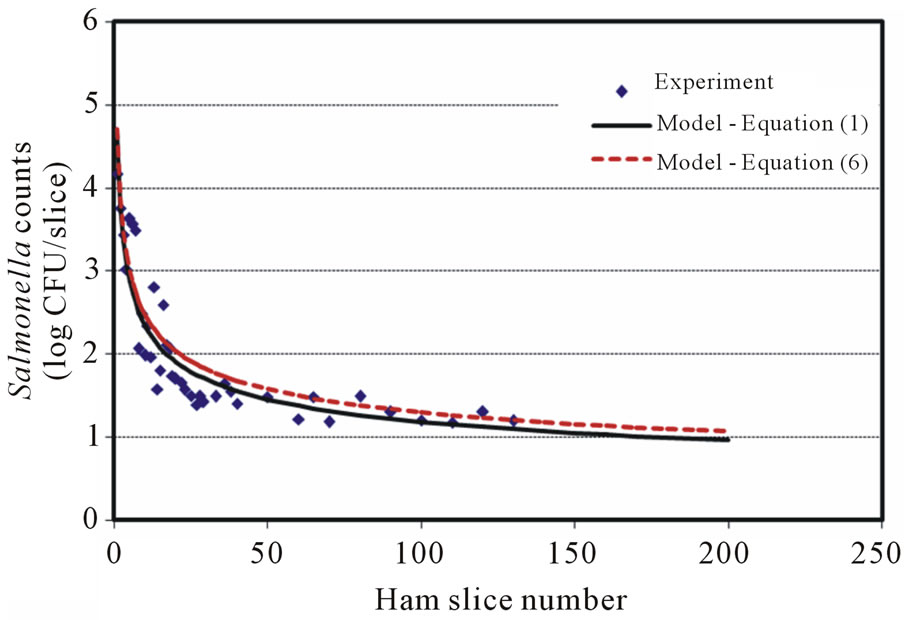 (b)
(b)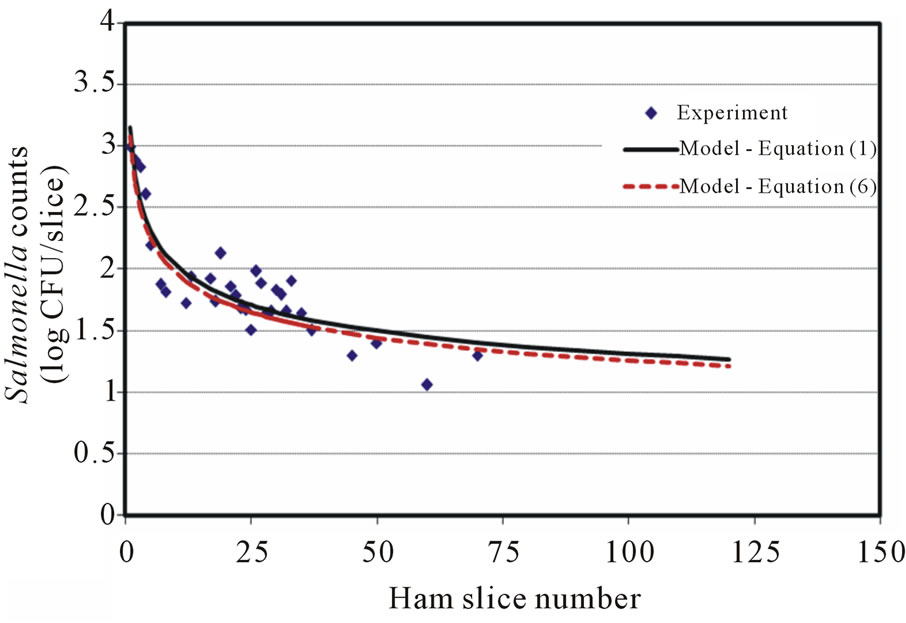 (c)
(c)
Figure 1. Salmonella surface transfer during slicing (contaminated blade to ham) at three different log CFU levels. (a) Salmonella at 8 log CFU level; (b) Salmonella at 7 log CFU level; (c) Salmonella at 5 log CFU level.
level inocula (≤4 log CFU), the surface transferred Salmonella may be reduced to 0 - 2 log CFU per sliced ham, which renders difficult to attain the meaningful cell enumeration counts for model development purposes. An enrichment test, which may detect the very low count, was not performed, since the results will not facilitate the transfer model development.
3.2. Models for Direct Blade Inoculation
The equation, which satisfies the model selection criteria, for surface transfer by direct blade inoculation at several inoculation levels is the Power equation and is shown below:
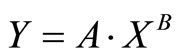 (1)
(1)
where Y is the log CFU count of the Salmonella per ham slice and X is the slice number (an integer) in one slicing series. A and B are constants derived from the regression analysis. Table 1 lists the A and B values for three different levels of inoculation on the blade edge (rim).
The model selection criteria mentioned previously were carefully examined and fulfilled. The R2 values ranged from 0.81 to 0.90 for low (5 log CFU) to high (8 log CFU) inoculation levels. The models predict that Y values approach zero at large X value without singularity. Figures 1(a)-1(c) also show a slight oscillation transfer pattern (in microbial counts) when slicing proceeds, which is similar to those observed in the surface transfer of Listeria [19,20] and E. coli [22]. A round blade operated at a certain rpm could contribute to this kind of microbial surface transfer results. The reason why the cells recovered from the first few slices were always 2 - 3 logs below the inoculum level on the blade was not clear, but it was probably caused by the adhesion of Salmonella onto the blade/food surface and/or death during slicing (shear stress impact—a harsh environmental condition).
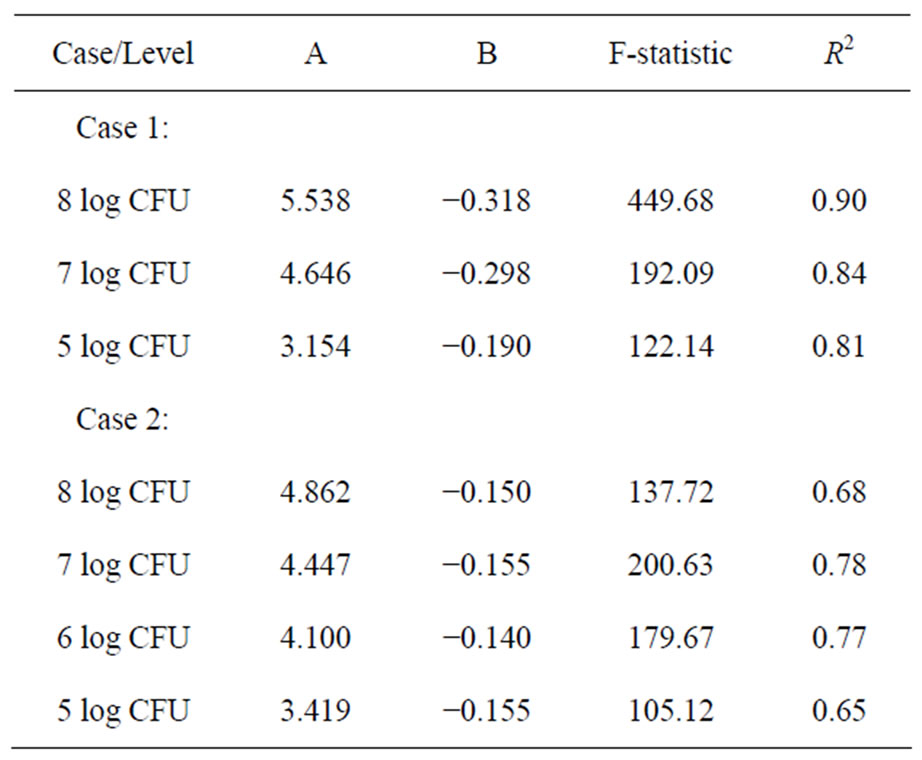
Table 1. Coefficients A and B in Equation (1) for 1) directblade inoculation to ham case and 2) contaminated-ham to blade to ham case of the Salmonella spp. surface transfer model.
3.3. Transfer of Salmonella from Inoculated Ham to Non-Contaminated Ham via Clean Blade
Ham surfaces pre-inoculated with Salmonella levels of 8, 7, 6, 5 and 4 log CFU were used to examine the cross surface transfer via a clean blade and model development. The results showed that the initial transfer (first slice) of Salmonella cells were about 0.5 log CFU/slice lower compared with those from the direct blade contamination at the same Salmonella inoculation level. It is reasonable to assume that only a portion of Salmonella cells attached to the blade, blade cover, and other slicer surfaces (e.g., meat holding plate) through the contaminated ham slicing. However, a significant amount of Salmonella transferred to the blade/slicer surface was observed based on the transfer results of Salmonella counts (Figure 2(a) to 2(d)). The results showed a similar decreasing pattern of Salmonella transferred onto the sliced ham in this scenario compared with the direct blade inoculation. The decreasing rate was slower and the cross-contamination lasts longer (more contaminated ham slices observed). With high inoculation levels at 8 (7.8 ± 0.2), 7 (6.8 ± 0.2) and 6 (5.8 ± 0.2) log CFU, a near-constant decreasing transfer was observed on the first 25 - 30 slices. Cells of Salmonella may loosely attach on the contaminated ham surface, which easily transferred to the blade, and other slicer surfaces, then to the un-contaminated ham surface.
Another possibility to cause the Salmonella more “transferable” was that a thin-film of ham residuals might have coated on the slicer surface (including blade) during the blade contamination procedure. When the attachment between Salmonella and ham was not as strong as between Salmonella and metal blade surface, the transfer became more feasible and a higher potential of crosscontamination might have occurred. The transfer results of 4 log CFU Salmonella inoculation showed a random mode (in all three replicates). There was no satisfactory model found to represent this type of transfer pattern.
 (a)
(a)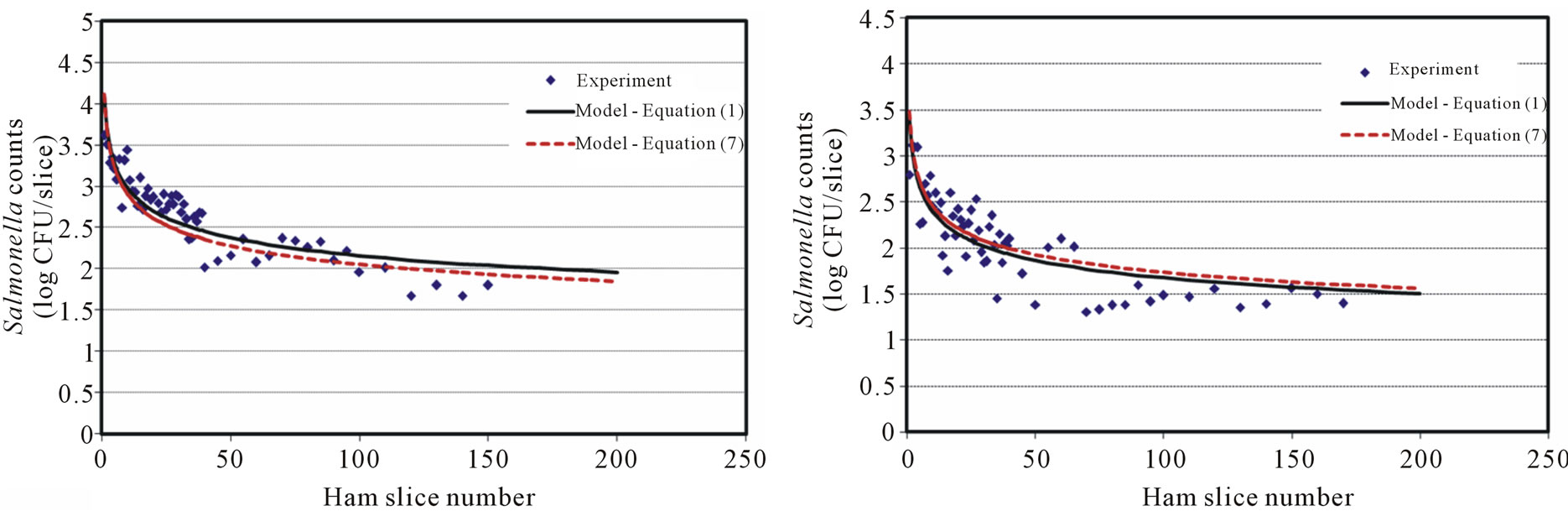 (b)
(b)
Figure 2. Salmonella surface transfer during slicing (inoculated ham to blade to ham) at four different log CFU levels. (a) Salmonella at 8 log CFU level; (b) Salmonella at 7 log CFU level; (c) Salmonella at 6 log CFU level; (d) Salmonella at 5 log CFU level.
3.4. Models for Transfer of Salmonella from Inoculated Ham to Non-Contaminated Ham via Clean Blade
The decaying Power equation, which satisfies the model selection criteria for surface transfer, is the same as Equation (1). Figures 2(a)-2(d) show the number of cells recovered from the slices at different levels of initial inoculum (8 to 5 log CFU) of Salmonella. The model coefficients (A and B) are shown in Table 1 with regression F-statistic and R2 values. When the model was used to predict an initial transfer count of the first slice (X = 1) at each level, the results were lower than the experimental data. For example, the predicted initial slice shows 4.86, 4.45, 4.10 and 3.42 for 8, 7, 6, and 5 log CFU levels, respectively. The observed data (X = 1) were 4.20, 4.10, 3.62 and 3.15 log CFU, which were about 0.3 - 0.7 log CFU below the estimated values. Although the models slightly over or under estimate the transfer counts, the application is to predict the potential cross-contamination in a longer process/operation time frame (i.e. at higher X value), which can be attained by using these models.
The general model, Equation (1), developed to describe the surface transfer of Salmonella on ham during slicing may be used to predict the possibility of Salmonella occurrence when approaching a large number of slices, e.g., 1000. By inserting 1000, 2000, and 5000 into Equation 1 of 5 log CFU, the Y value will be 1.20, 1.08, and 0.94 log CFU, respectively. The smaller the predicted number, the lower possibility the pathogen could exist. The predicted number may be further combined with a probability concept or model, if developed and not covered in this study, to estimate the potential risk of crosscontamination in a continuous process operation. Since there are other parameters in mechanical slicing (e.g., shear and surface roughness), which may impact microbial surface transfer at low food pathogen levels, the possible correlations between a potential distribution function and the model predicted data require more in-depth research to establish.
3.5. Transfer Models as Inoculation Level (n) Dependent
The values of coefficient A and B in Table 1 indicate that a simple relationship may exist between those coefficients and inoculation level (n). For both surface transfer scenarios in cross-contamination, the dependency of transfer count, Y, on inoculation level, n, was derived using the Microsoft Excel spreadsheet program and is shown below:
Direct blade inoculation:
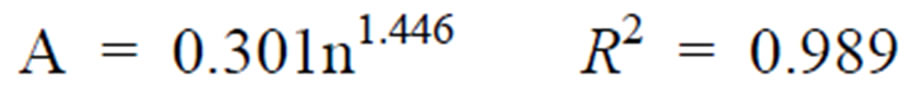 (2)
(2)
 (3)
(3)
Contaminated ham-blade-clean ham:
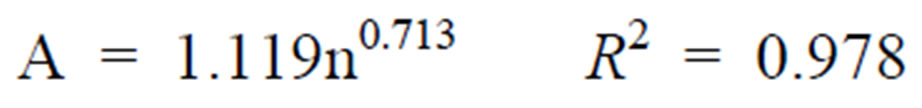 (4)
(4)
 (5)
(5)
By incorporating A (Equation (2)) and B (Equation (3)) into Equation (1), the transfer model for direct blade inoculation becomes:
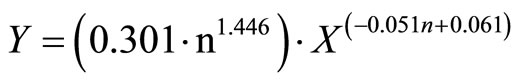 (6)
(6)
And, A (Equation (4)) and B (Equation (5)) into Equation (1), for the contaminated ham-blade-clean ham,
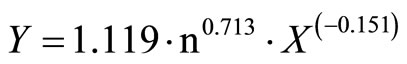 (7)
(7)
Using the non-linear regression procedures available in SAS® System [24] to examine Equations (6) and (7), the results showed both types of models satisfied the criteria (5) with the F-statistic result P > F (<0.0001). This result agreed with the finding by Montville and Schaffner [25] who studied the cross contamination between surfaces as influenced by inoculum size and concluded that the inoculum size and the amount of bacteria transferred must both be considered to accurately determine bacterial transfer rates.
3.6. Model Prediction Comparisons
Figures 1(a)-1(c) show the transfer predicting curves of direct blade inoculation using Equation (1) and Equation (6) at 8 to 5 log CFU levels, respectively. The two predictions agree well at the beginning, then, Equation (6) tends to have 0.1 log CFU predicted values difference (higher or lower) compared with those from Equation (1). Figures 2(a)-2(d) show the predicted surface transfer data for the contaminated ham to blade to ham case with Equation (1) and Equation (7) at 8 to 5 log CFU levels, respectively. The simulation comparisons between models, Equation (1) vs. Equation (6), and Equation (1) vs. Equation (7), showed similar results at different assigned levels of cross-contamination. Therefore, Equation 6 and Equation (7) can be applied to any Salmonella level of surface transfer during ham slicing.
3.7. Models and Predictions for the Microbial Surface Transfer—L. monocytogenes, E. coli O157:H7 and Salmonella
The available surface transfer models for food pathogens during slicing showed that L. monocytogenes (Lm) follows the exponentially decaying trend, which is similar to the first-order kinetic reaction model in both directblade-inoculation to meat and inoculated-meat to blade to meat cases [19]. However, the models for E. coli O157: H7 (Ec) were a power equation and an exponential equation for direct-blade-inoculation to meat transfer and inoculated-meat to blade to meat transfer case, respectively [22]. In this study, Salmonella (Sal) surface transfers were governed by the decaying Power law. The surface transfer complexities and other factors may have different degree of influence during slicing and/or surface shearrelated operations. The models that describe the surface cross-contamination could be microbial and process dependent. Figures 3 and 4 showed the surface transfer predictions and comparison for contaminated blade to RTE meat using the published models and the currently developed models for L. monocytogenes (Sheen) [19], E. coli O157:H7 (Sheen and Hwang) [22] and Salmonella. It is clearly demonstrated that the transfer patterns are different among those three foodborne pathogens. Figures 5 and 6 further demonstrated the surface transfers (contaminated meat to blade to meat) were foodborne pathogen and contamination level dependent.
When a slicing rate is known, the models may be converted to a time-dependent function using a similar ma-
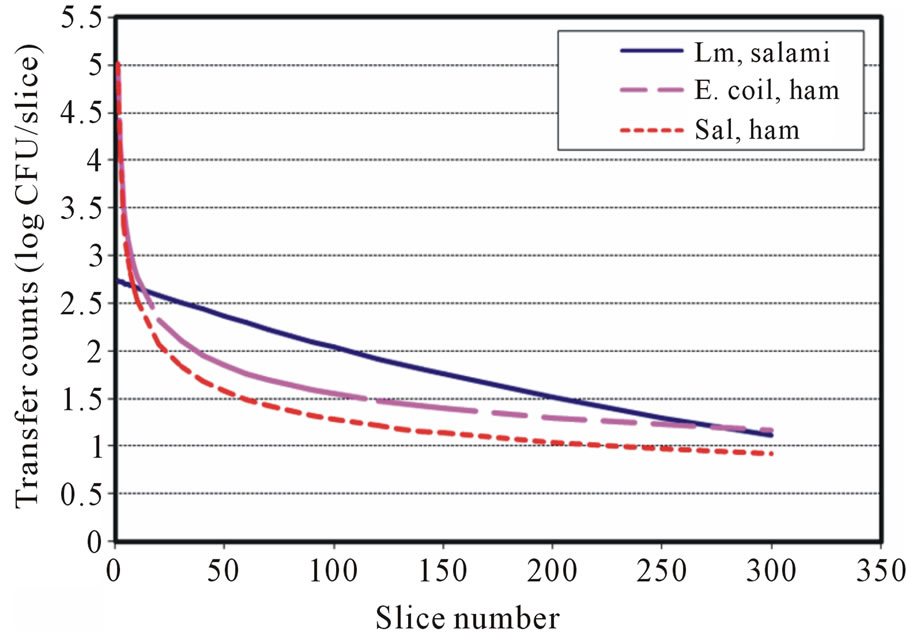
Figure 3. Transfer predictions using models with inoculation level at 7 log CFU for Lm, Ec and Salmonella (contaminated blade to RTE meat).
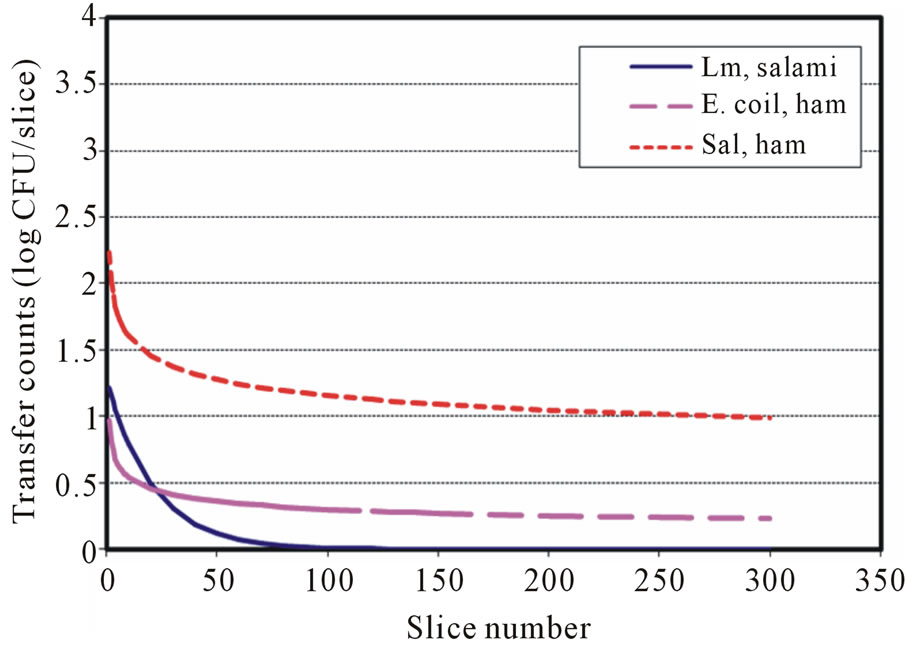
Figure 4. Transfer predictions using models with inoculation level at 4 log CFU for Lm, Ec and Salmonella (contaminated blade to RTE meat).
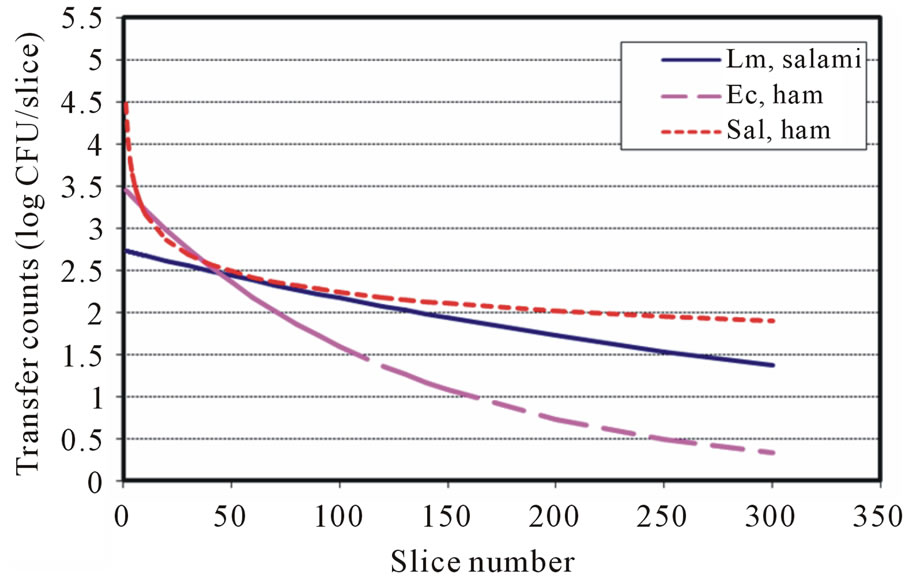
Figure 5. Transfer predictions using models with inoculation level at 7 log CFU for Lm, Ec and Salmonella (Inoulated RTE meat to blade to RTE meat).
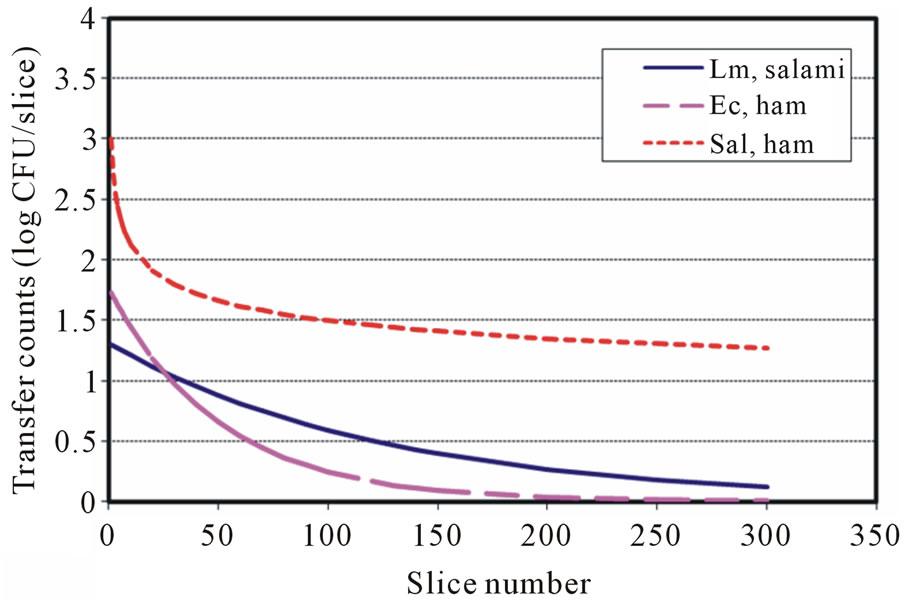
Figure 6. Transfer predictions using models with inoculation level at 4 log CFU for Lm, Ec and Salmonella (Inoulated RTE meat to blade to RTE meat).
thematical procedure reported by Sheen and Hwang [20]. Also, the models were presented in microbial counts per ham slice; a simple conversion may yield the results in microbial counts per gram or per cm2 with weight and surface area of sliced ham available which are 12 g and 270 cm2 (15 cm × 9 cm × 2 side) in the current study, respectively.
4. Conclusions
The surface transfer of Salmonella cells from slicer to ham (RTE deli meat) during slicing operation was simulated and modeled for two cross-contamination routes. The transfer was found to be significantly affected by the microbial inoculation level and contamination route. In general, the higher the initial contamination levels of Salmonella on the blade, the larger the number of ham slices that were contaminated with Salmonella during slicing. Salmonella cells that were introduced onto the blade by ham resulted in a longer time cross-contamination than cells that were inoculated directly onto the blade. The surface transfer pattern can be described using the Power law model, which is a function of inoculation level (n) and slicing number index (X). By using the models, surface transfers at any cross-contamination levels may be estimated or predicted. Surface transfer patterns of three foodborne pathogen were also presented. Although they all showed decreasing trends, however, the describing models were different and important for predictions. The empirical models may also provide a useful tool in the development of foodborne pathogen risk assessment for RTE deli meat.
5. Acknowledgements
The authors recognize the valuable and dedicated laboratory work of Sonya Costa and Peggy Williamson of the Food Safety and Intervention Technologies Research Unit, ERRC/ARS/USDA, Wyndmoor, PA.
REFERENCES
- S. Ojha and M. Kostrzynska, “Approaches for Reducing Salmonella in Pork Production,” Journal of Food Protection, Vol. 70, No. 11, 2007, pp. 2676-2694.
- K. E. Smith, C. Medus, S. D. Meyer, D. J. Boxrud, F. Leano, C. W. Hedberg, K. Elfering, C. Braymen, J. B. Bender and R. N. Danila, “Outbreaks of Salmonellosis in Minnesota (1998 through 2006) Associated with Frozen, Microwaveable, Breaded, Stuffed Chicken Products,” Journal of Food Protection, Vol. 71, 2008, pp. 2153-2160.
- Centers for Disease Control and Prevention, “CDC Reports 1 in 6 Get Sick from Foodborne Illnesses Each Year—Online Newsroom,” 2010.
- K. J. Vought and S. R. Tatini, “Salmonella Enteritidis Contamination of Ice Cream Associated with A 1994 Multistate Outbreak,” Journal of Food Protection, Vol. 61, No. 1, 1998, pp. 5-10.
- R. Shapiro, M. L. Ackers, S. Lance, M. Rabbani, L. Schaefer, J. Daugherty, C. Thelen and D. Swerdlow, “Salmonella Thompson Associated with Improper Handling of Roast Beef at A Restaurant in Sioux Falls, South Dakota,” Journal of Food Protection, Vol. 62, No. 2, 1999, pp. 118-122.
- J. McLaughlin, L. J. Castrodale, M. J. Gardner, R. Ahmed and B. D. Gessner, “Outbreak of Multidrug-Resistant Salmonella Typhimurium Associated with Ground Beef Served at a School Potluck,” Journal of Food Protection, Vol. 69, No. 3, 2006, pp. 666-670.
- C. J. Sauer, J. Majkowski, S. Green and R. Eckel, “Foodborne Illness Outbreak Associated with A Semi-Dry Fermented Sausage Product,” Journal of Food Protection, Vol. 60, No. 12, 1997, pp. 1612-1617.
- S. L. Burnett, E. R. Gehm, W. R. Weissinger and L. R. Beuchat, “Survival of Salmonella in Peanut Butter and Peanut Butter Spread,” Journal of Applied Microbiology, Vol. 89, No. 3, 2000, pp. 472-477. doi:10.1046/j.1365-2672.2000.01138.x
- US Food and Drug Administration, “Safety: Major Product Recalls,” 2011. http://www.fda.gov/Safety/Recalls/MajorProductRecalls/default.htm
- K. L. Winthrop, M. S. Palumbo, J. A. Farrar, J. C. Mohle-Boetani, S. Abbott, M. E. Beatty, G. Inami and S. B. Werner, “Alfalfa Sprouts and Salmonella Lottubs Infection: A Multistate Outbreak Following Inadequate Seed Disinfection with Heat and Chlorine,” Journal of Food Protection, Vol. 66, No. 3, 2003, pp. 13-17.
- S. K. Sagoo, C. L. Little, L. Ward, I. A. Gillespie and R. T. Mitchell, “Microbiological Study of Ready-to-eat Salad Vegetables from Retail Establishments Uncovers A National Outbreak of Salmonellosis,” Journal of Food Protection, Vol. 66, No. 3, 2003, pp. 403-409.
- S. Isaacs, J. Aramini, B. Ciebin, J. A. Farrar, R. Ahmed, D. Middleton, A. U. Chandran, L. J. Harris, M. Howes, E. Chan, A. S. Pichette, K. Campbell, A. Gupta, L. Y. Lior, M. Pearce, C. Clark, F. Rodgers, F. Jamieson, I. Brophy and A. Ellis, “An International Outbreak of Salmonellosis Associated with Raw Almonds Contaminated with a Rare Phage Type of Salmonella enteritidis,” Journal of Food Protection, Vol. 68, No. 1, 2005, pp. 191-198.
- US Food and Drug Administration, “Bad Bug Book: Foodborne Pathogenic Microorganisms and Natural Toxins Handbook Salmonella spp.,” 2011. http://google2.fda.gov/search?q=major+product+recalls%2C+2009&client=FDAgov&site=FDAgov&lr=&proxystylesheet=FDAgov&output=xml_no_dtd&getfields=*&x=6&y=10
- Centers for Disease Control and Prevention, “Investigation Update: Multistate Outbreak of Human Salmonella Montevideo Infections,” 2010.
- G. Y. Miller, X. Liu, P. E. McNamara and D. A. Barber, “Influence of Salmonella in Pigs Preharvest and During Pork Processing on Human Health Costs and Risks from Pork,” Journal of Food Protection, Vol. 68, No. 9, 2005, pp. 788-1798.
- D. L. Gallager, E. D. Ebel and J. R. Kause, “FSIS Risk Assessment for Listeria monocytogenes in Deli Meats,” Food Safety and Inspection Service, US Department of Agriculture, 2003. http://www.fsis.usda.gov/OPPDE/rdad/FRPubs/97-013F/ListeriaReport.pdf
- E. D. den Aantrekker, R. M. Boom, M. H. Zwietering and M. van Schothorst, “Quantifying Recontamination through Factory Environments—A Review,” International Journal of Food Microbiology, Vol. 80, No. 2, 2002, pp. 117- 130.
- K. Aarnisalo, S. Sheen, L. Raaska and M. Tamplin, “Modelling Transfer of Listeria Monocytogenes during Slicing of ‘Gravad’ Salmon,” International Journal of Food Microbiology, Vol. 118, No. 1, 2007, pp. 69-78. doi:10.1016/j.ijfoodmicro.2007.06.017
- S. Sheen, “Modeling Surface Transfer of Listeria Monocytogenes on Salami during Slicing,” Journal of Food Science, Vol. 73, No. 6, 2008, pp. E304-311. doi:10.1111/j.1750-3841.2008.00833.x
- S. Sheen and C. A. Hwang, “Modeling Transfer of Listeria monocytogenes from Slicer to Deli Meat during Mechanical Slicing,” Foodborne Pathogens and Diseases, Vol. 5, No. 2, 2008, pp. 135-146. doi:10.1089/fpd.2007.0049
- F. Pérez-Rodríguez, A. Valero, E. C. D. Todd, E. Carrasco, R. M. Carcía-Gimeno and G. Zurera, “Modeling Transfer of Escherichia coli O157:H7 and Staphylococcus Aureus during Slicing of a Cooked Meat Product,” Meat Science, Vol. 76, No. 4, 2007, pp. 692-699. doi:10.1016/j.meatsci.2007.02.011
- S. Sheen and C. A. Hwang, “Mathematical Modeling the Cross-contamination of Escherichia coli O157:H7 on the Surface of Ready-to-Eat Meat Product While Slicing,” Food Microbiology, Vol. 27, No. 1, 2010, pp. 37-43. doi:10.1016/j.fm.2009.07.016
- C. Lin, K. Takeuchi, L. Zhang, C. B. Dohm, J. D. Meyer, P. A. Hall and M. P. Doyle, “Cross-Contamination between Processing Equipment and Deli Meats by Listeria monocytogenes,” Journal of Food Protection, Vol. 69, No. 1, 2006, pp. 71-79.
- R. J. Freund and R. C. Littell, “SAS System for Regression,” 2nd Edition, SAS Institute Inc., Cary, 1991.
- R. Montville and D. W. Schaffner, “Inoculum Size Influences Bacterial Cross Contamination between Surfaces,” Applied and Environmental Microbiology, Vol. 69, No. 12, 2003, pp. 7188-7193. doi:10.1128/AEM.69.12.7188-7193.2003
NOTES
*Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.

