American Journal of Plant Sciences
Vol.5 No.5(2014), Article ID:43556,7 pages DOI:10.4236/ajps.2014.55067
Development of a Genetic Transformation Method for Seabuckthorn (Hippophae rhamnoides L.)
Sridevy Sriskandarajah*, David Clapham, Per-Olof Lundquist
Department of Plant Biology, Uppsala BioCenter, Linnean Centre of Plant Biology in Uppsala, Swedish University of Agricultural Sciences, Uppsala, Sweden
Email: *Sridevy.Sriskandarajah@slu.se
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 19 November 2013; revised 16 January 2014; accepted 12 February 2014
ABSTRACT
Seabuckthorn (Hippophae rhamnoides L.) is a dioecious plant with berries containing high amounts of several bioactive compounds with nutritional and medicinal traits. It is also planted to control soil erosion. A genetic transformation procedure will facilitate studies of the control of plant development and interactions with symbionts and pathogens, and will provide a tool for plant breeding. Here, we present a particle bombardment method for transforming seabuckthorn. The early stages of induced adventitious shoots from roots were chosen as a novel target tissue for the transformation procedure. The root system was bombarded with gold particles coated with plasmid pRT99gus containing genes for plant kanamycin resistance and for β-glucuronidase expression, and shoots were regenerated under kanamycin selection. PCR analysis of the regenerated transformed lines confirmed the presence of a 603 bp gus (uidA) gene fragment and a 1.5 kb fragment from the 35S promoter in three shoots from independent transformation events.
Keywords:Hippophae rhamnoides; Particle Bombardment; Transformation

1. Introduction
Seabuckthorn (Hippophae rhamnoides L.) and its relatives in the family Eleaegnaceae are shrubs and small trees having berries containing compounds with high nutritional and medicinal value [1] [2] . These plants are also able to form symbiotic N2-fixing root nodules and explore infertile soils. The main geographical distribution of seabuckthorn is in the temperate regions of Asia but is also found in Europe mainly in river and seashore habitats. Seabuckthorn has since long been used in China, Russia and India for nutritional and medicinal purposes. In recent decades, there is an increasing interest to use the berries for pharmaceutical, cosmetic and nutritional applications. Owing to the clonal growth habit with sucker-forming root systems seabuckthorn is also successfully applied to control soil erosion [1] .
Expected increased cultivation of seabuckthorn is enhancing demands for genetic improvement [2] . Breeding goals will be those requested by growers, breeders and consumers to meet problems with pathogens, quality, yield and cultivation. Targeted gene studies will be needed to understand several aspects of seabuckthorn growth and development e.g. fruit and root development and interactions with symbiotic N2-fixing root nodule-inducing Frankia, as well as interactions with pathogenic organisms such as Fusarium sporotrichioides causing dried-shrink disease [3] . Transgenic approaches will be powerful tools in these areas.
To date there is no published study of stably transformed plants of Hippophae spp or other genera within Elaeagnaceae. Recently, root transformation of seabuckthorn by the use of the wild-type strain Agrobacterium rhizogenes 15834 resulting in transformed hairy root systems in chimeric plants was presented [4] . Among actinorhizal plants, i.e. plants having a N2-fixing root nodule symbiosis with the actinomycete bacterium Frankia, Agrobacterium tumefaciens-mediated transformation has been successful for Casuarina glauca and Allocasuarina verticillata [5] [6] .
Regeneration is an essential step during a transformation procedure. Previously, we have established an efficient regeneration protocol for Hippophae rhamnoides [7] . Particle bombardment has been shown to be effective in woody plants including Picea abies, where lignin content has been reduced by transforming with an antisense construct for an enzyme of lignin synthesis [8] [9] , Pinus roxburghii [10] and Curcuma longa [11] . In the present study we report, for the first time, a method to transform Hippophae rhamnoides by particle bombardment of shoots regenerating from roots after hormonal induction, enabling production of transgenic shoots.
2. Materials and Methods
2.1. Plant Material
Seeds of Hippophae rhamnoides L. ssp. rhamnoides cultivar Gold Rain (Prozrachnaya) [12] were obtained from a field in Sweden cultivated with the male H. rhamnoides L. ssp. rhamnoides cultivar Lord [12] as the only male within long distance. Seedlings were established in vitro as described by Sriskandarajah and Lundquist [7] . Two to three week old seedlings grown on woody plant medium (WPM) salts [13] were transferred to Petri dishes containing WPM salts, 4.4 µM 6-benzyladenine (BA), 0.29 µM gibberellic acid (GA3), 57 µM indole-3-acetic acid (IAA) and 30 g·l−1 sucrose (W4 medium). Two to three seedlings were grown in each Petri dish, and the culture conditions were same as described earlier [7] . After two weeks of culturing, at the stage when meristematic activity was visible along the root system, the seedlings were subjected to particle bombardment treatments.
2.2. Determination of Kanamycin Dosage for Selection Criteria
Seedlings grown for 3 weeks in WPM medium were transferred to W4 medium containing 0, 25, 50, 75 or 100 mg·l−1 kanamycin (Sigma). The seedlings were allowed to grow for 6 weeks, and the effect of kanamycin dosage on regeneration was determined.
2.3. Transformation by Particle Bombardment
2.3.1. Plasmid for Transformation
Plasmid pRT99gus [see 14 for map] was used for transformation by particle bombardment. The plasmid is a pUC18 derivative, total size 6170 bp, containing an NPT II gene coding for kanamycin resistance and a uidA (i.e. GUS) gene coding for β-glucuronidase enzyme activity, each flanked by the promoter and terminator sequences of the cauliflower mosaic virus 35S RNA. The host bacterium HB101 was grown overnight in Terrific Broth [15] containing 50 mg·l−1 ampicillin and the plasmid was purified by a modified polyethylene glycol method [16] .
2.3.2. Coating of Gold Particles
Particles for bombardment were coated essentially after Clapham et al. [9] and Sivamani et al. [17] . To a suspension of 10 mg gold particles (1.5 - 3.0 µm diameter, Aldrich) in 100 µl water, was added sequentially 20 µl of plasmid DNA (1 mg·ml−1) and 100 µl of freshly prepared protamine solution (1 mg·ml−1). While gently vortexing the above mixture 100 µl of 2.5 M CaCl2 was slowly added. The final mixture was incubated for 10 minutes and then pelleted at 10,000 g for one minute. The supernatant was discarded and the pellet was suspended in a final volume of 1.1 ml ice cold 100% ethanol.
2.3.3. Preparation of Plant Material for Bombardment with the Particle Inflow Gun
Seedlings grown as described above and having well developed root system were immersed in liquid WPM medium containing 0.25 M myo-inositol for 1.5 hours to induce plasmolysis. The seedlings were quickly blotted using sterile filter papers and a single seedling was placed at each bombardment occasion on a Petri dish of 5 cm diameter with the root system confined to the Petri dish. A piece of sterile wire mesh was placed on top of the root system to restrict movement during shooting. Then the Petri dish with seedling was placed on the platform below the particle inflow gun, described in [8] . This is an inexpensive, easily constructed gun, into which the DNA-coated gold particles are loaded directly as a suspension. Subsequently the particles are blown into the tissue under partial vacuum with a puff of helium gas. The particle inflow gun differs from commercial guns where the particles are dried on plastic squares, and the squares are inserted into the machine.
2.3.4. Bombardment
About 25 µl of the suspension of coated gold particles was dispensed onto the centre of the metal sieve plate in a ‘Swinney’-type filter holder (Millipore, Eschborn, Germany) and mounted to the gun above the vacuum chamber. Pressure was reduced to 7.5 cm mercury and conditions for bombardment were as described [8] . Ten seedlings were bombarded in each of the two trials conducted. Each seedling was bombarded three times, with different positioning of the Petri dish. For control seedlings, bombardment was omitted. Control and bombarded seedlings were placed in W4 medium containing 0.25 M myo-inositol in Petri dishes (9 cm) and cultured under light and temperature conditions as described above. After seven days the seedlings were transferred to fresh W4 medium with 0.125 M myo-inositol and 100 mg·l−1 kanamycin. The seedlings were then subcultured monthly on fresh W4 medium without myo-inositol but with kanamycin.
2.4. Purification of Plant DNA
DNA was isolated from small leaves of shoots developing from bombarded roots by a combined protocol derived from two methods [18] [19] . About 100 mg of leaf samples from small individual shoots growing on the root systems on selection medium were put in eppendorf tubes, quickly frozen in liquid nitrogen and kept in a −80˚C freezer. Steel balls, 2.5 mm diameter, baked at 160˚C - 180˚C for 2 - 3 hours, and cooled down to −80˚C were added to the eppendorf tubes which contained leaf tissue (one ball per tube) before inserting the tubes in the cold (−80˚C) Tissuelyser II (Qiagen, Hilden, Germany) blocks. The tissues were ground for 30 seconds at speed 20. Grinding was repeated once more after changing the sides of the tubes. Final extraction buffer was prepared beforehand [19] , and 1.2 ml of this buffer was added to each sample at room temperature. After adding 2 µl RNaseA (10 mg/ml, Fermentas) to each sample, the tubes were kept at 37˚C for 15 min and then incubated at 42˚C for 10 min and at 65˚C for 30 min. The contents were transferred to fresh 2 ml eppendorf tubes and 1 ml of CHISAM (chloroform/isoamylalcohol 24:1) added to each tube and mixed. Chloroform extraction was repeated twice, and the upper aqueous phase was transferred to 1.5 ml “LoBind” eppendorf tubes. Finally, DNA was precipitated and purified according to the method described by [18] . The A260/A280 ratio of the purified DNA was 1.8 - 2.1.
2.5. Polymerase Chain Reaction (PCR)
To amplify a fragment of 603 bp from the GUS gene, the forward primer was gus1 (5’-TTTGCAAGTGGTGAATCCGCACCT) and the reverse primer was gus2 (5’-AGTTTAGGCGTTGCTTCCGCCAGT). To amplify a fragment of about 1.5 kb from the 3’-end of the 35S promoter into the GUS gene, the forward primer was 5’-CCACTATCCTTCGCAAGACCCTTC and the reverse primer was gus2. The reaction mixture for PCR contained 2 µl of 10x PCR buffer, 1 U TrueStart™ Hot Start Taq DNA Polymerase (Fermentas), 2 pmol of each primer, 2 mM dNTPs and 100 ng DNA in a final volume of 20 µl. After an initial 3 minutes denaturation at 94˚C, thirty to thirty-five cycles of 94˚C (30 s) 55˚C (30 s) and 72˚C (60 s), followed by a final 3 min period of elongation at 72˚C were employed for amplification. PCR products were run out on 1.5% agarose electrophoresis gels to check for size of PCR products. MassRuler DNA Ladder Mix (Thermo Fisher Scientific Inc., Sweden) was used as DNA size markers, range 80 to 10,000 bp.
3. Results
Kanamycin concentrations greater than 50 mg·l−1 affected adventitious shoot regeneration from the roots, which was completely inhibited at 100 mg·l−1. Therefore, kanamycin at 100 mg·l−1 was used in selection media for all transformation experiments.
Swelling and cracking at the base of the lateral root initials began after about 2 weeks in W4 medium. The bombarded seedlings began producing shoots after 6 - 7 weeks while the control seedlings produced shoots after 3 - 4 weeks (Figure 1). A total of four shoots grew from the root systems of 10 seedlings in the first bombardment trial; six shoots grew from the root systems of 10 seedlings in the second bombardment trial.
PCR analysis confirmed the presence of the 603 bp gus gene fragment (Figure 2) and the presence of the 1.5 kb fragment from the 35S promoter to the position of the gus2 primer in the gus gene (Figure 3) in three shoots from independent transformation events. The bands were not amplified from DNA extracted from unbombarded controls (Figures 2, 3). The band size was confirmed by PCR amplification from the plasmid pRT99gus (Figures 2, 3). The identity of the gus gene fragment amplified by one of the transformants was further confirmed by DNA sequencing (data not shown).
4. Discussion
A useful transformation procedure normally requires the regeneration of plantlets from transformed totipotent cells that are selected in vitro. In many reports, Seabuckthorn has been mentioned as being difficult to cultivate in tissue culture, e.g. [20] . Recently, however, we have developed rapid and efficient methods for production of adventitious shoots from the roots of both juvenile and adult plants of seabuckthorn [7] . By using various media, pre-treatments and different plant growth regulator combinations including the two synthetic cytokinins TDZ and CPPU, it was possible to discover interesting regeneration pathways for this plant. One of which was the induction of shoots from roots of young seedlings within a short period. In the present study, the initial induction stages of these adventitious shoots arising from root systems were chosen to be the target for transformation.

Figure 1. Several shoots forming from the root system of a Control seedling after 5 weeks in W4 medium (left), and a few adventitious shoots forming in a bombarded seedling after 7 weeks in selection medium (right). Bar is 1 cm.
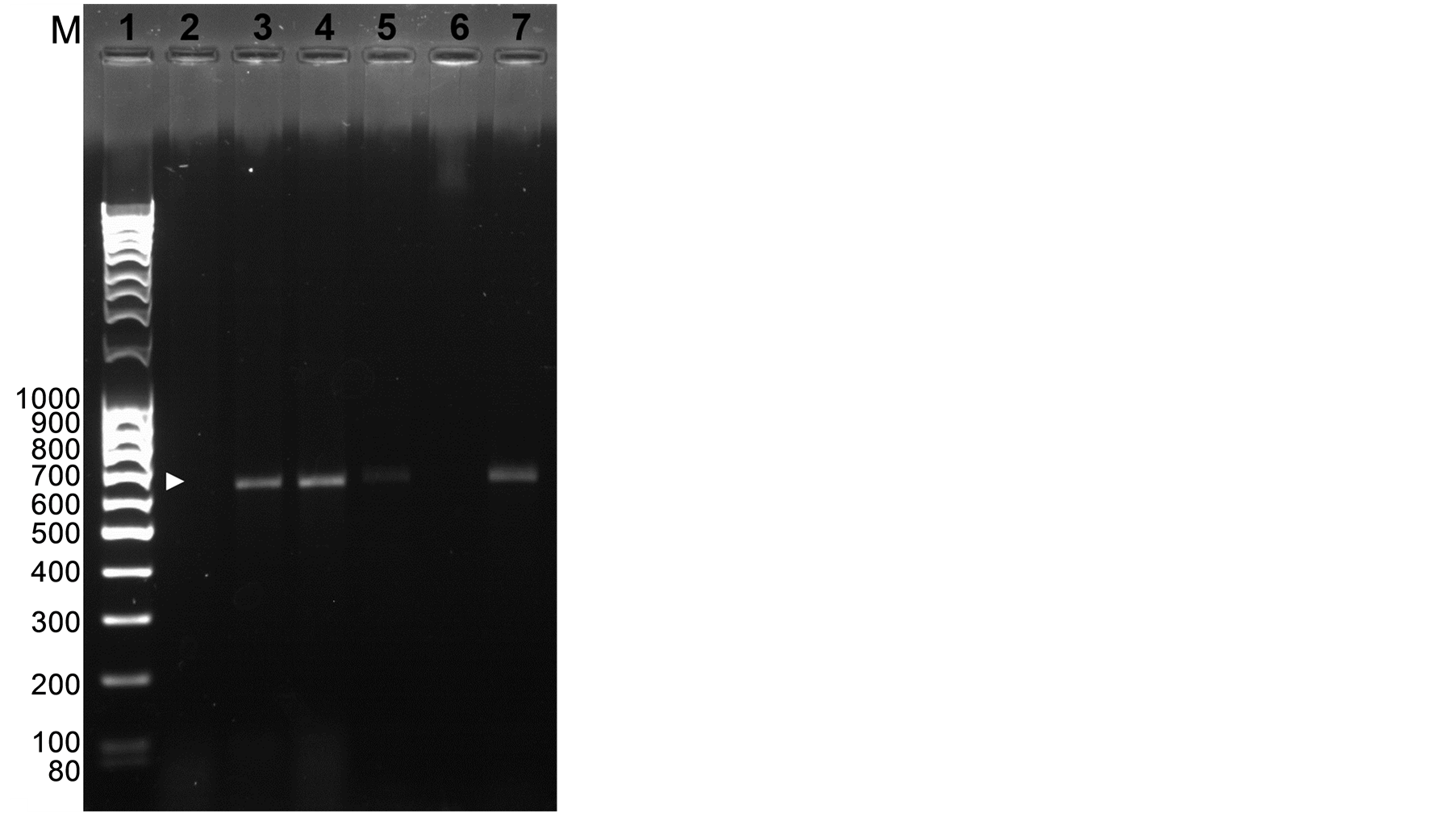
Figure 2. PCR analysis of leaves from regenerated shoots. PCR fragments amplified from the gus1 and gus2 primers. Lane 1: molecular weight makers; Lane 2: untransformed shoot; Lanes 3 - 5: transformed shoots; Lane 6: empty; Lane 7: DNA from plasmid positive control. Expected PCR product (603 bp) is indicated by an arrow.
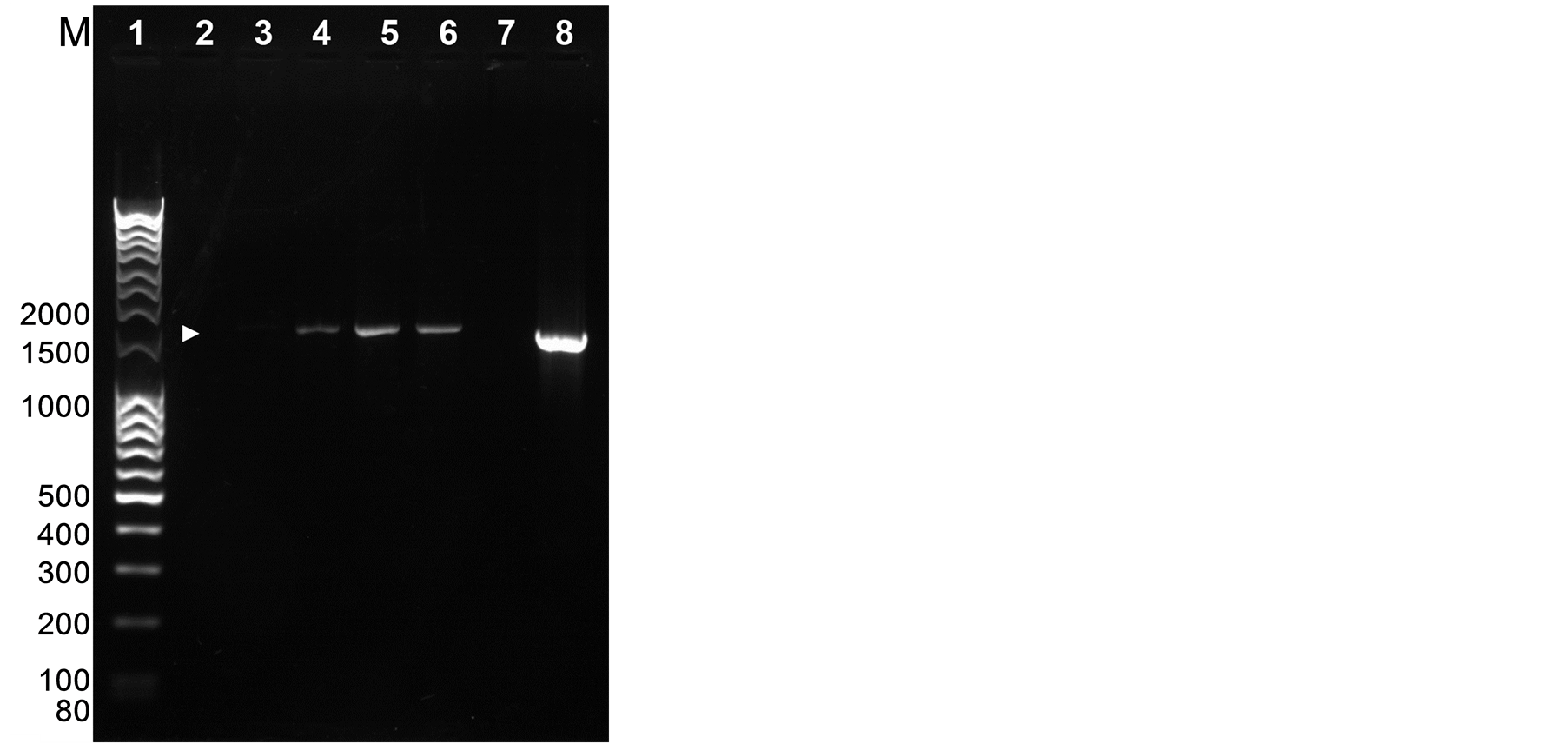
Figure 3. PCR analysis of leaves from regenerated shoots. PCR fragments after amplifying from the 35S and gus2 primers. Lane 1: molecular weight makers; Lane 2: untransformed control; Lane 3: empty; Lanes 4-6: transformed regenerated shoots; Lane 7: empty; Lane 8: plasmid pRT99gus. Expected PCR product (1.5 kb) is indicated by an arrow.
We present a first method to transform Hippophae rhamnoides by particle bombardment using the shoot regeneration system that we established. Transformed shoots were selected on kanamycin medium. Experiments in which similarly treated root material were co-cultivated with one of two strains either of Agrobacterium tumefaciens (A281and EHA105) or A. rhizogenes (A4 and 15834) gave no positive results, so we changed to particle bombardment.
Transient expression of the gus gene in roots or shoots after bombardment was weak (data not shown), probably because of the high content of phenolics in the Hippophae tissue; phenolics in woody tissue often inhibit the activity of the β-glucuronidase enzyme [21] . To confirm that the shoots selected on antibiotic media were transformed, we isolated DNA from the leaf tissue of shoots and amplified fragments of the plasmid DNA by PCR. Hippophae shoots are rich in phenolics, and phenolics often interfere with DNA extraction. However, a combination method that we used for extracting DNA yielded high quality DNA.
Products of the expected size of 603 bp were obtained by amplifying a part of the gus gene using the primers gus1 and gus2, and products of the expected size of 1.5 kb were obtained by amplifying the fragment extending from the 35S promoter to the position of primer gus2. The amplification result with the 35S promoter primer thus confirmed the gus primer amplification and supported that the construct was introduced by the bombardment. Also, the results showed that the gus gene fragment was not amplified from any endophytic bacteria carrying the gene since the bacterial gene would not be controlled by the viral 35S promoter. An advantage that particle bombardment has over agrobacterial methods for transformation is that there is no co-cultivation step with bacteria that can leave residual DNA that can interfere with the PCR analysis.
In the present study, protamine-mediated DNA coating was used and may have helped in transformation. It has been shown that the protamine method [17] for coating the gold particles enhanced transient gus expression for rice and maize. The present study is a first study on transformation of Hippophae and future improvements should be able to increase transformation frequency and regeneration and provide valuable tools for the study of Hippophae plant development.
Acknowledgements
Funding from Stiftelsen Oscar och Lili Lamms Minne to P-O L is gratefully acknowledged.
References
- Singh, V (2005) Sea buckthorn (Hippophae L.). A Multipurpose Wonder Plant. Vol. 2, Singh V. Daya Publishing House; New Delhi.
- Kalia, R.K., Singh, R., Rai, M.K., Mishra, G.P., Singh, S.R. and Dhawan, A.K. (2011) Biotechnological Interventions in Seabuckthorn (Hippophae L.): Current Status and Future Prospects. Trees, 25, 559-575. http://dx.doi.org/10.1007/s00468-011-0543-0
- Sun, Y.-L. and Hong, S.-K. (2012) Effect of Chitinase on Resistance to Fungal Pathogens in Sea Buckthorn, Hippophae rhamnoides, and Cloning of Class I and III Chitinase Genes. Biochemical Genetics, 50, 600-615. http://dx.doi.org/10.1007/s10528-012-9504-6
- Vershinina, Z.R., Baimiev, A.K. and Chemeris, A.V. (2010) Symbiotic Reactions of Sea-Buckthorn Roots Transformed with the Pea Lectin Gene. Russian Journal of Plant Physiology, 57, 101-109. http://dx.doi.org/10.1134/S1021443710010140
- Le, Q., Bogusz, D., Gherbi, H., Lappartient, A., Duhoux, E. and Franche, C. (1996) Agrobacterium tumefaciens Gene Transfer to Casuarina glauca, a tropical nitrogen-fixing tree. Plant Science, 118, 57-69. http://dx.doi.org/10.1016/0168-9452(96)04386-5
- Franche, C., Diouf, D., Le, Q., Bogusz, D., N’Diaye, A., Gherbi, H., Gobé, C. and Duhoux, E. (1997) Genetic Transformation of the Actinorhizal Tree Allocasuarina verticillata by Agrobacterium tumefaciens. The Plant Journal, 11, 897-904. http://dx.doi.org/10.1046/j.1365-313X.1997.11040897.x
- Sriskandarajah, S. and Lundquist, P.-O. (2009) High Frequency Shoot Organogenesis and Somatic Embryogenesis in Juvenile and Adult Tissues of Seabuckthorn (Hippophae rhamnoides L.). Plant Cell, Tissue and Organ Culture (PCTOC), 99, 259-268. http://dx.doi.org/10.1007/s11240-009-9597-8
- Clapham, D., Elfstrand, M., Sabala, I., Von Arnold, S., Demel, P. and Koop, H.-U. (2000) Gene Transfer by Particle Bombardment to Embryogenic Cultures of Picea abies and the Production of Transgenic Plantlets. Scandinavian Journal of Forest Research, 15, 151-160. http://dx.doi.org/10.1080/028275800750014957
- Wadenbäck, J., von Arnold, S., Egertsdotter, U., Walter, M.H., Grima-Pettenati, J., Goffner, D., Gellerstedt, G., Gullion, T. and Clapham, D. (2008) Lignin Biosynthesis in Transgenic Norway Spruce Plants Harboring an Antisense Construct for Cinnamoyl CoA Reductase (CCR). Transgenic Research, 17, 379-392. http://dx.doi.org/10.1007/s11248-007-9113-z
- Parasharami, V.A., Naik, V.B., von Arnold, S., Nadgauda, R.S. and Clapham, D.H. (2006) Stable Transformation of Mature Zygotic Embryos and Regeneration of Transgenic Plants of Chir Pine (Pinus roxburghii Sarg). Plant Cell Reports, 24, 708-714. http://dx.doi.org/10.1007/s00299-005-0019-z
- Shirgurkar, M.V., Naik, V.B., Von Arnold, S., Nadgauda, R.S. and Clapham, D. (2006) An Efficient Protocol for Genetic Transformation and Shoot Regeneration of Turmeric (Curcuma longa L.) via Particle Bombardment. Plant Cell Reports, 25, 112-116. http://dx.doi.org/10.1007/s00299-005-0033-1
- Bruvelius, A. (2003) Sea Buckthorn Cultivation in Baltic States. Proceeding of the 1st Congress of the International Seabuckthorn Association, Berlin, 14-18 September 2003, 64-66.
- Lloyd, G. and McCown, B. (1981) Commercially Feasible Micropropagation of Mountain Laurel, Kalmia latifolia, by Use of Shoot Tip Culture. Combined Proceedings—International Plant Propagator’s Society, 30, 421-427.
- Töpfer, R., Schell, J. and Steinbiss, H. (1988) Versatile Cloning Vectors for Transient Gene Expression and Direct Gene Transfer in Plant Cells. Nucleic Acids Research, 16, 8725. http://dx.doi.org/10.1093/nar/16.17.8725
- Tartoff, K.D. and Hobbs, C.A. (1987) Improved Media for Growing Plasmid and Cosmid Clones. Bethesda Research Laboratories Focus, 9, 12.
- Sambrook, J., Fritsch, E. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
- Sivamani, E., DeLong, R.K. and Qu, R. (2009) Protamine-Mediated DNA Coating Remarkably Improves Bombardment Transformation Efficiency in Plant Cells. Plant Cell Reports, 28, 213-221. http://dx.doi.org/10.1007/s00299-008-0636-4
- Doyle, J.J. and Doyle, J.L. (1990) A Rapid Total DNA Preparation Procedure for Fresh Plant Tissue. Focus, 12, 13-15.
- Azevedo, H., Lino-Neto, T. and Tavares, R. (2003) An Improved Method for High-Quality RNA Isolation from Needle of Adult Maritime Pine Trees. Plant Molecular Biology Reporter, 21, 333-338. http://dx.doi.org/10.1007/BF02772582
- Lummerding, L. (2001) Agri-Food Innovation Fund Project. Prairie Plant Systems, Saskatoon.
- Serres, R., McCown, B. and Zeldin, E. (1997) Detectable β-Glucuronidase Activity in Transgenic Cranberry Is Affected by Endogenous Inhibitors and Plant Development. Plant Cell Reports, 16, 641-646.
Abbreviations
BA: 6-benzyladenine; GA3: gibberellic acid; IAA: indole-3-acetic acid.
NOTES

*Corresponding author.

