American Journal of Plant Sciences
Vol.4 No.5(2013), Article ID:32110,8 pages DOI:10.4236/ajps.2013.45119
Molecular Analysis of Genetic Fidelity in Micropropagated Plants of Stevia rebaudiana Bert. Using ISSR Marker
![]()
1National Center for Natural Product Research, Research Institute of Pharmaceutical Sciences, School of Pharmacy, The University of Mississippi, Oxford, USA; 2Department of Pharmacognosy, School of Pharmacy, The University of Mississippi, Oxford, USA.
Email: *ikhan@olemiss.edu, hlata@olemiss.edu
Copyright © 2013 Hemant Lata et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received March 28th, 2013; revised April 30th, 2013; accepted May 15th, 2013
Keywords: Asteraceae; Genetic Fidelity; HPLC; ISSR; Micropropagation; Stevia rebaudiana; Stevioside
ABSTRACT
Inter-simple sequence repeat (ISSR) markers were used to evaluate the genetic fidelity of in vitro propagated and hardened plants of Stevia rebaudiana Bert. Nodal segments containing axillary buds were used as explant and inoculated on Murashige and Skoog’s (MS) medium containing 3% (w/v) sucrose, 0.8% (w/v) agar supplemented with various concentrations of benzyladenine (BA), kinetin (Kn) and thidiazuron (TDZ) ranging from 0.20 to 2.00 mg·L−1. Maximum multiple shoots (93%) were obtained in MS medium supplemented with 0.20 mg L−1 TDZ. The best in vitro root induction (87%) was achieved on half strength MS medium without any growth regulator. The rooted plantlets were successfully established in soil and grown to maturity at the survival rate of 96% in the indoor grow room. For ISSR analysis, total genomic DNA was extracted from 20 mg fresh leaves of mother and randomly selected in vitro propagated plants. Out of fifteen arbitrary primers tested, each produced clear and scorable amplification products ranged in size from about 216 bp in UBC 811 to 1917 bp in (GGGGT)3M with an average of 4.5 products per primer. A total of 45 bands (number of plantlets analyzed multiplied by number of bands with all primers) were generated by the ISSR method. All the ISSR profiles from micropropagated plants were monomorphic and comparable to mother plants, confirming the genetic stability among micropropagated plants and mother plant. Chemical analysis, using high-performance liquid chromatography (HPLC), was done to further confirm the existence of qualitative and quantitative differences in the major secondary metabolites (rebaudioside A, stevioside and steviolbioside) between the mother plant and in vitro propagated plants. Our results clearly show similar chemical profiles and insignificant differences in the major secondary metabolites between the two types of plants. These results suggest that the micropropagation protocol followed in this study is appropriate and applicable for clonal mass propagation of true-to-type elite Stevia rebaudiana plants.
1. Introduction
Stevia rebaudiana Bertoni (Asteraceae) is a sweet, perennial herb and an important medicinal plant native from northeastern Paraguay and Southern Brazil. This species is now being cultivated on a large scale in many other countries including Japan, Taiwan, Korea, Thailand and Indonesia for medicinal purposes and for use as a natural sweetener in food products [1]. Stevia leaves contain a non-caloric natural sugar (diterpenoid steviol-glycosides), alternative to artificially produced sugar substitutes and is 300 times sweeter than the sugar (sucrose) obtained from sugarcane [2-4]. Many researchers have demonstrated that S. rebaudiana may be used in different therapeutic treatments, such as, for hypoglycemic activity [5], hypotension [6], and non-ariogenic sucrose substitute [7]. In addition, it has been used for patients suffering from obesity, heart disease, and dental maladies [4].
The seeds of Stevia show a very less vigour and low germination percentage [8]. Propagation by seeds does not allow the production of homogeneous populations, resulting in great variability of important features like sweetening levels and composition [9-11]. Vegetative propagation is too slow and limited by the lower number of individuals that can be obtained simultaneously from a single plant due to pathogen accumulation in the tissues [12-14]. Therefore, to overcome all these obstacles, tissue culture is the plausible alternative for rapid mass propagation of Stevia plants.
Plant tissue culture technology is an important biotechnological tool used for regenerating and conserving elite, valuable plant genetic resources. It has also been successfully used for the large scale propagation of the clonal propagules for the commercial planting of the desired genotype. However, in vitro techniques are known to induce somaclonal variations [15] which can seriously limit the broader utility of micropropagation system [16, 17]. Although there are several reports of micropropagation of S. rebaudiana [8,18-21], there was a need to develop an improved and efficient mass propagation protocol for the production of chemically characterized plantlets. Therefore, we have established an efficient high frequency plant regeneration protocol for S. rebaudiana using tissue culture technology [22]. Since the sustainability of the regeneration system depends upon the maintenance of metabolic uniformity and genetic integrity of micropropagated plants, it is therefore imperative to confirm the quality of the plantlets for its commercial utility.
At present, molecular techniques are powerful and valuable tools for the analysis of genetic fidelity of in vitro propagated plants. These are not influenced by environmental factors and generate reliable and reproducible results. Among the markers, inter simple sequence repeat (ISSR) [23] has been mostly favored and have been successfully applied to detect genetic similarities or differences in tissue-cultured materials of various plants [24-26]. Since ISSR markers amplify different regions of the genome, their use allows better analysis of genetic stability/variation of the plantlets. The present investigation was aimed to test the genetic fidelity of in vitro regenerated plantlets of an elite and selected genotype of S. rebaudiana using ISSR profiles. The information gained on genetic stability/variation will be valuable for quality control of the large-scale propagation of S. rebaudiana.
2. Material and Methods
2.1. Plant Material and Culture/Hardening Conditions
High-frequency shoot regeneration using nodal segments from a one-year-old high-yielding S. rebaudiana mother plant (screened and selected based on HPLC analysis) was achieved on Murashige and Skoog (MS) medium [27] containing 3% (w/v) sucrose and 0.8% (w/v) type-E agar supplemented with 0.2 mg·L−1 thidiazuron (TDZ) and best root induction was achieved on half strength MS medium without any growth regulator, by following Lata et al. 2013 [22]. Sub culturing was done every three weeks on sterile medium dispensed (25 ml) in glass culture vessels (4 cm diameter × 9.5 cm high, glass jars with magenta B caps). All cultures were incubated at 25˚C ± 2˚C with 16 h photoperiod under fluorescent light with a photon flux of ~52 µmol m−2·s−1.
The well rooted plantlets obtained on half strength MS medium without any growth regulators were further used for acclimatization. Plantlets were carefully taken out of the medium and washed thoroughly in running tap water to remove all traces of medium attached. Plantlets were preincubated in coco natural growth medium (Canna Continental, USA) in thermocol cups for 10 days. The cups were covered with polythene bags to maintain humidity and kept in an indoor growing room and later acclimatized in sterile potting mix-fertilome (Canna Continental, USA) in large pots. All these plantlets were kept under similar and controlled environmental conditions in an indoor cultivation facility at National Center for Natural Product Research, The University of Mississippi. Light (700 ± 24 μmol m−2·s−1 at plant canopy level, 16 hour photoperiod) was provided with full spectrum 1000 watts HID (high density discharge) lamps (Sun Systems, USA) through the top of plants. Temperature and relative humidity of growing room was maintained at 25˚C - 30˚C and 60% respectively. Plants were watered regularly and individually to maintain sufficient moisture content in the pots. After three months, out of sixty micropropagated and hardened plants, six healthy plants were randomly selected and their mother plant were used for the analysis of genetic stability using ISSR markers.
2.2. DNA Extraction
A 20-mg fresh leaf sample was frozen in liquid nitrogen and ground in a 2.0 ml micro-centrifuge tube using Mixer Mill MM 2000 (Retsch, USA). The total genomic DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, USA) and resuspended in 50 ul dilution buffer. The purified total DNA was quantified and its quality verified by using Nano DropTM 1000 Spectrophotometer (Thermo Scientific, USA).
2.3. PCR Amplifications
Fifteen arbitrary primers (eleven from The University of British Columbia and four of our design) were used to screen polymorphism among mother and daughter plants through PCR amplification. The reactions were performed in a 25 μl final volume comprising 0.1 μM each primer, 1 unit of Platinum Taq DNA Polymerase (Invitrogen, USA), 200 μM of each dNTP (Promega Corporation, USA), 1.5 mM of MgCl2, 20 ng of template DNA, and 1 × PCR buffer. Amplifications were carried-out in heated lid M&J Research Gradient Cycler PTC-225 programmed at 94˚C for 3 min for initial denaturation, followed by 94˚C for 30 sec, 50˚C for 30 sec and 72˚C for 3 min for 45 cycles followed by a final step of extension at 72˚C for 7 min. After amplification, each PCR reaction was analyzed by electrophoresis on a 2% TAE Agarose gel and visualized under UV light. The sizes of the PCR products were compared to the molecular size standard 1kb plus DNA ladder (Invitrogen, USA). Gels were also scanned with the Bio-Rad Gel Imaging System and analyzed using Quantity One analysis software version 4.3.0 (Bio-Rad Laboratories Inc., USA).
2.4. Amplified DNA Marker Scoring
Only the discrete bands in size range of 0.1 - 3.0 kb with high intensity were scored as being present or absent for ISSR markers. Data was scored as 1 for being present and 0 for the absence of DNA band in each micropropagated and mother plant.
2.5. HPLC Profiling and Stevioside Content
Dry powdered samples (0.2 g) were weighed and sonicated in 2.5 mL of 80% methanol (methanol/water = 80:20, v/v) at room temperature for 30 min followed by centrifugation for 10 min at 4000 rpm. The supernatant was transferred to a 10 mL volumetric flask. The procedure was repeated three times and respective supernatants combined. The final volume was adjusted to 10 mL with 80% methanol and mixed thoroughly. Prior to injection, an adequate volume (ca. 2 mL) was passed through a 0.45 µm nylon membrane filter. The first 1.0 mL was discarded and the remaining volume was collected in a LC sample vial. Standards rebaudioside A, stevioside and steviolbioside were isolated at NCNPR, the identity and purity was confirmed by chromatographic (TLC, HPLC) methods and by the analysis of the spectroscopic data (IR, 1Dand 2D-NMR, HR-ESI-MS). All HPLC analyses were applied on Waters Alliance 2695 HPLC system (Waters Corp., USA) and a computerized data station using Waters Empower 2 software. A reverse phase Gemini C18 column (250 × 4.6 mm i.d., 5 µm particle size) from Phenomenex (USA) was used as the stationary phase and the temperature was maintained at 40˚C. The column was equipped with a 2 cm LC-18 guard column (Phenomenex, USA). The mobile phase consisted of water (A), acetonitrile/methanol (B), both containing 0.1% phosphoric acid. Analysis was performed using the following linear gradient elution at a flow rate of 0.7 mL/minute: 0 - 15 min, 58% A/42% B to 42% A/52% B; 15 - 20, 42% A/52% B to 100% B; 20 - 25 min, 100% B. The total run time was 25 min. Each run was followed by a 5 min wash with 100% acetonitrile and an equilibration period of 15 min. Ten microliters of sample was injected and detective wavelength was 205 nm. Peaks were assigned by spiking the samples with standard compounds, comparison of UV spectra and the retention times.
3. Results and Discussion
In vitro propagated S. rebaudiana plantlets were produced according to the protocol described by Lata et al. (2013). Nodal segments containing axillary buds were used as explant and inoculated on Murashige and Skoog (MS) medium containing 3% (w/v) sucrose, 0.8% (w/v) agar supplemented with various concentrations of benzyladenine (BA), kinetin (Kn) and thidiazuron (TDZ) ranging from 0.20 to 2.00 mg·L−1. Of all the cytokinins tested, the quality and quantity of regenerants was best in the MS medium supplemented with 0.20 mg·L−1 TDZ (Table 1 and Figure 1). Elongated shoots were transferred to full strength MS medium without growth regulators, half strength MS medium without growth regulators and full strength MS medium supplemented with different concentrations of IAA, IBA or NAA ranging from 0.1 mg·L−1 to 0.5 mg·L−1, for root induction. The best rooting (87%) was achieved in half strength MS medium without any growth regulator as compared to other media (Table 1 and Figure 1). The rooted plantlets were successfully established in soil and grown to maturity at the survival rate of 96% in the climate controlled indoor grow room conditions.
A total of 15 ISSR primers were initially screened with the DNA of single donor plant of S. rebaudiana and 6 daughter plants as templates. Based on a criterion of the generation of distinct bands that were completely reproducible between the samples, all the 15 primers were selected as suitable primers S. rebaudiana, and thus used for the present study (Table 2).
Out of sixty micropropagated plants, six randomly selected plants and mother plant were subjected to ISSR analysis at maturity. These plants showed no discernible differences among them and as compared to the mother plant in the ISSR analysis. The general morphology of the plants grown under the controlled grow room conditions was also similar. Each tested primer produced clear and scorable amplification products ranging in size from about 216 bp in UBC 811 to 1917 bp in (GGGGT)3M (Table 1) with an average of 4.5 products per primer. A total of 45 bands (number of plantlets analyzed multiplied by number of bands with all primers) were generated by the ISSR method, giving rise to monomorphic patterns across all 7 plantlets analyzed. No ISSR polymorphism was observed in the micropropagated plants (Figure 2).
Based on our results, using ISSR markers, the number of the primers (15) used in this study, as well as total number of bands (45) together with the observed normality and homogeneity of the plants generated in this study, strongly suggest that the regeneration from nodal segments is a reliable approach for germplasm conservation of S. rebaudiana. Our results corroborate with reports of
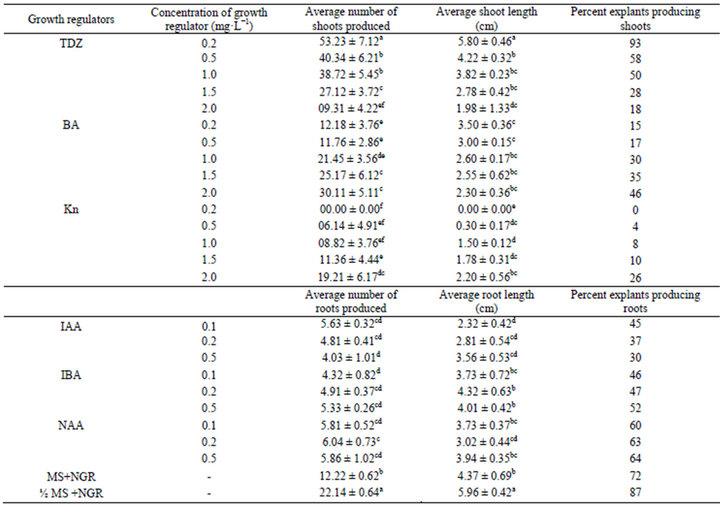
Table 1. The effect of different concentrations of growth regulators on multiplication, shoot proliferation and root induction of in vitro propagated plants of Stevia rebaudiana.
Values are mean ± SE. n = 27. Values followed by the same letter do not differ statistically at p < 0.05. MS - Murashige and Skoog medium, ½MS: Half strength MS medium, NGR: No growth regulator.

Table 2. ISSR Banding pattern of micropropagated and mother plant of Stevia rebaudiana.

Figure 1. Micropropagation of Stevia rebaudiana using nodal segment. (a) Mother plant, bar 15 cm; (b) in-vitro shoot multiplication, bar 1.60 cm; (c) well rooted in-vitro propagated plants, bar 1.25 cm; (d, bar 2.38 and e, bar 4.00 cm) rooted plants under acclimatization and (f) fully grown in-vitro propagated Stevia rebaudiana plant, bar 12 cm.
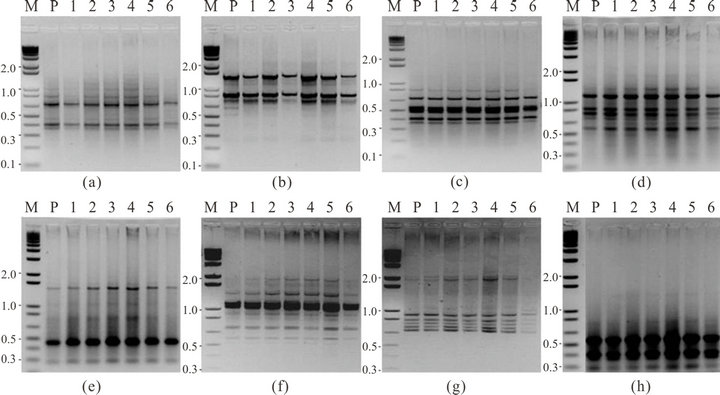
Figure 2. Inter-simple sequence repeats (ISSR) amplification pattern obtained for mother plant (P) and daughter plants of S. rebaudiana (1-6); using (a) primer (GGC)6W, (b) primer (AAG)6Y, (c) primer UBC 812, (d) primer UBC 836, (e) primer UBC 826, (f) primer (GGAT)4H, and (g) primer (GGGGT)3M, (h) primer UBC 811.
genetic stability of in vitro derived plants based on ISSR analysis of cauliflower [28], Populus tremuloides [29], Swertia chirayita [30], Dictyospermum ovalifolium [31], Nothapodytes foetida [32], and Cannabis sativa [33,34].
The genetic control of biochemical characteristics is equally important to guarantee stability in the drug yield, for the commercial production of elite medicinal plant S. rebaudiana by tissue culture. Therefore, evaluation of
| 2.00 |
| 4.00 |
| 6.00 |
| 8.00 |
| 10.00 |
| 12.00 |
| 14.00 |
| 16.00 |
| 18.00 |
| 20.00 |
| 1 |
| 2 |
| 3 |
| 0.00 |
| SM |
| 4.00 |
| 6.00 |
| 8.00 |
| 10.00 |
| 12.00 |
| 14.00 |
| 16.00 |
| 18.00 |
| 20.00 |
| 1 |
| 2 |
| 0.00 |
| 0.50 |
| MP |
| 4.00 |
| 6.00 |
| 8.00 |
| 10.00 |
| 12.00 |
| 14.00 |
| 16.00 |
| 18.00 |
| 20.00 |
| 1 |
| 2 |
| 0.50 |
| 0.00 |
| IVPP |
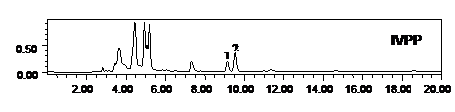
Peak 1, Rebaudioside A Peak 2, Stevioside Peak 3, Steviolbioside

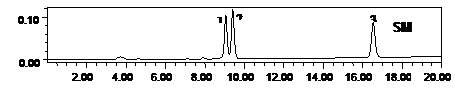
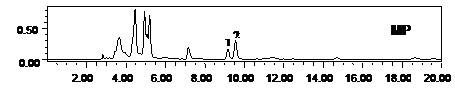
Figure 3. Metabolic profiling of mother plant (MP) and a randomly selected in vitro propagated plant (IVPP) of Stevia rebaudiana. SM: standard of rebaudioside A (peak 1, MF-C44H70O23 and MW-967.01), stevioside (peak 2, Stevioside: MFC38H60O18, MW-804.87) and steviolbioside (peak 3, Steviolbioside: MF-C32H50O13, MW-642.734), MP: mother plant and IVPP. a randomly selected in vitro propagated plant derived from mother plant.
drug yielding potential of micropropagated plants was made through HPLC analysis for the mass production of the propagules. Our results on HPLC analysis also showed homogeneity in the chemical profile and, rebaudioside A and stevioside content of the mother plant and the randomly selected fully hardened micropropagated clones of S. rebaudiana (Figure 3). The level of rebaudioside A in the mature leaves of tissue culture raised clones of S. rebaudiana was comparable in all the clones (3.26% ± 0.05%, n = 24) and to that of the mother plant (3.16% ± 0.25 %, n = 3). Similar to rebaudioside A, the stevioside profile and content in tissue culture raised plants was also identical and comparable to each other (5.25% ± 0.05%, n = 24) and that of the mother plant (5.10% ± 0.09%, n = 3). These results confirm that the biochemical mechanism followed to micropropagate the S. rebaudiana clones maintains the same metabolic profile of the mother plant and, therefore, can be used for the mass propagation of true-to-type plants for commercial pharmaceutical use. High uniformity of the drug yield in the micropropagted plants has been reported by several authors using HPLC analysis [35-38].
Since no variability was detected among in vitro propagated plants and the mother plant, we conclude that the elite germplasm of S. rebaudiana Bert. can be conserved without the risk of genetic instability and can be used for the mass production of chemically characterized Stevia crop using the micropropagation protocol followed in this study. The study is particularly of high significance since the worldwide demand for high-potency natural sweeteners is constantly increasing.
4. Acknowledgements
This research was partially funded by the United States Department of Agriculture, Agricultural Research Service (USDA-ARS), Specific Cooperative Agreement No. 58-6408-6-067.
REFERENCES
- R. V. Sreedhar, L. Venkatachalam, R. Thimmaraju, N. Bhagyalakshmi, M. S. Narayan and G. A. Ravishankar, “Direct Organogenesis from Leaf Explants of Stevia rebaudiana and Cultivation in Bioreactor,” Biologia Plantarum, Vol. 52, No. 2, 2008, pp. 355-360. doi:10.1007/s10535-008-0073-9
- A. S. Richman, M. Gijzen, A. N. Starralt, Z. Yang and J. E. Brandle, “Diterpene Synthesis in Stevia rebaudiana Recruitment and Up Regulation of Key Enzymes from the Gibberellins Biosynthetic Pathway,” The Plant Journal, Vol. 19, No. 4, 1999, pp. 411-421. doi:10.1046/j.1365-313X.1999.00531.x
- N. Bondarev, “Peculiarities of Propagation and Development of Stevia rebaudiana Bertoni Plants in Vitro,” Proceedings of 9th International Conference of Horticulture, Lednice, 3-6 September 2001, pp. 431-434.
- S. D. Singh and G. P. Rao, “Stevia: The Herbal Sugar of 21st Century,” Sugar Tech, Vol. 7, No. 1, 2005, pp. 17-24. doi:10.1007/BF02942413
- N. Lailerd, V. Saengsirisuwan, J. A. Sloniger, C. Toskulkao and E. J. Henriksen, “Effects of Stevioside on Glucose Transport Activity in Insulin-Sensitive and InsulinResistant Rat Skeletal Muscle,” Metabolism, Vol. 53, No. 1, 2004, pp. 101-107. doi:10.1016/j.metabol.2003.07.014
- P. B. Jeppesen, S. Gregersen, S. E. Rolfsen, M. Jepsen, M. Colombo, A. Agger, J. Xiao, M. Kruhøffer, T. Orntoft and K. Hermansen, “Antihyperglycemic and Blood PressureReducing Effects of Stevioside in the Diabetic Goto-Kakizaki Rat,” Metabolism, Vol. 52, No. 3, 2003, pp. 372- 378. doi:10.1053/meta.2003.50058
- J. M. Geuns, “Stevioside,” Phytochemistry, Vol. 64, No. 5, 2003, pp. 913-921. doi:10.1016/S0031-9422(03)00426-6
- A. Mitra and A. Pal, “In Vitro Regeneration of Stevia rebaudiana (Bert.) from Nodal Explants,” Journal of Plant Biochemistry and Biotechnology, Vol. 16, No. 1, 2007, pp. 59-62. doi:10.1007/BF03321930
- S. Nakamura and Y. Tamura, “Variation in the Main Glycosides of Stevia (Stevia rebaudiana),” Japanese Journal of Tropical Agriculture, Vol. 29, 1985, pp. 109-116.
- L. Sivaram and U. Mukundan, “In Vitro Culture Studies on Stevia rebaudiana,” In Vitro Cellular and Developmental Biology of Plant, Vol. 39, No. 5, 2003, pp. 520- 523. doi:10.1079/IVP2003438
- M. A. Shatnawi, R. A. Shibli, S. M. Abu-Romman, M. S. Al-Mazra’awi1, Z. I. Al Ajlouni, W. A. Shatanawi and W. H. Odeh, “Clonal Propagation and Cryogenic Storage of the Medicinal Plant Stevia rebaudiana,” Spanish Journal of Agricultural Research, Vol. 9, No. 1, pp. 213- 220.
- M. Sakaguchi and T. Kan, “Japanese Researches on Ste- - via rebaudiana,” Ci Cult, Vol. 34, 1982, pp. 235- 248.
- M. Debnath, “Clonal Propagation and Antimicrobial Activity of an Endemic Medicinal Plant Stevia rebaudiana,” Journal of Medicinal Plants Research, Vol. 2, No. 2, 2008, pp. 45-51.
- P. K. Mishra, R. Singh, U. Kumar and V. Prakash, “Stevia rebaudiana: A Magical Sweetener,” Global Journal of Biotechnology and Biochemstry, Vol. 5, No. 1, 2010, pp. 62-74.
- P. J. Larkin and W. R. Scowcroft, “Somaclonal VariationA Novel Source of Variability from Cell Cultures for Plant Improvement,” Theoretical and Applied Genetics, Vol. 60, No. 4, 1981, pp. 197-214. doi:10.1007/BF02342540
- N. D. Salvi, L. George and S. Eapen, “Plant Regeneration from Leaf Base Callus of Turmeric and Random Amplified Polymorphic DNA Analysis of Regenerated Plants,” Plant Cell Tissue and Organ Culture, Vol. 66, No. 2, 2001, pp. 113-119. doi:10.1023/A:1010638209377
- M. Moktaduzzaman and S. M. M. Rahman, “Regeneration of Stevia rebaudiana and Analysis of Somaclonal Variation by RAPD,” Biotechnology, Vol. 8, No. 4, 2009, pp. 449-455.
- I. A. Ibrahim, M. I. Nasr, B. R. Mohammed and M. M. El-Zefzafi, “Nutrient Factors Affecting in Vitro Cultivation of Stevia rebaudiana,” Sugar Tech, Vol. 10, No. 3, 2008, pp. 248-253. doi:10.1007/s12355-008-0044-7
- B. Janarthanam, M. Gopalakrishnan, G. Lakshmisai and T. Sekar, Plant Regeneration from Leaf Derived Callus of Stevia rebaudiana Bertoni,” Plant Tissue Culture and Biotechnology, Vol. 19, No. 2, 2009, pp. 133-141. doi:10.3329/ptcb.v19i2.5430
- M. Kalpana, M. Anbazhagan and V. Natarajan, “Utilization of Liquid Medium for Rapid Micropropagation of Stevia rebaudiana Bertoni,” Journal of Ecobiotechnology, Vol. 1, No. 1, 2009, pp. 16-20.
- A. Das, S. Gantait and N. Mandal, “Micropropogation of an Elite Medicinal Plant (Stevia rebaudiana Bertoni),” International Journal of Agriculture Research, Vol. 6, No. 1, 2011, pp. 40-48. doi:10.3923/ijar.2011.40.48
- H. Lata, S. Chandra, Y. H. Wang, V. Raman and I. A. Khan, “TDZ-Induced High Frequency Plant Regeneration through Direct Shoot Organogenesis in Stevia rebaudiana Bertoni: An Important Medicinal Plant and a Natural Sweetener,” American Journal of Plant Sciences, Vol. 4, 2013, pp. 117-128. doi:10.4236/ajps.2013.41016
- E. Zietkiewicz, A. Rafalski and D Labuda, “Genome Fingerprinting by Simple Sequence Repeat (SSR)-Anchored Polymerase Chain Reaction Amplification,” Genomics, Vol. 20, No. 2, 1994, pp. 176-183. doi:10.1006/geno.1994.1151
- H. Yang, Y. Tabei, H. Kamada, T. Kayano and F. Takaiwa, “Detection of Somaclonal Variation in Cultured Rice Cells Using Digoxigenin-Based Random Amplified Polymorphic DNA,” Plant Cell Reports, Vol. 18, 1999, pp. 520-526.
- W. L. Guo, Y. Li, L. Gong, F. Li, Y. Dong and B. Liu, “Efficient Micropropagation of Robinia ambigua and Detection of Genomic Variation by ISSR Markers,” Plant Cell Tissue and Organ Culture, Vol. 84, No. 3, 2006, pp. 343-351. doi:10.1007/s11240-005-9043-5
- J. B. Thomas, D. Vijayan, S. D. Joshi, S. J. Lopez and R. R. Kumar, “Genetic Integrity of Somaclonal Variants in Tea (Camellia sinensis (L.) O Kuntze) as Revealed by Inter Simple Sequence Repeats,” Journal of Biotechnology, Vol. 123, No. 2, 2006, pp. 149-154. doi:10.1016/j.jbiotec.2005.11.005
- T. Murashige and F. Skoog, “A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures,” Physiologia Plantarum, Vol. 15, No. 43, 1962, pp. 473- 497. doi:10.1111/j.1399-3054.1962.tb08052.x
- X. Leroy, K. Leon, G. Charles and M. Branchard, “CauliFlower Somatic Embryogenesis and Analysis of Regenerants Stability by ISSRs,” Plant Cell Report, Vol. 19, No. 11, 2000, pp. 1102-1107. doi:10.1007/s002990000252
- M. H. Rahman and O. P. Rajora, “Microsatellite DNA Somaclonal Variation in Micropropagated Trembling Aspen (Populus tremuloides),” Plant Cell Report, Vol. 20, No. 6, 2001, pp. 531-536. doi:10.1007/s002990100365
- P. Joshi and V. Dhawan, “Assessment of Genetic Fidelity of Micropropagated Swertia chirayita Plantlets by ISSR Marker Assay,” Biologia Plantarum, Vol. 51, No. 1, 2007, pp. 22-26. doi:10.1007/s10535-007-0005-0
- M. Chandrika, Thoyajaksha, V. Ravishankar Rai and K. Ramachandra Kini, “Assessment of Genetic Stability of in Vitro Grown Dictyospermum ovalifolium,” Biologia
- Plantarum, Vol. 52, No. 4, 2008, pp. 735-739. doi:10.1007/s10535-008-0142-0
- M. Chandrika, V. Ravishankar Rai and Thoyajaksha, “ISSR Marker Based Analysis of Micropropagated Plantlets of Nothapodytes foetida,” Biologia Plantarum, Vol. 54, No. 3, 2010, pp. 561-565. doi:10.1007/s10535-010-0100-5
- H. Lata, H. S. Chandra, N. Techen, I. Khan and M. A. ElSohly, “Assessment of Genetic Stability of Micropropagated Cannabis sativa Plants by ISSR Markers,” Planta Medica, Vol. 76, No. 1, 2010, pp. 97-100. doi:10.1055/s-0029-1185945
- H. Lata, S. Chandra, N. Techen, I. Khan and M. A. ElSohly, “Molecular Analysis of Genetic Fidelity in Cannabis sativa L. Plants Grown from Synthetic Seeds Following in Vitro Storage Conditions,” Biotechnology Letter, Vol. 33, No. 12, 2011, pp. 2503-2508. doi:10.1007/s10529-011-0712-7
- K. Hatano, K. Kamura, Y. Shoyama and I. Nishioka, “Clonal Propagation of Aconitum carmichaeli by Tip Tissue Culture and Alkaloid Contents of Clonally Propagated Plants,” Planta Medica, Vol. 54, No. 2, 1988, pp. 152-155. doi:10.1055/s-2006-962375
- S. Ahuja, B. B. Mandal, S. Dixit and P. S. Srivastava, “Molecular, Phenotypic and Biosynthetic Stability in Dioscorea floribunda Plants Derived from Cryopreserved Shoot Tips,” Plant Science, Vol. 163, No. 5, 2002, pp. 971-977. doi:10.1016/S0168-9452(02)00246-7
- C. Z. Liu, M. Gao and B. Guo, “Plant Regeneration of Erigeron breviscapus (vant.) Hand. Mazz. and Its Chromatographic Fingerprint Analysis for Quality Control,” Plant Cell Report, Vol. 27, No. 1, 2008, pp. 39-45. doi:10.1007/s00299-007-0466-9
- H. R. Shilpashree and R. Rai, “In Vitro Plant Regeneration and Accumulation of Flavonoids in Hypericum mysorense,” International Journal of Integrative Biology, Vol. 8, No. 1, 2009, pp. 43-49.
NOTES
*Corresponding author.

