Pharmacology & Pharmacy
Vol.05 No.11(2014), Article ID:50506,9 pages
10.4236/pp.2014.511111
Proteomic Analysis of Salt-Induced Changes in Protein Expression in PPARα Null Mice
Patience O. Obih1*, Adebayo Oyekan2
1College of Pharmacy, Xavier University of Louisiana, New Orleans, USA
2College of Pharmacy and Health Sciences, Texas Southern University, Houston, USA
Email: *poobih@xula.edu
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 27 August 2014; revised 22 September 2014; accepted 8 October 2014

ABSTRACT
PPARs are ligand-activated nuclear transcription factors that regulate β-oxidation of fatty acids in the cardiovascular system and PPARα isoform is a putative target for regulation of cardiovascular function. High salt diet is an injurious stimulus to cardiovascular function but its effect on PPARα and PPARα–associated profile of proteins is unknown. Quantitative proteomics involving a two- dimensional electrophoresis (2D-DIGE) followed by LC-MS/MS technology was used to characterize the changes in protein expression profile in the kidney, heart, and blood vessels from PPARα null (KO) and wild type (WT) mice placed on normal (0.3%, NS) or high salt (4% NaCl, HS) diet. Initial biological variation analysis using DeCyder software (v. 6.0) revealed the presence of 20 upregulated proteins and 9 proteins that are downregulated in the kidney, aorta, and heart tissues from KO and WT mice. A multimodality comparison of the differentially expressed proteins showing ≥ 1.5-fold change, ≥20% appearance at P ≤ 0.05 between strains (WT vs KO) and treatment (NS vs HS) revealed that HS diet affected 20 proteins in WT mice and 17 proteins in KO mice. However, 9 proteins were altered between WT and KO placed on NS and 7 proteins were altered by HS between WT and KO mice. The identified proteins include but not limited to those involved in fatty acid oxidation (FAO), mitochondrial electron transport chain, amino acid metabolism, stress response, DNA synthesis, and programmed cell death. HS diet led to upregulation of FAO enzymes viz: acyl-coenzyme A dehydrogenase, transketolase, and electron-transferring-flavoprotein dehydrogenase to different extents in WT and KO mice. These data showed differential and protein-spe- cific responses to HS diet in PPARα WT and KO mice that probably reflect the functional capacities of PPARα as a means to limiting any salt-induced injury to the heart, kidney, and blood vessels.
Keywords:
PPARα, β-Oxidation, Salt Diet, Proteome

1. Introduction
Peroxisome proliferator-activated receptors (PPARs) are ligand-dependent nuclear transcription factors that form a subfamily of the nuclear receptor belonging to the steroid-thyroid hormone superfamily. PPARs comprise 3 isoforms, viz., α, β/δ, and γ that exhibit tissue-specific distribution and ligand-specific effects [1] . PPARα is expressed in tissues with very active fatty acid metabolism such as the heart, kidney, liver, endothelium, and vascular smooth muscle cells [2] [3] [4] [5] . PPARα is involved not only in peroxisomal and mitochondrial fatty acid oxidation, but also in microsomal fatty acid hydroxylation by cytochrome P450 enzymes, lipoprotein, bile and amino acid metabolism, glucose homeostasis, biotransformation, inflammation control, hepato-carcinoge- nesis and other pathways [1] [6] .
In animal cells, mitochondria and peroxisomes oxidize fatty acids via β-oxidation, with long chain fatty acids (LCFAs) and very long chain fatty acids (VLCFAs) being preferentially oxidized by peroxisomes [7] [8] . On the other hand, mitochondrial β-oxidation involves metabolism (dehydrogenation reactions) by a family of four chain-length-specific acyl-CoA dehydrogenases, which include very long chain, long chain, medium chain, and short chain acyl-CoA dehydrogenases (VLCAD, LCAD, MCAD, and SCAD respectively) [7] [8] . Evidence for the importance of PPARα in fatty acid metabolism was provided in studies that demonstrated a reduction in the constitutive levels of mitochondrial fatty acid β-oxidation though the constitutive or basal oxidation of VLCFAs by peroxisomal β-oxidation system appears unaffected by PPARα deficiency [9] [10] . LCFAs constitute the bulk of fatty acids in animals, and their abundance makes them the only significant source of metabolic fuel for mitochondrial β-oxidation system [8] . Under normal physiologic conditions, mitochondrial β-oxidation is the dominant metabolic pathway, whereas the extramitochondrial fatty acid oxidation occurring within peroxisomes and endoplasmic reticulum assumes a minor role [7] [8] . Accordingly, decreased mitochondrial fatty acid β- oxidation has been considered one of the major mechanisms underlying the disturbances in lipid metabolism in liver leading to steatosis [11] [12] . However, oxidation of fatty acids by mitochondria depends not only upon the capacity of β-oxidation enzymes and substrate concentration but is also influenced by other factors such as changes in carnitine content and oxidative phosphorylation [11] which are affected in turn by salt status and ATP-dependent Na+ transport [13] [14] .
The above information is consistent with emerging data that demonstrated physiologic and pathological roles for PPARα in tissues that depend on oxidation of fatty acids for energy production (see [6] [13] [15] ), a requirement for meeting the metabolic needs of such tissues. We therefore tested in this study, the hypothesis that PPARα plays a major role in triggering fatty acid utilization for adaptive responses to changes in dietary salt in the kidney, heart, and blood vessels. The goal of the present study was to evaluate the role of PPARα and identify the proteins that are differentially expressed in response to high salt diet in PPARα wild type (WT) and knock out (KO) mice. In this study, using MS-based proteomics methodologies to characterize the proteome of mouse tissues harvested from animals given a high salt diet, we identified various candidate proteins including but not limited to those involved in fatty acid metabolism or biogenesis, isoprenoid biosynthesis, collagen synthesis, amino acid metabolism, stress response, and DNA synthesis.
2. Materials and Methods
2.1. Experimental Design
This study was approved by the Animal Care Committee of the Texas Southern University and conforms to the institutional guidelines on animal care and use. Age- and weight-matched adult (18 - 22 gm) male PPARα knockout (−/−; KO) mice (Jackson Laboratory, Bar Harbor, ME) or their wild type (129S1/Sv; +/+; WT) littermates were used for the study. The animals were maintained in a 12:12-hr light:dark cycle at 23˚C - 25˚C. They were randomly divided into groups (n = 5 - 7 per group) and assigned to normal rat chow (0.3% NaCl diet [normal salt (NS) diet], Harlan Tekland, Houston, USA) or high salt (HS) (4.0% NaCl) diet. These resulted in the following experimental groups:
Group 1 PPARα KO mice (n = 5) placed on normal salt (0.3% NaCl) diet
Group 2 PPARα KO mice (n = 6) placed on high salt (4% NaCl) diet
Group 3 PPARα WT mice (n = 6) placed on normal salt (0.3% NaCl) diet
Group 4 PPARα WT mice (n = 5) placed on high salt (4% NaCl) diet
Tap water was provided ad libitum and the mice were placed on the respective diets for 3 weeks after which they were anesthetized (sodium pentobarbital, 60 mg/kg i.p.). Tissues including the kidney, the heart and the blood vessels were collected from each animal and stored at −80˚C until subjected to proteomic analysis. Unless specified otherwise in the text, all chemicals used in this study were obtained from Sigma-Aldrich (St Louis, MO, USA) and are of the highest analytical grade.
2.2. Identification of Differentially Expressed Proteins
A comparative proteomics approach using the two-dimensional difference in gel electrophoresis (2D-DIGE) technology was adopted to identify proteins that showed differential expression as a function of the strains of the mice and salt content of the diet. Briefly, total protein was isolated from each tissue using the protocol described in ToPI-DIGE kit protein isolation kit (ITSI Biosciences, Johnstown, PA). The ToPI-DIGE lysis buffer contained 7 M urea, 2 M thiourea, 30 mM TRIS, 4% CHAPS, 5 mM magnesium acetate tetrahydrate and 1% Nonylphenylpolyethylene Glycol (NP40). Proteins were quantified with the Total Protein Assay kit (ToPA) a Bradford-based protein assay reagent (ToPA, ITSI Biosciences, Johnstown, PA) using bovine serum albumin (BSA) as standard. 50 µg of each sample was labeled with 200 pmol of either Cy3 or Cy5 fluorescent dye and an equal amount of each tissue sample was combined in a tube and labeled with Cy2 dye at the same concentration as above. The Cy2 labeled sample served as the universal internal standard to allow normalization and multiple gel comparisons. Equal amounts of the Cy3, Cy5 and Cy2 labeled samples were mixed and subjected to 2D-DIGE as previously described (Somiari et al., 2003). For iso electric focusing (IEF), the labeled samples were loaded on a 24 cm, pH 3 - 11 NL immobilized pH gradient (IPG) strips. The strips were re-hydrated for 12 hours with the sample buffer consisting of 7 M urea, 2 M thiourea, 30 mM TRIS, 4% CHAPS, 5 mM magnesium acetate tetrahydrate and 1% NP40 and focused for 65,000 vhrs. Following IEF, the strips were equilibrated in SDS equilibration buffer (6 M Urea, 30% glycerol, 15 mM TRIS and 2%) and placed on a 24 cm × 20 cm SDS- PAGE gel (12.5%) for second dimension separation. The second dimension separation was carried out at 15 watts/gel for approximately 4.5 hours. The 2D gels were scanned with the Typhoon Trio Variable Mode Imager (GE Healthcare) at there wavelengths to generate Cy2, Cy3 and Cy5 signals. Images were imported into DeCyder version 6.0 (GE Healthcare) and analyzed with the Difference In-gel Analysis (DIA) and Biological Variation Analysis (BVA) modules. Candidate protein spots on the gel that showed statistically significant difference in abundance between the “test” e.g. Cy3 labeled sample and “reference” e.g. Cy5 labeled sample were automatically identified and selected for picking and identification by tandem mass spectrometry.
2.3. Sequencing of Differentially Expressed Proteins by Tandem Mass Spectrometry
The candidate protein spots of interest were picked from preparatory gels with a spot-picking robot (Ettan Spot Picker, GE Healthcare) and in-gel digested using the Ettan Digester (GE Healthcare) proteomics grade trypsin (Promega, Madison, WI) as previously described [16] . Digested peptides were reconstituted with ultra pure water and sequenced by tandem mass spectrometry to identify the proteins using nano flow capillary Liquid Chromatography Mass Spectrometry (LC/MS/MS). The LC/MS/MS was carried out with 10 cm nanobore electrospray column that is constructed from 360 mm o.d., 75 mm i.d. fused silica capillary with the column tip tapered to a 15 mm opening, and packed with 200 Å 5 μm C18 beads (Michrom BioResources, Auburn, CA). The mobile phase used for gradient elution consisted of (A) 0.3% acetic acid, 99.7% water and (B) 0.3% acetic acid, 99.7% acetonitrile. The flow through the column was split pre-column to achieve a constant flow rate of 350 nL/min. All the tandem mass spectra were acquired on a Thermo LTQ ion trap mass spectrometer (Thermo Corp., San Jose, CA). Needle voltage was set at 3 kV and ion signals above a predetermined threshold automatically triggered the instrument to switch from MS to MS/MS mode for generating fragmentation spectra. The obtained MS/MS spectra were searched against the NCBI non-redundant protein sequence database using the SEQUEST computer algorithm to establish the protein identity [17] . In order to evaluate the interactions of salt and PPARα as well as the global effect of PPARα on genes that are regulated by salt, multimodal comparisons were made for each organ (kidney, heart, or aorta) for genes that are differentially but commonly expressed in the kidney, heart, and the aorta in four groups of WT or KO mice placed on NS or HS diet: 1) WT (NS vs HS diet); 2) WT vs KO (NS diet); 3) WT vs KO (HS diet); 4) KO (NS vs HS diet).
2.4. Analysis
Multimodality analysis was used to identify proteins that were expressed in the kidney, heart, or blood vessels of WT and KO mice. Criteria for differential expression were a P < 0.05, a difference > 1.5-fold, and ≥20% appearance of the spots on the gel.
3. Results
3.1. Proteomic Results on Kidney, Heart, and Liver
Proteomic profiles were compared in the kidney, heart, and aorta of PPARα WT and KO mice fed 0.3% NaCl (NS) or 4% NaCl (HS) diet for 3 weeks. A summary of the throughput of the proteomic analysis is shown in Table 1. Note that a protein might be identified in multiple spots, and changes in individual spots can occur with no change in the total amount of a given protein. Additional information for each differentially expressed protein spot, including standardized gene name, mass, isoelectric points, number of peptide hits, accession numbers (NCBI data base) are also shown.
Table 1. Proteins identified by MS/MS spectra.
The table depicts the particulars of proteins identified by using MS/MS spectra searched against NCBI proteins sequence data base based on the SEQUEST computer algorithm. The master spot numbers that showed significance on the gels, the name of the protein, the mass, isoelectric point (PI), are accession # and GenBank # are also shown.
Based on the criteria of ≥1.5-fold change in expression, ≥20% appearance on the gel, and a P value ≤ 0.05, at least 24 proteins were positively identified from 14 master spots (Table 1). 8 of the master spots contained more than 1 protein, namely, spot #s 646, 648, 672, 678, 740, 883, 1027, and 1764. The proteins identified include those that participate in fatty acid oxidation and mitochondrial electron transport chain e.g. acyl-CoEnzyme A dehydrogenase, transketolase, electron-transferring-flavoprotein dehydrogenase, NADH-ubiquinone oxidoreductase. The other proteins identified include those involved in other cellular activities not directly related to fatty acid metabolism, e.g. prolyl 4-hydroxylase, program cell death 8, stress-induced-phosphoprotein 1, etc., reflecting the pleiotropic nature of the PPARα gene. Based on the criterion that proteins identified in more than one master spot are considered unique, there were 4 unique proteins, namely; acyl-CoEnzyme A dehydrogenase, electron-transferring-flavoprotein dehydrogenase, 5-aminoimidazole-4-carboxamide ribonucleotideformyltransferase, and NADH flavoprotein dehydrogenase/ubiquinone-oxidoreductase. Of these, the most unique is acyl- CoEnzymeA dehydrogenase which appeared in three spots, namely; spot numbers 646, 648, and 684.
3.2. Comparison of the Proteomic Profile in PPARα KO and WT Mice on Normal Diet
Table 2 shows that in KO and WT mice placed on normal salt diet, proteomic profiling of the kidney, heart, and aorta harvested from these animals revealed a significant differential expression in 4 of the 14 master spots analyzed, namely; spot #s 646, 648, 672, and 678. Compared to tissues from PPARα KO mice, there were increases in proteins in WT mice in spot #s 672 (1.68-fold increase) and 678 (1.95-fold increase). The proteins associated with these spots are: electron-transferring-flavoprotein dehydrogenase (EFTDH) and malate dehydrogenase. On the other hand, there was a decrease in proteins associated with spot #s 646 (−2.41) and 648 (−1.53) namely; acyl-Coenzyme A dehydrogenase and transketolase, respectively.
3.3. Effect of High Salt Diet on Proteomic Profile in PPARα WT Mice
When the expression of the different proteins was compared in the kidney, heart, and aorta of PPARα WT mice placed on high or normal salt diet, there was a significant difference in the expression in 9 of the 14 master spots, Table 3. Compared to PPARα WT mice on normal salt, HS diet elicited an increase in proteins associated with 8 of the master spots: 4 of these spots, namely; spot #s 672, 1746, 1764, and 1840 contained proteins involved in fatty acid metabolism, namely; electron-transferring-flavoprotein dehydrogenase, NADPH-ubiquinone oxidoreductase, enoyl-coenzyme hydratase, and NADH dehydrogenase (ubiquinone) flavoprotein 2. However, there was a down regulation of proteins associated with master spot # 740 which contained proteins that are not involved in fatty acid metabolism.
3.4. Proteomic Profile in PPARα KO Mice on High Diet
When the expression of the different proteins was compared in the kidney, heart, and aorta of PPARα KO mice placed on high versus normal salt diet, Table 4 shows that HS diet in KO mice led to a significant difference in the expression of the proteins in 11 of the 14 master spots. The three spots that did not show significance in expression were spot #s 672, 883, and 1764. Compared to PPARα KO mice placed on normal salt diet, HS diet elicited an increase of proteins associated with 10 of the master spots, namely; spot #s 646, 648, 678, 684, 740, 828, 1027, 1591, 1746, 1840, and 1855. Only 5 of these spots, namely: spot #s 646, 648, 684, 1746, and 1840 contained proteins that are directly involved in fatty acid metabolism. These are: acyl-Coenzyme A dehydrogenase (spot #s 646, 648, 684), NADPH-ubiquinone oxidoreductase, and NADH dehydrogenase (ubiquinone) flavoprotein 2. There was a decrease in the level of proteins associated with spot #678 i.e. electron transferring- flavoprotein dehydrogenase.
3.5. Comparison of the Effect of High Salt Diet on Proteomic Profile in PPARα KO and WT Mice
Comparison of the effect of high salt diet on proteomic profile in PPARα KO and WT mice
When the expression profile of proteins was compared between tissues collected from PPARα KO and WT mice placed on high salt diet, Table 5 illustrates that only 3 of the master spots showed a significant difference in expression, namely; spot #s 648, 740, and 1591. Compared to tissues from PPARα KO mice, tissues from WT mice placed on HS diet showed that 2 of these spots were up regulated, namely; spot #s 648 (1.61-fold increase)
Table 2. Spots in tissues from WT or KO mice placed on 3% NaCl.
Spots that showed significant differences in protein levels in the gel samples in tissues harvested from WT or KO mice placed on 0.3% NaCl (normal salt) diet. The average ratio of the levels of the proteins in KO relative to WT mice is presented: + indicates increase; − indicates decrease; # indicates proteins that are directly involved in fatty acid oxidation.
Table 3. Spots in tissues from WT mice.
The spots that showed significant differences in protein levels in tissues harvested from WT mice were placed on 0.3 % (normal, NS) or 4% NaCl (high salt, HS) diet. The average ratio of the levels of the protein in mice placed on HS diet is presented: + indicates increase; − indicates decrease; # indicates proteins that are directly involved in fatty acid oxidation.
Table 4. Spots in tissues from KO mice.
The spots that showed significant differences in tissues from KO mice placed on 0.3% (normal, NS) or 4% NaCl (high salt, HS) diet. The average ratio of the levels of the proteins in mice placed on HS diet is presented: + indicates increase; − indicates decrease; # indicates proteins that are directly involved in fatty acid oxidation.
Table 5. Spots in tissues from WT or KO mice placed on 4% NaCl.
The spots that showed significant differences in protein levels in tissues from WT or KO mice placed on 4% NaCl in (high salt, HS) diet. The average ratio of the levels of the proteins in mice placed on HS diet is presented: + indicates increase; − indicates decrease; # indicates proteins that are directly involved in fatty acid oxidation.
and 740 (2.78-fold). The only protein of significance in relation to fatty acid metabolism is acyl-Coenzyme A dehydrogenase (spot # 648).
4. Discussion
The results of this study demonstrate potentially important interactions between high salt diet and PPARα as they affect fatty acid oxidation enzymes and other proteins involved in important cellular functions. The differentially expressed proteins include but not limited to those primarily associated with fatty acid oxidation phenotype, reflecting the pleiotropic nature of PPARα gene and its activation. Thus, apart from fatty acid oxidation, the proteins identified include those involved in such cellular functions or structures such as signal transduction, energy metabolism, and cytoskeleton. These include heat shock protein e.g. HSP70/90, mitochondrial enzymes, transcriptional factors, structural proteins such as prolyl 4-hydroxylase and others.
PPARα is well characterized in cardiovascular tissues/organs including the heart, blood vessel, and kidney. A protective role has long been established for PPARα in the heart and blood vessel [2] [18] but only recently in the kidney where studies demonstrated that PPARα KO mice were more prone to renal ischemia-reperfusion (I/R) injury than their WT counterparts [19] . In a follow up study, increased renal fatty acid oxidation was suggested to be a cytoprotective mechanism during cisplatin-mediated nephrotoxicity [20] . In a recent study, we established a role for PPARα in renal function and provided evidence that PPARα gene is salt-responsive in as much as PPARα KO mice had an impaired ability to excrete a saline load [21] . The current study evaluated the effect of high salt diet and the resulting differential in the profile of proteins in organs from PPARα KO and WT mice as a means to providing an insight into the molecular mechanisms involved in salt-sensitivity as related to the status of PPARα expression and/or fatty acid oxidation.
Fatty acid oxidation was long known to be important in organs with high metabolism such as the heart, liver, blood vessels [2] . Considering the widely recognized protective role for PPARα in the cardiovascular system [2] [18] , we hypothesized that increased salt load will result in adaptive changes in proteins associated with increased fatty acid oxidation and mitochondrial electron transport chain as a means to handling an excessive sodium load and thereby limiting the injury that may result from a greater than normal sodium intake. Thus, proteins associated with fatty acid oxidation will respond with a greater increase to HS diet in WT and KO mice. A priori, reduced fatty acid oxidation compromises the body’s ability to adequately carry out normal functions and by extension the body’s ability to mitigate against the effects of deleterious injury as seen in exacerbation of renal I/R injury [22] and impaired ability to excrete a saline load [21] in PPARα null mice. Proteomic analysis of the kidney, heart, and aorta of animals exposed to high salt diet in this study revealed a differential expression of 24 proteins. On account of their appearance in more than 1 spot, four of these proteins are considered unique These are: acyl-CoEnzyme A dehydrogenase (MCAD) which is involved in the first step of beta oxidation of fatty acid metabolism during which acyl-coA is converted to enoyl-coA, electron-transferring-flavoprotein dehydrogenase (EFTDH), a third entry point to the electron transport chain which accepts electrons from electron- transferring-flavoprotein in the mitochondrial matrix and uses them to reduce ubiquinone in the inner mitochondrial membrane [23] thereby helping to build the electrochemical potential used to produce ATP. The other unique proteins are: 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) formyltransferase, a bifunctional enzyme that catalyzes the last two steps in the de novo purine biosynthetic nucleotide pathway, NADPH-ubi- quinone-oxidoreductase and NADH dehydrogenase (also known as NADH-coenzyme Q oxidoreductase or complex I), the first protein in the electron transport chain that is involved in the two electron reduction by NADH of coenzyme Q10 or ubiquinone [24] .
The present study focused on proteins that are directly associated with fatty acid oxidation. Thus, comparison between tissues from WT and KO mice placed on NS diet revealed a surprising decrease in protein levels of acyl-Coenzyme A dehydrogenase and transketolase in WT mice, an observation at odds with our hypothesis and inconsistent with an intact PPARα gene in WT mice to effect metabolism of long chain fatty acids. By contrast, there were increases in the levels of malic enzyme, EFTDH, and AICAR in KO mice as expected. Malic enzyme is involved in the oxidative decarboxylation of malate to produce pyruvate and CO2 coupled with the reduction of NAD+ or NADP+. An increase in its level and that of ETFDH in KO mice could be explained on the necessity to compensate for depleted fatty acid oxidation proteins due to lack of PPARa gene. When placed on HS diet, tissues from WT mice showed increases in protein levels of ETFDH, NADPH ubiquinone-oxidoreductase, NADH flavoprotein dehydrogenase, and enoyl coenzyme A hydratase, the enzyme involved the second step of mitochondrial beta oxidation of short chain fatty acid metabolism. Consistent with our hypothesis and the known protective role of PPARα [2] [18] via increased fatty acid oxidation [20] , we interpret these data to mean that HS diet induced fatty acid oxidation enzymes to a greater extent in these animals to ameliorate salt-induced injury to the kidney, heart, and blood vessels. Surprisingly, the proteomic profile in KO mice placed on high salt diet, when compared with that obtained in those placed on NS diet revealed a significant increase in protein levels of acyl-coenzyme A dehydrogenase, NADH-ubiquinone oxidoreductase and transketolase. This observation raises an issue regarding the requirement for an intact gene for the activity and/or expression of these enzymes thus arguing in support of the observation showinga reduction in the constitutive levels of mitochondrial fattyacid β-oxidation but not constitutive or basal oxidationof VLCFAs by peroxisomal β-oxidation system with PPARα deficiency [9] . Unlike these proteins, the protein level of enoyl coenzyme A hydratase was not significantly altered while that of AICAR formyltransferase, ETFDH, and malic enzyme was decreased in KO mice, indicating the coupling of these other proteins to the PPARα gene. Comparing the spectrum of the proteomic profile of WT and KO mice placed on HS diet, Table 5, acyl-Coenzyme A dehydrogenase was the only fatty acid oxidation-related protein that was increased in response to HS diet. This suggests a pivotal role for this enzyme involved in the 1st step of fatty acid oxidation as the frontline enzymatic reaction protecting the body from injurious stimuli.
The increase in FAO enzyme in response to HS diet in this study is in agreement with the findings that HS diet affected a number of proteins in the Dahl salt-sensitive rat kidney including fumarate hydratase and other proteins involved in the metabolism of fatty acids [25] - [27] . The similarity in the profile of these proteins in the present study and the studies by Tian et al. [25] suggests that these differentially expressed proteins might contribute to the differences in the susceptibility to injurious stimuli in PPARα KO mice as it does in Dahl salt-sen- sitive rats. Proteomic studies also have revealed changes in the heart of diet-induced pre-diabetic mice [28] . The significance of our study is that it demonstrated a role for PPARα in the body’s adaptive (protective) responses to changes in dietary salt in organs such as the heart, liver and kidney that depend on FAO for energy production. High salt diet is known to increase blood pressure in people that have salt-sensitive hypertension and our previous study showed that absence of PPARα gene leads to lack of its renal protective effect rendering the kidneys more injury-prone and interfering with the normal reabsorptive capacity of the kidney. This therefore leads to reduced sodium excretion and higher blood pressure in PPARα KO mice [21] . The significance of this proteomic study is the possible use of gene therapy (PPARα gene) in the management of cardiovascular disease.
5. Conclusion
PPARα appears to be a salt-responsive gene controlling multiple gene targets through changes in the expression of different proteins that mitigate any possible injury to the heart, kidney, and the blood vessel. These multiple proteins and/or targets involve fatty acid oxidation, lipid and amino acid metabolism, and other cellular pathways that serve critical protective roles in the body.
References
- Lemberger, T., Desvergne, B. and Wahli, W. (1996) Peroxisome Proliferator Activated Receptors: A Nuclear Receptor Signaling Pathway in Lipid Physiology. Annual Review of Cell and Developmental Biology, 12, 335-363. http://dx.doi.org/10.1146/annurev.cellbio.12.1.335
- Bishop-Bailey, D. (2000) Peroxisome Proliferator-Activated Receptors in the Cardiovascular System. British Journal of Pharmacology, 129, 823-834. http://dx.doi.org/10.1038/sj.bjp.0703149
- Yang, T., Michele, D.E., Park, J., Smart, A.M., Lin, Z., Brosius III, F.C., Schnermann, J.B. and Briggs, J.P. (1999) Expression of Peroxisomal Proliferator-Activated Receptors and Retinoid X Receptors in the Kidney. American Journal of Physiology, 277, 966-973.
- Guan, J. (2002) Targeting Peroxisome Proliferator-Activated Receptors (PPARα) in Kidney and Urologic Disease. Minerva Urologica e Nefrologica, 54, 65-79.
- Guan, Y., Zhang, Y., Davis, L. and Breyer, M.D. (1997) Expression of Peroxisome Proliferator-Activated Receptors in Urinary Tract of Rabbits and Humans. American Journal of Physiology, 273, 1013-1022.
- Djouadi, F. and Bastin, J. (2001) PPARα Gene Expression in the Developing Rat Kidney: Role of Glucocorticoids. Journal of the American Society of Nephrology, 12, 1197-1203.
- Hashimoto, T. (1999) Peroxisomal β-Oxidation Enzymes. Neurochemical Research, 24, 551-563. http://dx.doi.org/10.1023/A:1022540030918
- Reddy, J.K. and Mannaerts, G.P. (1994) Peroxisomal Lipid Metabolism. Annual Review of Nutrition, 14, 343-370. http://dx.doi.org/10.1146/annurev.nu.14.070194.002015
- Aoyama, T., Peters, J.M., Iritani, N., Nakajima, T., Furihata, K., Hashimoto, T. and Gonzalez, F.J. (1998) Altered Constitutive Expression of Fatty Acid-Metabolizing Enzymes in Mice Lacking the Peroxisome Proliferator-Activated Receptor Alpha (PPAR). Journal of Biological Chemistry, 273, 5678-5684. http://dx.doi.org/10.1074/jbc.273.10.5678
- Hashimoto, T., Fujita, T., Usuda, N., Cook, W., Qi, C., Peters, J.M., Gonzalez, F.J., Yeldandi, A.V., Rao, M.S. and Reddy, J.K. (1999) Peroxisomal and Mitochondrial Fatty Acid Oxidation in Mice Nullizygous for Both Peroxisome Proliferator-Activated Receptor and Peroxisomal Fatty Acyl-CoA Oxidase. Journal of Biological Chemistry, 274, 19228-19236. http://dx.doi.org/10.1074/jbc.274.27.19228
- Ockner, R.K., Kaikaus, R. and Bass, N.M. (1993) Fatty-Acid Metabolism and the Pathogenesis of Hepatocellular Carcinoma: Review and Hypothesis. Hepatology, 18, 669-676. http://dx.doi.org/10.1002/hep.1840180327
- Formenty, B. and Pessayre, D. (1995) Inhibition of Mitochondrial Beta-Oxidation as a Mechanism of Hepatotoxicity. Pharmacology & Therapeutics, 67, 101-154. http://dx.doi.org/10.1016/0163-7258(95)00012-6
- Ghadiminejad, I. and Saggerson, D. (1992) Physiological State and the Sensitivity of Liver Mitochondrial Outer Membrane Carnitine Palmitoyltransferase to Malonyl-CoA. Correlations with Assay Temperature, Salt Concentration and Membrane Lipid Composition. International Journal of Biochemistry, 24, 1117-1124.
- Rachamim, N., Latter, H., Malinin, N., Asher, C., Wald, H. and Garty, H. (1995) Dexamethasone Enhances Expression of Mitochondrial Oxidative Phosphorylation Genes in Rat Distal Colon. American Journal of Physiology, 269, C1305- C1310.
- Wirthensohn, G. and Guder, W.G. (1983) Renal Lipid Metabolism. Mineral Electrolyte Metabolism, 9, 203-211.
- Somiari, R.I., Sullivan, A., Russell, S., Somiari, S., Hu, H., Jordan, R., George, A., Katenhusen, R., Buchowiecka, A., Arciero, C., Brzeski, H., Hooke, J. and Shriver, C. (2003) High-Throughput Proteomic Analysis of Human Infiltrating Ductal Carcinoma of the Breast. Proteomics, 3, 1863-1873. http://dx.doi.org/10.1002/pmic.200300560
- Yates, J.R.D., Eng, J.K., McCormack, A.L. and Schieltz, D. (1995) Method to Correlate Tandem Mass Spectra of Modified Peptides to Amino Acid Sequences in the Protein Database. Analytical Chemistry, 67, 1426-1436. http://dx.doi.org/10.1021/ac00104a020
- Braissant, O., Foufelle, F., Scotto, C., Dauca, M. and Wahli, W. (1996) Differential Expression of Peroxisome Proliferator-Activated Receptors (PPARα): Tissue Distribution of PPAR-α, -β, and -γ in the Adult Rat. Endocrinology, 137, 354-366.
- Portilla, D., Dai, G., Peters, J.M., Gonzalez, F.J., Crew, M.D. and Proia, A.D. (2000) Etomoxir-Induced PPAR
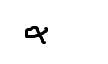 -Modulated Enzymes Protects during Acute Renal Failure. American Journal of Physiology—Renal Physiology, 278, F667-F675.
-Modulated Enzymes Protects during Acute Renal Failure. American Journal of Physiology—Renal Physiology, 278, F667-F675. - Li, S., Wu, P., Yarlagadda, P., Vadjunec, N.M., Proia, A.D, Harris, R.A. and Portilla, D. (2004) PPARα Ligand Protects during Cisplatin Induced Acute Renal Failure by Preventing Inhibition of Renal FAO and PDC Activity. American Journal of Physiology-Renal Physiology, 286, F572-F580. http://dx.doi.org/10.1152/ajprenal.00190.2003
- Obih, P. and Oyekan, A.O. (2008) Regulation of Blood Pressure, Natriuresis and Renal Thiazide/Amiloride Sensitivity in PPARα Null Mice. Blood Pressure, 17, 55-63.
- Negishi, K., Noiri, E., Sugaya, T., Li, S., Megyesi, J., Nagothu, K. and Portilla, D. (2007) A Role of Liver Fatty Acid- Binding Protein in Cisplatin-Induced Acute Renal Failure. Kidney International, 72, 348-358.
- Ramsay, R.R., Steenkamp, D.J. and Husain, M. (1987) Reactions of Electron-Transfer Flavoprotein and Electron- Transfer Flavoprotein: Ubiquinone Oxidoreductase. Biochemical Journal, 241, 883-892.
- Hirst, J. (2005) Energy Transduction by Respiratory Complex I—An Evaluation of Current Knowledge. Biochemical Society Transactions, 33, 525-529. http://dx.doi.org/10.1042/BST0330525
- Tian, Z., Greene, A.S., Usa, K., Matus, I.R., Bauwens, J., Pietrusz, J.L., Cowley Jr., A.W. and Liang, M. (2008) Renal Regional Proteomes in Young Dahl Salt-Sensitive Rats. Hypertension, 51, 899-904. http://dx.doi.org/10.1161/HYPERTENSIONAHA.107.109173
- Grussenmeyer, T., Meili-Butz, S., Roth, V., Dieterle, T., Brink, M., Matt, P., Carrel, T.P., Eckstein, F.S., Lefkpovits, I. and Grapow, M.T. (2011) Proteome Analysis in Cardiovascular Pathology Using Dahl Rat Model. Journal of Proteomics, 74, 672-682.
- Zheleznova, N.N., Yang, C., Ryan, R.P., Halligan, B.D., Liang, M., Greene, A.S. and Cowley Jr., A.W. (2012) Mitochondrial Proteomic Analysis Reveals Deficiencies in Oxygen Utilization in Medullary Thick Ascending Limb of Henle in Dahl Salt-Sensitive Rat. Physiological Genomics, 44, 839-842. http://dx.doi.org/10.1152/physiolgenomics.00060.2012
- Cruz-Topete, D., List, E.O., Okada, S., Kaider, B. and Kopchick, J.J. (2012) Proteomic Changes in the Heart of Diet- Induced Pre-Diabetic Mice. Journal of Proteomics, 74, 716-727. http://dx.doi.org/10.1016/j.jprot.2011.02.018

*Corresponding author.


