Open Journal of Apoptosis
Vol.3 No.2(2014), Article
ID:45074,16
pages
DOI:10.4236/ojapo.2014.32003
Staurosporine-Induced Cell Death in Trypanosoma brucei and the Role of Endonuclease G during Apoptosis
Torsten Barth1, Gustavo Bruges2, Andreas Meiwes1, Stefan Mogk1, Celestin N. Mudogo1, Michael Duszenko1,3*
1Interfakultäres Institut für Biochemie, University of Tübingen, Tübingen, Germany
2Insituto Venezolano de Investigaciones Científicas, Caracas, Venezuela
3Faculty of Medicine, Tongji University, Shanghai, China
Email: *michael.duszenko@uni-tuebingen.de
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 10 December 2013; revised 31 January 2014; accepted 10 February 2014
Abstract
Apoptosis in single-cell organisms like Trypanosoma or Leishmania was characterized in several studies in the last few years [1] -[4] . Cell death in these caspase lacking protozoa is still poorly understood and a conclusive apoptotic pathway has not been identified so far. In the work presented here, we studied the effects of prostaglandin D2 and staurosporine induced cell death in bloodforms of Trypanosoma brucei in a time dependent manner and focused on the role of a nuclease similar to endonuclease G of higher eukaryotes. We found that these parasites undergo apoptotic cell death as demonstrated by the appearance of several canonical hallmarks of apoptosis in higher eukaryotes, but that different stimuli induce remarkable differences in the way these cells die. We compared the effects of prostaglandin D2 and staurosporine in trypanosomes with and without endonuclease G overexpression by flow cytometric and electron microscopic methods with the result that endonuclease G overexpression led to a significant modification of intracellular organelles and accelerated apoptotic cell death in prostaglandin D2 or staurosporine treated cells. Our results demonstrate that different stimuli induce apoptosis even in these ancient organisms in different caspase-independent ways. Whereas central processes of apoptosis like ROS formation, loss of mitochondrial membrane potential, endonuclease G release, phosphatidylserine exposure and DNA fragmentation appeared in the same chronology during treatment with either one of both drugs, other effects like cell cycle arrest or change of cell shape occurred only in the case of prostaglandin D2 or staurosporine treatment. We conclude from these results that trypanosomes react to stimuli of apoptosis with the concerted action of cellular responses but cannot control the final outcome if additional stress, as in the case of staurosporine, is superimposed.
Keywords:Trypanosoma brucei, Endonuclease, EndoG, Staurosporine, Programmed Cell Death, Apoptosis

1. Introduction
Trypanosomatids form a group of mono-flagellated single cell parasites of insects. Some of these, like Trypanosoma brucei, Trypanosoma cruzi and various species of leishmania are responsible for human infecting diseases which concern some 445 million people all over the world [5] : Leishmaniasis affects some 350 million people primarily in the tropics; in Africa some 70 million people are at the risk of contracting human African trypanosomiasis (HAT or sleeping sickness) and in Latin America about 25 million people are at the risk of getting American trypanosomiasis also known as Chagas disease.
T. brucei is a heteroxenous parasite with a complex life cycle. It uses two hosts, the tsetse fly (Glossina spp.) and different vertebrate hosts including man. The parasite lives in the mid-gut of the fly before it migrates to the salivary gland. Here it differentiates first to the epimastigotic form and eventually to the metacyclic form, which is infectious for the vertebrate host. Following the bite of a tsetse fly, the parasite appears in the hemolymphatic system, where it transforms to the slender blood-form. In blood, this form divides by binary fission and, depending on cell density, may differentiate to the non-dividing stumpy blood-form, which was used throughout this study. Stumpy parasites will die if not taken up by another tsetse fly during a blood meal. As described previously, trypanosomes produce different prostaglandins (PGs) including PGF2α and PGD2. PGF2α is mainly produced by the slender form and may act as a growth factor, whereas PGD2 is mainly produced by the stumpy form and acts as a cell density regulator inducing apoptosis [6] .
In higher eukaryotes, apoptosis is a fundamental phenomenon in the homoeostasis of tissues and especially involved in embryonic development, morphogenesis, selectivity of immune cells, tissue atrophy and tumour regression. While it is thus a significant contributor to the functional development and maintenance of multicellular organisms, the advantages of apoptosis for unicellular organisms are much less evident. Nevertheless, apoptosis or apoptosis-like phenotypes have been described in many different single-cell eukaryotes, such as yeast [7] [8] , Tetrahymena [9] , Dictyostelium [10] , and different kinetoplastids like trypanosomes [3] and leishmania [1] [4] . It has also been suggested that apoptosis may even occur in bacteria [11] [12] . In T. brucei, apoptosis contributes to cell density regulation [3] and acts as a mechanism to maintain genetic stability and differentiation [13] .
Extensive analyses of protozoa genomes revealed that genes encoding typical proteins for apoptosis regulation in metazoa like caspases, members of the Bcl-2 family or a caspase-activated DNase (CAD) are missing. Interestingly, endonuclease G (EndoG), another nuclease active during apoptosis in higher eukaryotes [14] [15] , is, however, also expressed in kinetoplastids [16] [17] . This nuclease is encoded in nuclear DNA and is translocated into the mitochondrion, where it may be involved in replication and repair functions [18] . EndoG belongs to the ββα-metal superfamily of DNA/RNA non-specific nucleases, requires divalent cations like Mg2+ for activity, and is inhibited by moderate salt concentrations (100 - 150 mM NaCl) [19] [20] . After induction of apoptosis by different stimuli (including staurosporine), the mitochondrial membrane potential is lost and EndoG is translocated via the cytosol into the nucleus, where it participates in chromatin DNA degradation [14] [21] .
The trypanosomal EndoG molecule is significantly larger than the respective endonuclease in higher eukaryotes. While human or bovine EndoG consists of 297 or 299 amino acids with a molecular weight of 32.6 kDa or 32.3 kDa respectively, trypanosomal EndoG contains 506 amino acids leading to a predicted molecular weight of 55.8 kDa.
Nevertheless, comparison of trypanosomal EndoG with mammalian EndoG molecules showed several conserved amino acid residues and revealed some 30% identity with human or bovine EndoG on the protein level.
In the present study, we used prostaglandin D2 (PGD2) and staurosporine (STS) to investigate appearance of apoptotic hallmarks like cell cycle arrest, intracellular ROS production, loss of mitochondrial membrane potential, DNA fragmentation and phosphatidylserine exposure by flow cytometric analysis.
PGD2 is a prostanoid derived from arachidonic acid via the cyclooxygenase pathway. In T. brucei, PGD2 is produced and secreted by the stumpy bloodform and induces an apoptosis-like cell death which includes intracellular ROS formation, phosphatidylserine exposure, loss of mitochondrial membrane potential and DNA degradation [3] . The stumpy bloodform was also shown to be more sensitive to PGD2 than the slender bloodform, suggesting that the physiological function of PGD2 is related to the parasite’s cell density regulation and acts therefore in favour of a balanced parasite-host interrelationship [2] [22] .
Staurosporine is a microbial alkaloid and a strong broad spectrum protein kinase inhibitor which has been shown to induce apoptosis in uniand multicellular organisms [23] -[27] . The mechanism of STS-induced apoptosis is still unknown but it involves alteration of the phosphorylation state, alterations related to cell cycle control and DNA degradation [24] .
In L. major STS-induced apoptosis showed several cytoplasmic and nuclear effects, including cell shrinkage, phosphatidylserine exposure, maintenance of plasma membrane integrity, loss of mitochondrial membrane potential, cytochrome c release, nuclear chromatin condensation and DNA fragmentation [23] . In former studies, the anti-parasitic activity of STS against T. brucei was observed, but has not been evaluated yet [28] [29] . In addition, STS has been used to assess its effect on cellular signalling pathways associated with phospholipids in T. cruzi [30] .
Elucidation of the molecular mechanisms leading to apoptosis in trypanosomes will help to understand the in vivo role and to identify new target molecules for chemotherapeutic drug development, because diverse substances like pentostam (a pentavalent antimony compound), amphotericin B, flavonoids, H2O2, nitric oxide, quercetin, staurosporine, prostaglandin D2 and metabolites of the J2 series seem to induce apoptosis in trypanosomes [1] [3] [13] [31] [32] . To obtain a better understanding of how EndoG acts, we also cloned and expressed rTbEndoG in E. coli and in T. brucei, expressed it in T. brucei as a fusion protein containing eGFP, and overexpressed or knocked down the parasite’s enzyme in trypanosomes.
2. Material and Methods
2.1. Parasites
For RNAi and overexpression experiments, a clonal single marker blood-form line (SMB BF) derived from BF 221 (MiTat1.2) [33] that constitutively expresses T7 RNA-polymerase (Plasmid pHD328) and Tn10 Tet repressor (Plasmid pLEW114hyg5’) was used.
2.2. Cloning, Protein Expression and Purification of rTbEndoG
T. brucei EndoG (Tb427.9.4040) was amplified by PCR using genomic DNA from T. brucei MiTat1.2 as template and the following primers for either recombinant expression of rTbEndoG in E. coli, TbEndoG overexpression in T. brucei, or TbEndoG-eGFP expression in T. brucei:
1) Primer for rTbEndoG in pProEx HTa in E. coli fw-EcoRI-TbEndoG: 5’-GAATTCAGTGGACGGAAGGACCTCATAG-3’
rev-NotI-TbEndoG: 5’-GCGGCCGCCCCTGGAAAGTTACAAATAAGG-3’
2) Primer for TbEndoG in pLew100v5-Hyg in T. brucei fw-HindIII-TbEndoG: 5’-ACGGAAGCTTATGCATCGCATCACCGTAC-3’
rev-BamHI-TbEndoG: 5’-ACTTGGATCCTTAACCGGTGTCGTTGGTC-3’
3) Primer for TbEndoG-eGFP in pCO57 in T. brucei fw-HindIII-TbEndoG: 5’-AAGCTTATGCATCGCATCACCGTA-3’
rev-PvuII-TbEndoG: 5’-CAGCTGACCGGTGTCGTTGGTCG-3’
The PCR reaction was run by using AccuPrime® Taq DNA polymerase high fidelity (Invitrogen) according to the manufacturer’s protocol.
TA Cloning Kit (Invitrogen) was used for subcloning the PCR products into pCR2.1 plasmids. Plasmids were transformed into TOP10 competent E. coli cells (Invitrogen) and grown in Luria-Bertani Broth. Plasmid pCR2.1-EndoG was purified by QIAprep spin miniprep kit (Qiagen) and then digested with the necessary restriction enzymes. DNA fragments extracted from agarose gels were cloned into expression plasmids.
pProEx HTa (Invitrogen) was used as expression vector in E. coli. The plasmid was transformed into BL21(DE3) competent E. coli cells (Invitrogen) and expressed by IPTG induction using a concentration of 0.5 mM for 5 h at 37˚C. The protein was expressed and appeared within inclusion bodies. Cells were washed with inclusion bodies wash buffer (20 mM Tris-HCl, pH 7.5, containing 10 mM EDTA and 1% Triton X-100). Re-suspended cells were incubated for 15 min at 30˚C in the presence of 100 µg/ml lysozyme. Cells were sonicated on ice using a sonifier with an appropriate tip size (3 × 1 min at 50% power). Immediately following cell lysis, a protease inhibitors cocktail (Thermo Scientific) was added. Inclusion bodies were collected by a 10 min centrifugation step at 10.000 g and solubilized in solubilisation buffer (20 mM Tris-HCl, pH 11, containing 1% N-lauroylsarcosine) for 15 min at RT and 4˚C overnight. Solubilized inclusion bodies were clarified by centrifuging at 10.000 g for 10 min at 4˚C. Inclusion bodies were then dialysed against dialysation buffer (20 mM Tris-HCl, pH 7.5, containing 300 mM NaCl and 0.1% Triton X-100) and purified by affinity chromatography.
For this purpose, solubilized inclusion bodies were loaded onto a Ni-NTA column (Qiagen), washed with 20 mM Tris-HCl, pH 7.5, containing 300 mM NaCl, 10 mM imidazole and 0.1% Triton X-100, and eluted with an imidazole gradient (0 to 500 mM) in the same buffer system. Eluted peak fractions were dialysed against cleavage dialysis buffer (20 mM Tris-HCl, pH 7.2, containing 100 mM KCl, 10% glycerol, 0.1% Triton X-100 and 0.5 mM DTT) using dialysis tubes with a cut off of 14 kDa. The obtained His-tagged protein was cleaved using AcTEV Protease (Invitrogen) by incubating it for 4 h at RT and overnight at 4˚C.
The cleavage reaction mixture was diluted 1:1 with wash buffer and loaded again onto a Ni-NTA column pre-equilibrated with wash buffer. Peak fractions of the purified protein were pooled and subsequently concentrated using a 10 kDa NMWL centricon (Millipore) and dialysed overnight against dialysis buffer.
2.3. Knock down and Overexpression of TbEndoG
For TbEndoG knock down experiments the tetracycline-inducible RNAi vector p2T7-TA blue was used. Synthesis of dsRNA was induced by adding 10 µg/ml doxycycline to cell cultures. Instead of tetracycline, doxycycline was used in this work.
For overexpression of TbEndoG, the tetracycline-inducible vector pLEW100v5-Hyg was used. Following a NotI linearization, both vectors were inserted into SMB blood-form cells by electroporation before these cells were cultivated in HMI-9 medium. Selection was obtained by adding 2 µg/ml G418 and 2.5 µg/ml hygromycin B to the culture medium.
pCO57 was used as expression vector for TbEndoG-eGFP expression in T. brucei. The plasmid was transfected into SMB blood-form trypanosomes by electroporation and selected using 2 µg/ml G418 and 2.5 µg/ml phleomycin.
2.4. Nuclease Cleavage Assay
One microgram of plasmid DNA was incubated for 30 min at 37˚C with recombinant wild-type T. brucei EndoG (either rTbEndoG with His-tag or rTbEndoG without His-tag and 200 ng purified protein) in assay buffer consisting of 20 mM HEPES, pH 7.5, containing 3 mM MgCl2 and 0.1% Triton X-100. The cleavage products were resolved on a 1% agarose gel and stained with ethidium bromide.
2.5. Growth Curves
SMB trypanosomes were grown axenically in HMI-9 medium. Stabilates of parasites from liquid nitrogen were thawed, seeded at a cell density of 2 × 105 cells/ml and grown at 37˚C and 5% CO2 for 20 h. Afterwards, cultures were diluted to 2 × 105 cells/ml using fresh culture medium to start the experiments. PGD2 was purchased from Cayman Chemical Co., reconstituted in ethanol and diluted to the respective concentrations using culture medium. Staurosporine from Streptomyces sp. was purchased from Sigma-Aldrich and dissolved in DMSO. The corresponding volume of solvent was added to the untreated control cells.
2.6. esiRNA
For TbEndoG gene silencing we used Mission® esiRNA from Sigma Aldrich. Endoribonuclease-prepared siRNAs or esiRNA are a mixture of siRNAs with an average length of 21 bp resulting from cleavage of long double-stranded RNA (dsRNA) with Escherichia coli RNase III. We added 5 µl (1 µg) of esiRNA derived from the target sequence 5’-CTCATGCCCACTGATACGTGCACTGTCATCCCACTTACTCCTTCTATTACAC TTTGTGG-3’ to TbEndoG-over and SMB control cells. Cell cultures were prepared as described before and cells were grown under normal growth conditions.
2.7. qRT-PCR
SMB trypanosomes were grown axenically in HMI-9 medium. TbEndoG expression was induced by adding 10 µg/ml doxycycline. After 24 h, cells were treated with 10 µM PGD2 and harvested after another 24 h under culture conditions. Total RNA was isolated using RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol, solved in RNase free water and stored at −20˚C. qRT-PCR experiments were set up using Power SYBR® Green RNA-to-CT™ 1-Step Kit (Invitrogen) and analyzed in a LightCycler® 480 Real-Time PCR System (Roche).
2.8. Detection of Apoptotic Hallmarks by Flow Cytometry
Characterization of apoptotic hallmarks was performed by using different fluorescence marker. Cells were analyzed using a FACSCantoII flow cytometer (BD Biosciences).
2.8.1. Intracellular ROS
Intracellular reactive oxygen species (ROS) were detected with 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA), 3’-(p-aminophenyl)fluorescein (APF) or dihydroethidium (DHE), all purchased from Sigma Aldrich (Germany). All reagents were dissolved in DMSO and stored at −20˚C until further use. Following a 24 h incubation with 10 µM PGD2 or 10 nM staurosporine to induce apoptosis, cells were washed with Ringer’s solution (147 mM NaCl, 4 mM KCl, 2.3 mM CaCl2, 1 M NaHCO3, pH 7.4) and incubated in the dark for 10 min at 37˚C using one of the respective ROS markers. Afterward cells were transferred on ice for performing flow cytometric analysis.
2.8.2. Mitochondrial Membrane Potential (Ψm)
Loss of mitochondrial membrane potential (ΔΨm) was detected using tetramethylrhodamine ethyl ester (TMRE). For this purpose, TMRE was solved in DMSO and stored at −20˚C until further use. Following a 24 h incubation in the presence of PGD2 or staurosporine to induce apoptosis, cells were washed with Ringer’s solution and incubated in the dark for 10 min at 37˚C with 25 nM TMRE. Afterward cells were transferred on ice to perform flow cytometric analysis.
2.8.3. DNA Content
Detection of DNA within the nucleus was performed using propidium iodide (PI). For this purpose, 1 × 106 cells were washed in Ringer’s solution and lysed with 6 mM digitonin. Samples were vortexed and incubated for 30 min at RT. Nuclei were stained with a propidium iodide solution (10 mg/ml) 1 h before measurements using a FACSCantoII flow cytometer.
2.8.4. Phosphatidylserine Exposure
To detect phosphatidylserine exposure on the outer membrane of cells, annexin-V-fluos (Roche) was used. Cells were washed in Ringer’s solution and incubated for 15 min with annexin-V at 4˚C. Fluorescence was measured by flow cytometric analysis as described before.
2.9. Fluorescence Microscopy
To stain mitochondria, 1 × 106 trypanosomes were incubated in HMI-9 medium containing 50 nM MitoTracker® Red CMXRos (Invitrogen) for 20 min at 37˚C and 5% CO2 prior to fixation in 3.5% paraformaldehyde. Following fixation, parasites were settled onto poly-L-lysine-coated slides, incubated with 50 mM glycine in PBS for 15 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min. Nuclei were labelled using 4’,6-diamidino-2-phenylindole (DAPI). Cells were visualised using a Zeiss Cell Observer microscope.
2.10. Transmission Electron Microscopy (TEM)
1 × 108 trypanosomes were harvested and washed twice with PBS. Fixation was performed for 1 h at 4˚C using 2% glutaraldehyde dissolved in 0.2 M sodium cacodylate buffer containing 0.12 M sucrose. After washing four times (10 min each) and storage overnight in sodium cacodylate buffer, cells were post-fixed in 1.5% osmium tetroxide and stained in 0.5% uranyl acetate. After dehydration in ethanol and clearing in propylene oxide, embedding in Agar 100 was performed according to standard protocols [34] . Sections were stained in 5% uranyl acetate and 0.4% lead citrate.
2.11. Scanning Electron Microscopy (SEM)
For SEM, the same fixation as described above was applied. Cells were sequentially dehydrated in 50% and 70% ethanol. Samples were placed onto poly-L-lysine-coated slides and incubated for 8 h in 96% and 100% ethanol. After critical point drying, cells were metalized with Pd-Au. For SEM, a Cambridge Stereo Scan 250 Mk2 was used in the group of Oliver Betz (evolution biology of invertebrates, University of Tübingen).
3. Results
DNA sequence analysis of TbEndoG clearly revealed its affiliation with the endonuclease superfamily containing a ββα-structure, its structure binding domains of divalent metal ions with essential histidine and asparagine residues and the active site as present in other previously reported EndoG molecules. Like in other trypanosomatids the aspartate residue (D) is substituted by serine (S) in the trypanosomal DRGH motif (Figure 1) [17] [20] .
Since EndoG is a membrane associated protein that may even possess a small transmembrane domain, expression and purification as a soluble protein proved rather difficult. Thus rTbEndoG containing a His-tag was always expressed in bacterial inclusion bodies, despite our numerous modifications of the expression protocol. Consequently, the protein was re-solubilized from inclusion bodies using 1% N-lauroylsarcosine and then slowly refolded in 20 mM Tris-HCl (pH 7.5, containing 300 mM NaCl and 0.1% Triton X-100) on a Ni-NTA column. Figure 2(a) shows the purified protein as analysed by Western blotting using a commercially available anti-6xHis antibody. The aberrant size of the detected protein (~66 kDa) proved different as compared to the predicted size of 59.3 kDa (including the His-tag), but this has also been observed in previous studies [35] [36] .
After refolding and elution from the Ni-NTA column, nuclease activity was restored, as evaluated using an activity assay (Figure 2(b)). Both recombinant forms of the enzyme, i.e. with and without His-tag, showed nuclease activity with plasmid DNA as substrate. This activity was Mg2+-dependent. Obviously, the His-tag does not affect the correct refolding of rTbEndoG. As divalent metal ions act as cofactors for EndoG, we also tested different metal compounds. Similarly to other nucleases, rTbEndoG showed a higher activity with Mg2+ than with Mn2+ (data not shown). However, the enzyme was inactive in the presence of Zn2+, consistent with earlier reports showing that EndoG of other organisms were inhibited by Zn2+ and Fe2+ [35] .
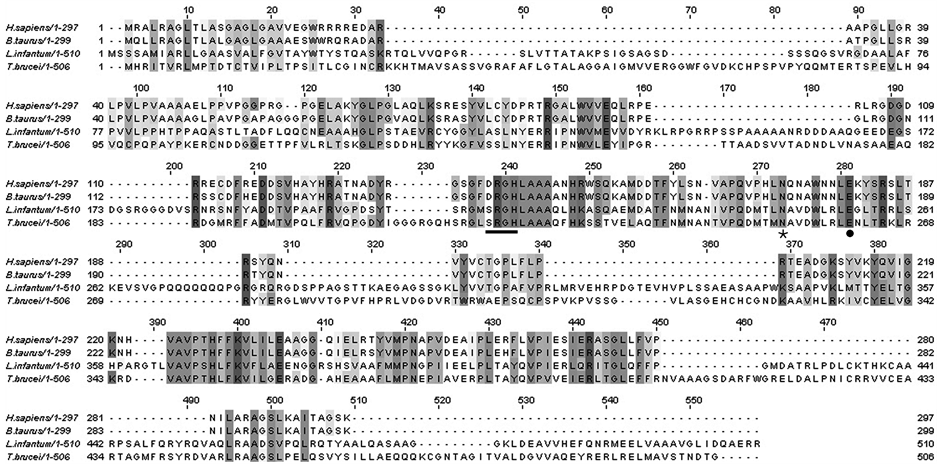
Figure 1. ClustalW alignment of T. brucei EndoG with mammalian and protozoan homologs. Shaded areas represent identical residues and dashes represents gaps. The catalytically important DRGHand SRGH-motif, respectively with the essential histidine is underlined. The cofactor binding asparagine (Ø) and glutamic acid (˜) are indicated.
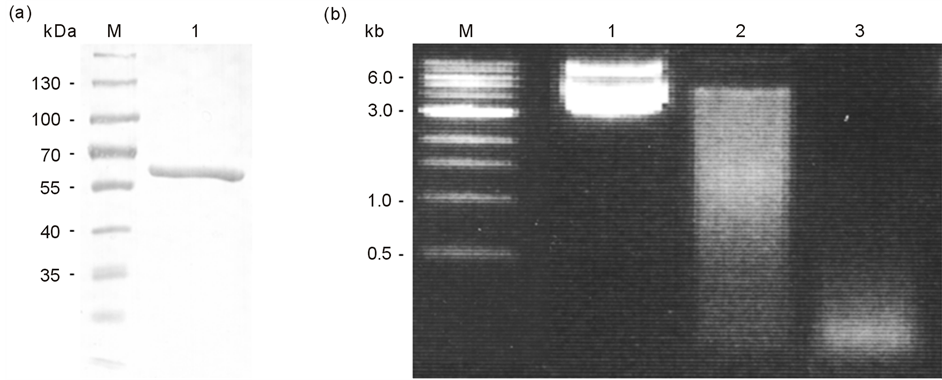
Figure 2. rTbEndoG purification and nuclease activity. a) Western blot of His-rTbEndoG after inclusion bodies isolation with anti-6xHis antibody. M) Proteinmarker PageRuler #SM0671 (Fermentas); 1) purified His-rTbEndoG. b) Nuclease activity of rTbEndoG. Agarose gel electrophoresis of 1 µg plasmid DNA incubated with 200 ng rTbEndoG at 37˚C in HEPES buffer for 4 h and 16 h. M) 1 kb DNA Ladder (NEB); 1) plasmid DNA without rTbEndoG; 2) plasmid DNA after 4 h incubation with 200 ng rTbEndoG; 3) plasmid DNA after 16 h incubation with 200 ng rTbEndoG.
Overexpression of TbEndoG in SMB trypanosomes showed no obvious phenotypic changes, but it significantly inhibited the parasite’s growth. Trypanosomes overexpressing TbEndoG (even when not induced by doxycycline) grew significantly slower with a generation doubling time of 24 h, as compared to 6 h of control SMB cells (Figure 3(a)). Induction of TbEndoG overexpression with 10 µg/ml doxycycline led to a fully inhibited growth. This growth inhibition was abrogated by adding esiRNA against TbEndoG (Figure 3(b)). In contrast, addition of esiRNA or knock down of TbEndoG using RNAi to control SMB cells showed no morphological alterations and no changes of the generation doubling time.
To investigate the role of TbEndoG during apoptosis, trypanosomes were treated with PGD2 or staurosporine. Measuring the intracellular levels of TbEndoG by qRT-PCR, TbEndoG overexpressing cells showed an approximately 7 folds higher mRNA level and thus probably a likewise increased enzyme level than the respective control SMB cells. Following apoptosis induction by addition of PGD2, the EndoG level increased 3 folds in control SMB cells, but remained constant in TbEndoG overexpressing cells. To control for loading differences, tubulin expression in respective cells was set to 1 (Figure 4). For additional loading control expression of GAP-DH was used.
Transmission electron microscopy of trypanosomes overexpressing TbEndoG and induced for 24 h by 10 µg/ml doxycycline revealed a significant increase in the number of glycosomes and densely packed acidocalcisomes as compared to control SMB cells (Figure 5).
In contrast to the known phenotypic changes after PGD2 treatment (occurrence of two or more flagella in one flagella pocket due to a cell cycle arrest in the G1 phase), in STS treated cells a considerable morphological change occurred. As shown by scanning electron microscopy, the cell body switched from a stretched to a ball-like structure (Figure 6(a)). Concomitantly, transmission electron microscopy revealed that this change was due to an extremely enlarged flagella pocket (Figure 6(b)), which was still in the process of taking up more vesicles.
There is a well-known set of different markers to characterize apoptosis in general [37] -[39] and caspase-free apoptosis in particular [40] . Beside microscopic techniques including fluorescence and electron microscopy, we used especially flow cytometry analysis to detect characteristic intracellular apoptosis markers. As described earlier, increased levels of reactive oxygen species (ROS) like H2O2 or hydroxyl radicals is indicative of caspase-independent apoptosis in trypanosomes [2] as well as in higher eukaryotes [41] . We here used different reagents to characterize which ROS is formed due to PGD2 or STS induced apoptosis. As a commonly used and rather unspecific ROS detection reagent we used 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA). Getting positive results with this reagent, indicative of ROS formation in general (Figure 7(a)), we applied 3’- (p-aminophenyl) fluorescein (APF), which is more specific and used to detect hydroxyl radicals (HO•),
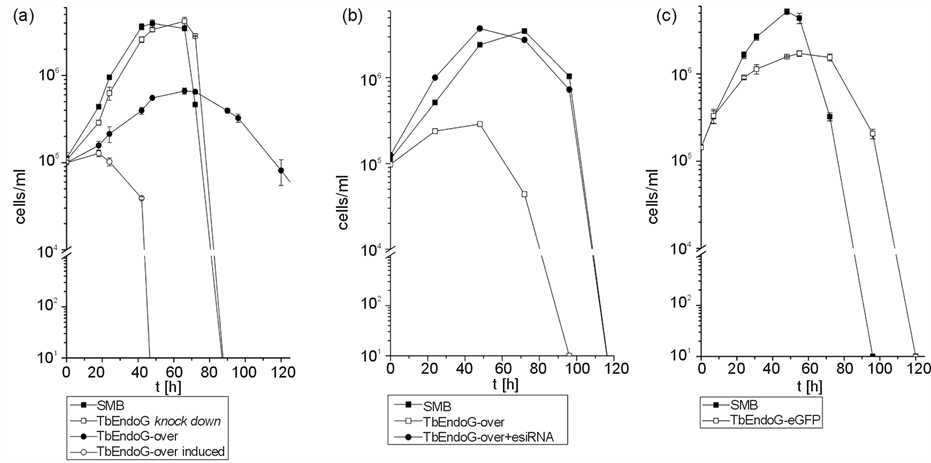
Figure 3. Growth curves of SMB and TbEndoG overexpressing trypanosomes. a) cultures of trypanosomes were induced with 10 µg/ml doxycycline at a cell density of 1 × 105 cells/ml and counted twice a day. Cell growth of TbEndoG knock down cells (£) was similar to SMB cells (¢). TbEndoG-over cells (˜) (even when not induced by doxycycline) showed a hindered growth behavior with a generation doubling time of 24 h, compared to 6 h of control SMB cells. Induction of TbEndoG overexpression with 10 µg/ml doxycycline (™) led to a fully inhibited growth. b) Addition of esiRNA to non-induced TbEndoG overexpressing cells could abrogate the growth inhibition. TbEndoG-over cells without esiRNA showed a hindered growth behavior with a generation doubling time of 24 h (£) whereas TbEndoG-over cells with 5 µl (1 µg) esiRNA (˜) grew similar to control SMB trypanosomes (¢). c) TbEndoG-eGFP cells (£) are inhibited in growth after addition of 10 µg/ml doxycycline with a generation doubling time of 11 h compared to 6 h of control SMB cells (¢).
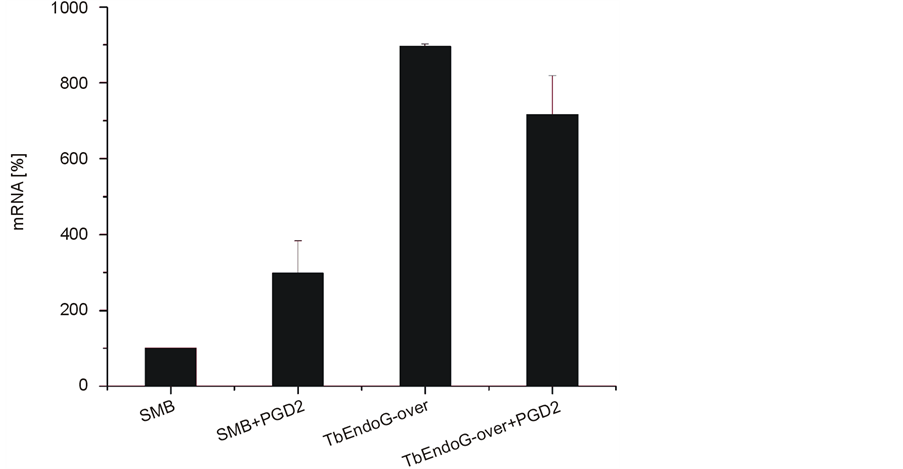
Figure 4. qRT-PCR of EndoG in SMB and TbEndoG overexpressing trypanosomes. SMB and TbEndoG overexpressing cells grown in HMI-9 culture medium were induced by adding 10 µg/ml doxycycline. Additionally, cells were incubated with 10 µM PGD2 for 24 h. After RNA-isolation 100 ng of RNA were added to each RT-PCR sample and analysed in a Roche Lightcycler 480. EndoG level in control SMB cells increased 3 fold after apoptosis induction by PGD2, but remained constant on a 7 folds increase in TbEndoG overexpressing cells.
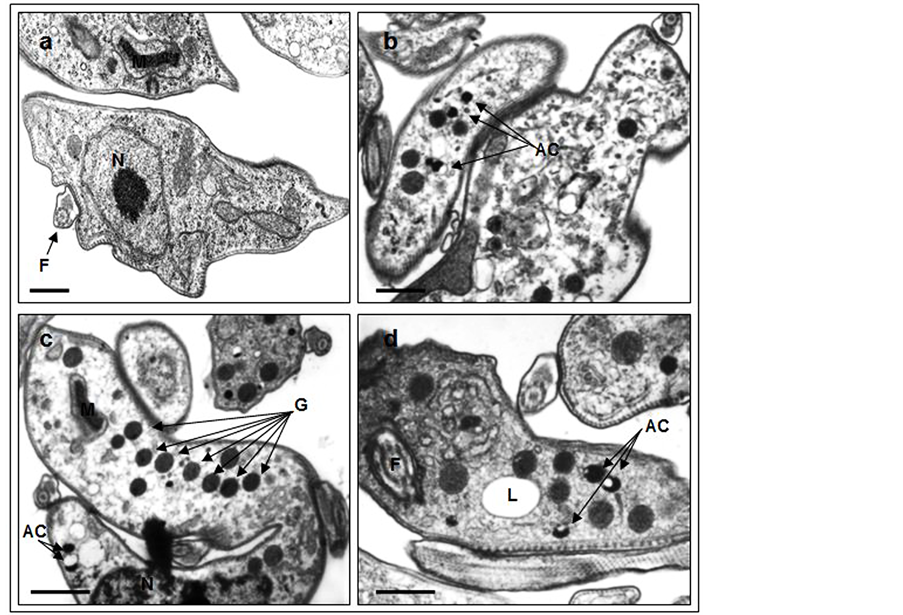
Figure 5. Transmission electron microscopic images of TbEndoG overexpressing trypanosomes. After induction of TbEndoG expression in SMB cells with 10 µg/ml doxycycline for 24 h a significant increase in glycosomes and acidocalcisomes enriched with phosphates could be observed (arrows in b-d) in contrast to control cells (a). AC: acidocalcisome; F: flagellum; G: glycosome; L: lysosome; M: mitochondrion; N: nucleus. bar: 0.5 µm.
 (a) (b)
(a) (b)
Figure 6. Scanning and transmission electron microscopic images of staurosporine treated trypanosomes. a) SMB cells treated with STS undergo a shape change in consequence of an enlarged flagellar pocket (b-d) in contrast to control cells a). bar: 2 µm; b) After induction with 10 nM STS trypanosomal flagellar pockets increase in size and volume (a-d). Vesicles and constrictions occure inside the flagellar pocket and the number of lysosomes increase. Displayed cells were incubated with 10 nM STS for 24 h before fixation. F: flagellum; FP: flagellar pocket; G: glycosome; L: lysosome; N: nucleus; V: vesicle. bar: 0.5 µm.
hypochlorite anions (–OCl) and peroxynitrite anions (ONOO–). Analysis of untreated and treated SMB cells stained with APF showed no fluorescence, thus omitting these ROS as participants in the apoptosis pathway in T. brucei. The next reagent used for ROS detection was dihydroethidium (DHE), which is rather specific for the superoxide anion (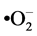 ). Compared with untreated SMB trypanosomes, DHE detected significantly increased levels of superoxide anions in PGD2 or STS treated SMB cells (Figure 7(b)), leading to the conclusion that this ROS is actively involved in the parasite’s apoptosis pathway. This is consistent with data reported for T. cruzi, where mitochondrial superoxide anions serve as a key mediator for the initiation of the apoptotic death process [42] .
). Compared with untreated SMB trypanosomes, DHE detected significantly increased levels of superoxide anions in PGD2 or STS treated SMB cells (Figure 7(b)), leading to the conclusion that this ROS is actively involved in the parasite’s apoptosis pathway. This is consistent with data reported for T. cruzi, where mitochondrial superoxide anions serve as a key mediator for the initiation of the apoptotic death process [42] .
As ROS may induce membrane ruptures, the predicted next steps in caspase-independent apoptosis are a breakdown of the inner mitochondrial membrane potential and the release of mitochondrial material into the cytosol. Amongst the latter are pro-apoptotic factors like AIF and EndoG.
Loss of mitochondrial membrane potential was confirmed by TMRE staining. While 97% of SMB control cells showed an intact mitochondrial membrane potential and were thus TMRE positive, 65% of PGD2 and 85% of STS treated cells proved to be TMRE negative after 24 h. This effect strongly increased in TbEndoG overexpressing cells, where 90% of SMB control cells showed an intact mitochondrial membrane and were thus TMRE positive, while 89% of PGD2 and 95% of STS treated cells were TMRE negative after 24 h (Figure 7(a)).
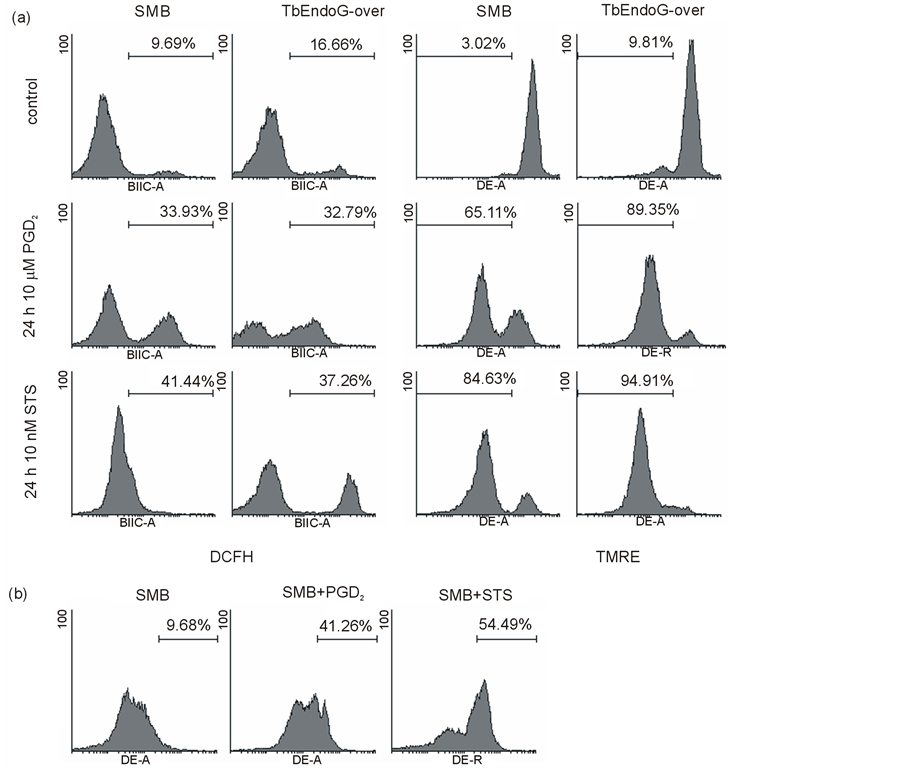
Figure 7. ROS, mitochondrial membrane potential and superoxide detection in apoptotic trypanosomes. Distribution of fluorescence in SMB and TbEndoG overexpressing cells treated with 10 µM PGD2 and 10 nM STS, respectively after 24 h. Prior to treatment with PGD2 and STS all cells were incubated 24 h with 10 µM doxycycline. a) Detection of ROS production with H2DCFH-DA. Intracellular ROS increased within the first few hours of apoptosis induction. After 24 h still 30% - 40% of treated cells are ROS positive (DCFH). Meanwhile mitochondrial membrane potential get lost (decrease of TMRE fluorescence). b) Cells treated with PGD2 or STS, respectively are positive for superoxide anions measured with dihydroethidium.
After a 24 h incubation of trypanosomes in the presence of 10 µM PGD2 or 10 nM STS, 21% or 27% of cell nuclei showed DNA fragmentation as confirmed by PI staining. Similarly, DNA fragmentation in TbEndoG overexpressing cells was 31% or 64%, respectively.
An often occurring sign of apoptosis is also a cell cycle arrest in the G1 phase. Therefore we lysed trypanosomes with 6 mM digitonin and measured the DNA content of trypanosomal nuclei. Treatment of SMB cells with PGD2 or STS caused a decrease of 21% or 31% of cells in the G1 phase, respectively. In STS but not in PGD2 treated cells an increase of 14% of cells in the G2 phase was observed after 24 h (Figure 8(a)).
Phosphatidylserine exposure on the outer membrane leaflet was detected using annexin-V staining. An increase of up to 65% annexin-V positive cells as compared to SMB control cells could be observed during apoptosis induction with PGD2 or STS (Figure 8(b)).
Expression of eGFP or TbEndoG-eGFP fusion protein, respectively, had no effect on the parasite’s growth upon induction of the expression of either protein by 10 µg/ml doxycycline. As shown in Figure 3(c), growth of TbEndoG-eGFP expressing cells was inhibited, while cells expressing only eGFP grew normally. We used TbEndoG-eGFP expressing cells in our studies to detect EndoG localization after induction of apoptosis by fluorescence microscopy. Evaluation of PGD2 or STS treated cells by fluorescence microscopy clearly showed release of TbEndoG out of the mitochondrion. It is not clear so far whether or not the EndoG trans-membrane domain is thereby cleaved. Nevertheless, an active translocation during apoptosis stimuli into the nucleus could not be observed. To avoid false positive immunofluorescence results we used a TbEndoG-eGFP fusion protein and Mitotracker Red CMXRos staining for mitochondrial localization and DAPI for nuclei localization (Figure 9). After PGD2 or STS treatment, TbEndoG-eGFP is distributed all over the cytosol and is also included in the nucleus (Figure 9(b)). Expression of eGFP was validated by Western blot analysis.

Figure 8. DNA content in apoptotic trypanosomal nuclei and annexin-V detection. a) DNA fragmentation of nuclei of SMBand TbEndoG overexpressing cells treated with 10 µM PGD2 and 10 nM STS, respectively after 24 h. Detected with propidium iodide (PI). Nuclei with less fluorescence than G1 peaks were defined as fragmentated. G2 peak possesses double amount of fluorescence (log. scale). Treated cells show DNA fragmentation between 21% and 64%. b) Phoshatidylserin exposure on the outer membrane leaflet detected with annexin-V. SMBand TbEndoG overexpressing cells were treated with 10 µM PGD2 and 10 nM STS, respectively for 24 h. In treated cells PS exposure is increased by 22% to 65% compared to control cells.
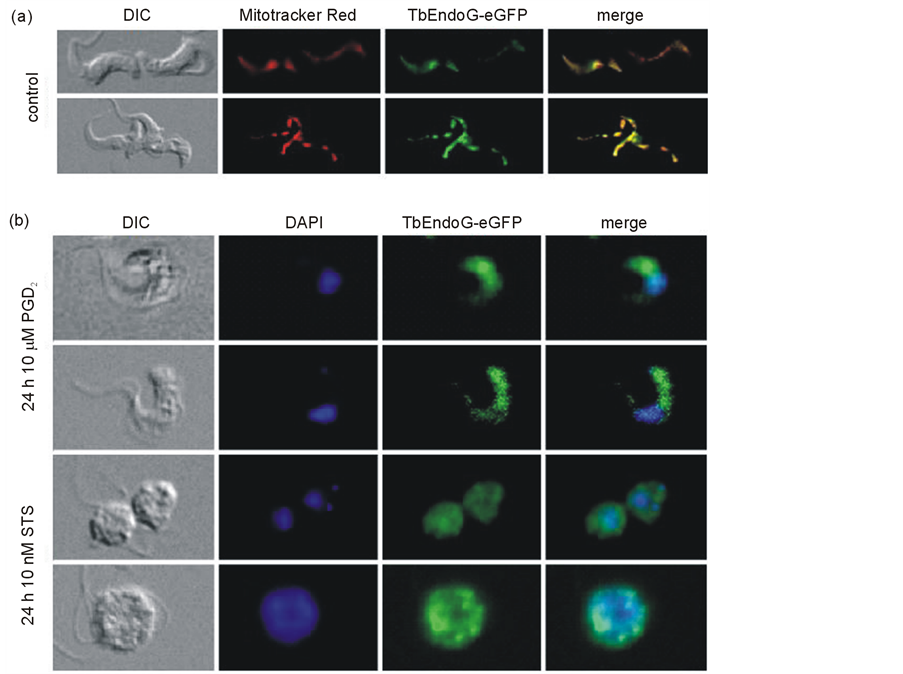
Figure 9. Fluorescence microscopic images of TbEndoG-eGFP cells. Cell cultures of TbEndoG-eGFP expressing trypanosomes were incubated with 50 nM Mitotracker Red CMXRos for 20 min at 37˚C and 5% CO2 before fixation with 3.5% paraformaldehyde to stain mitochondria. After fixation cells were incubated with DAPI to stain nuclei and kinetoplasts. Afterwards cells were coated on poly-L-lysin-coated slides and embedded with ProLong® Gold Antifade. a) TbEndoG-eGFP control cells; b) TbEndoG-eGFP cells, treated 24 h with 10 µM PGD2 (upper two rows) and 10 nM STS (lower two rows), respectively.
4. Discussion
The molecular mechanism of apoptosis in higher eukaryotes is well investigated and its necessity for the functional development and maintenance of multicellular organisms has been shown. During the last years, several orthologs of mammalian apoptotic proteins have been discovered and apoptotic processes similar to those in higher organisms were demonstrated in different single-cell organisms. However, the mechanisms and proteins involved in this pathway are still poorly described and not well understood.
Since endonuclease G has been identified as a protein involved in caspase-independent DNA fragmentation in metazoan and protozoan organisms [14] [17] [21] [36] we investigated its role during apoptosis in T. brucei induced by PGD2 or staurosporine, respectively.
The results presented in this work demonstrate that TbEndoG is an endonuclease which degrades DNA in the presence of Mg2+ or Mn2+ ions. Like other EndoGs of higher eukaryotes it localizes to the single mitochondrion of trypanosomes, probably because of an N-terminal signal sequence. However, in contrast to EndoGs from metazoa or leishmania, this mitochondrial localization sequence (mls) of TbEndoG seems considerably different, although it is recognized as a putative mls by prediction tools like MitoProtII or PrediSi. Firstly, it does not contain the classical sequence of an alternating pattern of hydrophobic and positively charged amino acids, and secondly it is about 30 amino acids longer than the respective leishmania counterpart. In addition, following the mitochondrial import, the mls is usually cleaved by a specific peptidase (like in AIF) to produce the mature form, which is inserted into the inner membrane via an N-terminal transmembrane domain. AIF release requires then the intrusion of a not yet identified protease, which cleaves AIF62 to a soluble AIF57. If this mechanism would also apply for TbEndoG release in trypanosomes, a putative nuclear targeting signal of 4 amino acids (recognized by prediction tools like PSORT) would also be cleaved off since it is part of the mls. Since data from another study indicate that the mls of TbEndoG might not been cleaved in T. brucei [36] , we favour the idea that TbEndoG is not translocated to the matrix, but to the intermembrane space of the mitochondrion. During permeabilization of the outer mitochondrial membrane in the course of apoptosis, TbEndoG may be released into the cytosol, and translocates most likely as part of a DNA-degradation complex together with other nucleases into the nucleus to cleave DNA [16] [21] .
Taken together, our results, together with those presented by other authors [36] , demonstrate that T. brucei expresses an ortholog of metazoan and protozoan EndoG that participates in the caspase-independent apoptosis pathway, which is triggered by different stimuli as shown here. In response to PGD2 or STS treatment, TbEndoG is released out of the mitochondrion into cytosol and nucleus. As shown here, T. brucei has the ability to undergo these processes in different kinds of manner. In the presence of either compound, overexpression of EndoG leads to a significant acceleration of apoptotic cell death. We thus conclude that, although this protein is not necessary for survival of trypanosomes (as reduced expression of TbEndoG by knock down had no effect on cell growth in vitro), it is involved in cell degradation processes. Our data also reveal that overexpression of EndoG leads to a significant inhibition of trypanosomal growth, indicating that EndoG is not only acting as a pro apoptotic enzyme or is only active following apoptosis induction, but is also responsible for the homeostasis of cells.
We conclude from our results that formation of intracellular ROS, especially superoxide anion formation is an early step in protozoan caspase-independent apoptosis, followed by mitochondrial membrane disruption and release of EndoG into the cytosol. In addition, phosphatidylserine exposure on the outer leaflet of plasma membrane occurs as it is the case in other organisms. In contrast to PGD2 treated cells, a shift from G1 to G2 phase occurs in STS treated cells. As a last detectable step, DNA fragmentation was detected in both cases of apoptosis induction.
Furthermore, we here demonstrate that STS is a potent apoptosis inducer in T. brucei which, compared to the established apoptosis inducer PGD2 (IC50 of 3.7 µM), is effective in very low concentrations (IC50 of 7.6 nM). However, one has to keep in mind that PGD2 binds very effectively to serum proteins, which are indispensable for axenic cultivation of the parasite [2] . We thus still consider PGD2 as a physiological apoptosis inducer for an effective cell density control especially during brain infection [22] . STS, in contrast, may act as a general inhibitor of phosphorylation, be involved in several processes of cellular regulation and thus lead to the observed differences of apoptotic progression.
EndoG is localized to the single mitochondrion of trypanosomes and is absent from the nucleus under normal growth conditions in control and SMB cells expressing TbEndoG-eGFP. Additionally, these cells were less inhibited in growth as compared to TbEndoG overexpressing cells in consequence of the used expression vector even after induction with doxycycline. Fluorescence microscopy confirmed localization in the mitochondrion as MitoTracker® Red CMXRos was used to stain mitochondria.
After PGD2 or STS treatment, TbEndoG-eGFP was released and distributed throughout the cytosol and within the nucleus. These observations are consistent with previously described effects of EndoG in other organisms. Under no circumstances, however, could we observe that EndoG was exclusively localized to the nucleus. One reason could be that overexpression of TbEndoG-eGFP caused a too high level of EndoG and thus excess quantities remained in the cytosol. Another reason could be that the mitochondrial localization sequence of TbEndoG is cleaved after uptake into the mitochondrion, thus leading to the concomitant loss of the nuclear translocation signal.
5. Conclusion
In conclusion, we show that trypanosomes possess an apoptotic mechanism to react on different stimuli in several ways. In general, apoptosis occurs as described with ROS formation at the beginning. As shown using flow cytometry, superoxide anion is the main ROS involved in apoptosis induction in T. brucei. Its increased formation in the mitochondrion leads probably to the rupture of the mitochondrial membrane and thus release of EndoG into the cytosol. Here, EndoG, together with other nucleases seems to form a DNA-degradation complex [16] [21] , which fragmentizes the parasite’s DNA in the last step of trypanosomal apoptosis. As trypanosomes belong to one of the most ancient diverging branches of the eukaryotic phytogenic tree and are amongst the first eukaryotes with a mitochondrion, our results offer an insight in apoptosis development and how complex these simple organisms can react on different stimuli. Moreover, as our results evinced, STS might be an interesting inducer of apoptotic cell death in T. brucei for further investigations on anti-parasitic drug development.
References
- Debrabant, A., Lee, N., Bertholet, S., Duncan, R. and Nakhasi, H.L. (2003) Programmed Cell Death in Trypanosomatids and Other Unicellular Organisms. International Journal for Parasitology, 33, 257-267. http://dx.doi.org/10.1016/S0020-7519(03)00008-0
- Figarella, K., Rawer, M., Uzcategui, N.L., Kubata, B.K., Lauber, K., et al. (2005) Prostaglandin D2 Induces Programmed Cell Death in Trypanosoma brucei Bloodstream Form. Cell Death & Differentiation, 12, 335-346. http://dx.doi.org/10.1038/sj.cdd.4401564
- Figarella, K., Uzcategui, N.L., Beck, A., Schoenfeld, C., Kubata, B.K., et al. (2006) Prostaglandin-Induced Programmed Cell Death in Trypanosoma brucei Involves Oxidative Stress. Cell Death & Differentiation, 13, 1802-1814. http://dx.doi.org/10.1038/sj.cdd.4401862
- Lee, N., Bertholet, S., Debrabant, A., Muller, J., Duncan, R., et al. (2002) Programmed Cell Death in the Unicellular Protozoan Parasite Leishmania. Cell Death & Differentiation, 9, 53-64. http://dx.doi.org/10.1038/sj.cdd.4400952
- Desjeux, P. (2004) Leishmaniasis: Current Situation and New Perspectives. Comparative Immunology Microbiology and Infectious Diseases, 27, 305-318. http://dx.doi.org/10.1016/j.cimid.2004.03.004
- Kubata, B.K., Duszenko, M., Kabututu, Z., Rawer, M., Szallies, A., et al. (2000) Identification of a Novel Prostaglandin f(2alpha) Synthase in Trypanosoma brucei. Journal of Experimental Medicine, 192, 1327-1338. http://dx.doi.org/10.1084/jem.192.9.1327
- Madeo, F., Frohlich, E., Ligr, M., Grey, M., Sigrist, S.J., et al. (1999) Oxygen Stress: A Regulator of Apoptosis in Yeast. Journal of Cell Biology, 145, 757-767. http://dx.doi.org/10.1083/jcb.145.4.757
- Frohlich, K.U. and Madeo, F. (2000) Apoptosis in Yeast—A Monocellular Organism Exhibits Altruistic Behaviour. FEBS Letters, 473, 6-9. http://dx.doi.org/10.1016/S0014-5793(00)01474-5
- Christensen, S.T., Kemp, K., Quie, H. and Rasmussen, L. (1996) Cell Death, Survival and Proliferation in Tetrahymena thermophila. Effects of Insulin, Sodium Nitroprusside, 8-Bromo Cyclic GMP, NG-Methyl-L-arginine and Methylene Blue. Cell Biology International, 20, 653-666. http://dx.doi.org/10.1006/cbir.1996.0087
- Arnoult, D., Tatischeff, I., Estaquier, J., Girard, M., Sureau, F., et al. (2001) On the Evolutionary Conservation of the Cell Death Pathway: Mitochondrial Release of an Apoptosis-Inducing Factor during Dictyostelium discoideum Cell Death. Molecular Biology of the Cell, 12, 3016-3030. http://dx.doi.org/10.1091/mbc.12.10.3016
- Engelberg-Kulka, H., Amitai, S., Kolodkin-Gal, I. and Hazan, R. (2006) Bacterial Programmed Cell Death and Multi-cellular Behavior in Bacteria. PLoS Genet, 2, Article ID: e135. http://dx.doi.org/10.1371/journal.pgen.0020135
- Bayles, K.W. (2003) Are the Molecular Strategies That Control Apoptosis Conserved in Bacteria? Trends in Microbiology, 11, 306-311. http://dx.doi.org/10.1016/S0966-842X(03)00144-6
- Welburn, S.C., Macleod, E., Figarella, K. and Duzensko, M. (2006) Programmed Cell Death in African Trypanosomes. Parasitology, 132, S7-S18. http://dx.doi.org/10.1017/S0031182006000825
- Li, L.Y., Luo, X. and Wang, X. (2001) Endonuclease G Is an Apoptotic DNase When Released from Mitochondria. Nature, 412, 95-99. http://dx.doi.org/10.1038/35083620
- Ohsato, T., Ishihara, N., Muta, T., Umeda, S., Ikeda, S., et al. (2002) Mammalian Mitochondrial Endonuclease G. Digestion of R-Loops and Localization in Intermembrane Space. European Journal of Biochemistry, 269, 5765-5770. http://dx.doi.org/10.1046/j.1432-1033.2002.03238.x
- BoseDasgupta, S., Das, B.B., Sengupta, S., Ganguly, A., Roy, A., Dey, S., Tripathi, G., Dinda, B. and Majumder, H.K. (2008) The Caspase-Independent Algorithm of Programmed Cell Death in Leishmania Induced by Baicalein: The Role of LdEndoG, LdFEN-1 and LdTatD as a DNA “Degradesome”. Cell Death and Differentiation, 15, 1629-1640. http://dx.doi.org/10.1038/cdd.2008.85
- Rico, E., Alzate, J.F., Arias, A.A., Moreno, D., Clos, J., Gagod, F., Morenoe, I., Domíngueze, M. and Jiménez-Ruiz, A. (2009) Leishmania infantum Expresses a Mitochondrial Nuclease Homologous to EndoG that Migrates to the Nucleus in Response to an Apoptotic Stimulus. Molecular and Biochemical Parasitology, 163, 28-38. http://dx.doi.org/10.1016/j.molbiopara.2008.09.007
- Cote, J. and Ruiz-Carrillo, A. (1993) Primers for Mitochondrial DNA Replication Generated by Endonuclease G. Science, 261, 765-769. http://dx.doi.org/10.1126/science.7688144
- Low, R.L. (2003) Mitochondrial Endonuclease G Function in Apoptosis and mtDNA Metabolism: A Historical Perspective. Mitochondrion, 2, 225-236. http://dx.doi.org/10.1016/S1567-7249(02)00104-6
- Schafer, P., Scholz, S.R., Gimadutdinow, O., Cymerman, I.A., Bujnicki, J.M., Ruiz-Carrillo, A., Pingoud, A. and Meiss, G. (2004) Structural and Functional Characterization of Mitochondrial EndoG, a Sugar Non-Specific Nuclease Which Plays an Important Role during Apoptosis. Journal of Molecular Biology, 338, 217-228. http://dx.doi.org/10.1016/j.jmb.2004.02.069
- Parrish, J., Li, L., Klotz, K., Ledwich, D., Wang, X.D. and Xue, D. (2001) Mitochondrial Endonuclease G Is Important for Apoptosis in C. elegans. Nature, 412, 90-94. http://dx.doi.org/10.1038/35083608
- Wolburg, H., Mogk, S., Acker, S., Frey, C., Meinert, M., Sch?nfeld, C., Lazarus, M., Urade, Y., Kubata, B.K. and Duszenko, M. (2012) Late Stage Infection in Sleeping Sickness. PLoS ONE, 7, Article ID: e34304. http://dx.doi.org/10.1371/journal.pone.0034304�
- Arnoult, D., Akarid, K., Grodet, A., Petit, P.X., Estaquier, J. and Ameisen, J.C. (2002) On the Evolution of Programmed Cell Death: Apoptosis of the Unicellular Eukaryote Leishmana Major Involves Cysteine Proteinase Activation and Mitochondrion Permeabilization. Cell Death and Differentiation, 9, 65-81. http://dx.doi.org/10.1038/sj.cdd.4400951
- Bruno, S., Ardelt, B., Skierski, J.S., Traganos, F. and Darzynkiewicz, Z. (1992) Different Effects of Staurosporine, an Inhibitor of Protein Kinases, on the Cell Cycle and Chromatin Structure of Normal and Leukemic Lymphocytes. Cancer Research, 52, 470-473.
- Belmokhtar, C.A., Torriglia, A., Counis, M.F., Courtois, Y., Jacquemin-Sablon, A. and Ségal-Bendirdjian, E. (2000) Nuclear Translocation of a Leukocyte Elastase Inhibitor/Elastase Complex during Staurosporine-Induced Apoptosis: Role in the Generation of Nuclear L-DNase II Activity. Experimental Cell Research, 254, 99-109. http://dx.doi.org/10.1006/excr.1999.4737
- Belmokhtar, C.A., Hillion, J. and Segal-Bendirdjian, E. (2001) Staurosporine Induces Apoptosis through both Caspase-Dependent and Caspase-Independent Mechanisms. Oncogene, 20, 3354-3362. http://dx.doi.org/10.1038/sj.onc.1204436
- Johansson, A.C., Steen, H., Ollinger, K. and Roberg, K. (2003) Cathepsin D Mediates Cytochrome c Release and Caspase Activation in Human Fibroblast Apoptosis Induced by Staurosporine. Cell Death and Differentiation, 10, 1253-1259. http://dx.doi.org/10.1038/sj.cdd.4401290
- Pimentel-Elardo, S.M., Kozytska, S., Bugni, T.S., Ireland, C.M., Moll, H. and Hentschel, U. (2010) Anti-Parasitic Compounds from Streptomyces sp. Strains Isolated from Mediterranean Sponges. Marine Drugs, 8, 373-380. http://dx.doi.org/10.3390/md8020373
- Gale Jr., M., Carter, V. and Parsons, M. (1994) Cell Cycle-Specific Induction of an 89 kDa Serine/Threonine Protein Kinase Activity in Trypanosoma Brucei. Journal of Cell Science, 107, 1825-1832.
- Malaquias, A.T. and Oliveira, M.M. (1999) Phospholipid Signalling Pathways in Trypanosoma cruzi Growth Control. Acta Tropica, 73, 93-108. http://dx.doi.org/10.1016/S0001-706X(99)00016-9
- Duszenko, M., Figarella, K., Macleod, E.T. and Welburn, S.C. (2006) Death of a Trypanosome: A Selfish Altruism. Trends in Parasitology, 22, 536-542. http://dx.doi.org/10.1016/j.pt.2006.08.010
- Debrabant, A. and Nakhasi, H. (2003) Programmed Cell Death in Trypanosomatids: Is It an Altruistic Mechanism for Survival of the Fittest? Kinetoplastid Biology and Disease, 2, 7. http://dx.doi.org/10.1186/1475-9292-2-7
- Wirtz, E., Leal, S., Ochatt, C. and Cross, G.A. (1999) A Tightly Regulated Inducible Expression System for Conditional Gene Knock-Outs and Dominant-Negative Genetics in Trypanosoma brucei. Molecular and Biochemical Parasitology, 99, 89-101.http://dx.doi.org/10.1016/S0166-6851(99)00002-X
- Glauert, A.M., Butterworth, A.E., Sturrock, R.F. and Houba, V. (1978) The Mechansim of Antibody-Dependent, Eosinophil-Mediated Damage to Schistosomula of Schistosoma Mansoni in Vitro: A Study by Phase-Contrast and Electron Microscopy. Journal of Cell Science, 34, 173-192.
- Widlak, P., Li, L.Y., Wang, X. and Garrard, W.T. (2001) Action of Recombinant Human Apoptotic Endonuclease G on Naked DNA and Chromatin Substrates: Cooperation with Exonuclease and DNase I. Journal of Biological Chemistry, 276, 48404-48409.
- Gannavaram, S., Vedvyas, C. and Debrabant, A. (2008) Conservation of the Pro-Apoptotic Nuclease Activity of Endonuclease G in Unicellular Trypanosomatid Parasites. Journal of Cell Science, 121, 99-109. http://dx.doi.org/10.1242/jcs.014050
- Steensma, D.P., Timm, M. and Witzig, T.E. (2003) Flow Cytometric Methods for Detection and Quantification of Apoptosis. Methods in Molecular Medicine, 85, 323-332.
- Elmore, S. (2007) Apoptosis: A Review of Programmed Cell Death. Toxicologic Pathology, 35, 495-516. http://dx.doi.org/10.1080/01926230701320337
- Riccardi, C. and Nicoletti, I. (2006) Analysis of Apoptosis by Propidium Iodide Staining and Flow Cytometry. Nature Protocols, 1, 1458-1461. http://dx.doi.org/10.1038/nprot.2006.238
- Jimenez-Ruiz, A., Alzate, J.F., Macleod, E.T., Luder, C.G., Fasel, N. and Hurd, H. (2010) Apoptotic Markers in Protozoan Parasites. Parasites & Vectors, 3, 104.
- Kroemer, G. and Martin, S.J. (2005) Caspase-Independent Cell Death. Nature Medicine, 11, 725-730. http://dx.doi.org/10.1038/nm1263
- Piacenza, L., Irigoin, F., Alvarez, M.N., Peluffo, G., Taylor, M.C., Kelly, J.M., Wilkinson, S.R. and Radi, R. (2007) Mitochondrial Superoxide Radicals Mediate Programmed Cell Death in Trypanosoma cruzi: Cytoprotective Action of Mitochondrial Iron Superoxide Dismutase Overexpression. Biochemical Journal, 403, 323-334. http://dx.doi.org/10.1042/BJ20061281
Abbreviations
CAD, caspase-activated DNase;
eGFP, enhanced green fluorescent protein;
EndoG, mitochondrial endonuclease G;
IPTG, isopropyl-beta-d-thiogalactopyranoside;
rTbEndoG, Trypanosoma brucei recombinant EndoG;
PGD2, prostaglandin D2;
SMB, single marker blood-stream form;
STS, staurosporine
NOTES

*Corresponding author.

