International Journal of Otolaryngology and Head & Neck Surgery
Vol.04 No.03(2015), Article ID:55783,9 pages
10.4236/ijohns.2015.43031
Prognostic and Predictive Protein Biomarkers in Laryngeal Squamous Cell Carcinoma―A Systematic Review
Matthew M. Kwok, Paul Goodyear
Department of Otolaryngology/Head and Neck Surgery, Western Health, Melbourne, Australia
Email: mattmkkwok@gmail.com
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 21 March 2015; accepted 10 April 2015; published 17 April 2015
ABSTRACT
Background: Despite recent advances in clinical management of laryngeal squamous cell carcinoma (LSCC), the overall 5-year survival continues to be poor. Consequently, biomarkers of treat- ment response will need to be identified. Proteomic strategies are one way to attempt to identify such biomarkers. Methods: The Medline, Embase and Cochrane Library databases were systematically searched until 1st March 2014 using the terms “larynx”, “squamous cell carcinoma”, “proteomic”, and “biomarker”. Articles which met inclusion criteria were assessed for the type of biomarker investigated, the proteomic technique used, and whether any validation had been performed. Results: Six studies identified biomarkers, including UCRP, ceramides, uPA, MT1-MMP, stratifin, transferrin, albumin, S100 calcium-binding protein A9, stathmin, enolase, PLAU, IGFBP7, MMP14, THBS1, and transthyretin. Transferrin was the only biomarker to appear in more than one study. Conclusions: Our review identified several potential biomarkers of outcome in LSCC. Well designed studies will need to further validate their use in the future.
Keywords:
Laryngeal, Squamous Cell Carcinoma, Cancer, Proteomics, Biomarker

1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is diagnosed in over 650,000 individuals annually worldwide and remains to be an important global health issue [1] . Laryngeal squamous cell carcinoma (LSCC) accounts for more than 150,000 of these cases, with an annual global mortality of approximately 83,000 individuals [1] . Although extensive research had been undertaken on LSCC in the past decade in an attempt to identify biomarkers of treatment response or outcome, there continued to be significant morbidity and mortality associated with this condition, to the extent that the overall 5-year survival had reduced between 1975 and 2010 in the USA [2] . The prognosis is especially poor for patients with metastatic disease, with a 5-year survival of less than 50% [3] . Consequently, a better understanding of biomarkers of outcome associated with LSCC may have clinical use in improved treatment stratification and prognostication and may even inform the development of future novel targeted therapies.
In recent years, biomarker development in other tumor types has resulted in treatment targeting and improved outcomes for biomarker derived subsets of patients. A notable example is the use of detection of the HER-2 receptor in breast cancer biopsy tissue [4] .
There are three main clinical applications for biomarkers in cancer―in the diagnosis and characterization of tumors, as well as the potential development of novel targeted therapy against certain cancers.
As a diagnostic tool, molecular biomarkers may aid in the diagnosis of occult metastasis as well as the early detection of local and regional spread [5] [6] . Biomarkers, such as desmoglein 3 and Tissue-Specific Mir-205, have been shown to be associated with metastatic head and neck cancer and hence have the potential to be diagnostic biomarkers for this condition [7] [8] .
In the characterization of tumors, they are used to better determine prognosis and treatment selection [5] . Epidermal growth factor receptor (EGFR) and p16 are well characterized biomarkers which are differentially expressed in a subset of LSCC [9] [10] , and may play a role in the carcinogenesis of LSCC [11] [12] . Various targeted therapies against EGFR positive HNSCC have been developed, including anti-EGFR antibodies such as cetuximab, which improve overall survival without adverse effects on quality of life [13] [14] .
Recent advances in proteomic technology and techniques have resulted in the discovery of new biomarkers in HNSCC [15] , using various proteomic techniques. Protein separation techniques, such as 2-dimensional (2D) differential in-gel electrophoresis, have allowed large numbers of proteins to be sampled in a reproducible manner [16] [17] . Moreover, the differential expression of protein biomarkers identified using proteomic techniques such as matrix-assisted laser desorption/ionization (MALDI) time of flight (ToF) mass spectrometry (MS) [18] , surface enhanced laser desorption/ionization (SELDI) ToF MS [19] , laser capture [20] , liquid chromatography mass spectrometry (LC-MS) [21] , as well as newer techniques such as isobaric tag for relative and absolute quantitation (iTRAQ) [22] has led to a better understanding of the carcinogenesis of certain types of cancers [23] [24] . Various types of tissue samples may be used for proteomic analysis, which include fresh frozen, formalin-fixed paraffin embedded, and cell lines [25] [26] . Slight differences may exist among results obtained from different types of tissue samples [26] . Although previous reviews have evaluated studies utilizing proteomic techniques to characterize salivary cancer biomarkers [27] [28] and colorectal cancer biomarkers [29] , no study has assessed the use of this technique in the identification of LSCC biomarkers.
This study aims to review the current literature to evaluate biomarkers in LSCC identified using proteomic techniques. In particular, this review will assess the type of biomarker and its potential use in LSCC, the proteomic technique used, and whether or not any validation of the biomarker has been performed.
2. Methods
A literature search for relevant articles was performed electronically by two independent reviewers―MK, a surgical HMO and PG, an otolaryngologist/head and neck surgeon. The Medline, Embase and Cochrane Library databases were searched using the following terms in combination: [laryngeal or larynx or glottis or glottis or supraglottic or supraglottis or subglottic or subglottis (all fields)] and [“squamous cell carcinoma” or SCC or cancer (all fields)] and [biomarker (all fields)] and [proteomic or proteome or electrophoresis or Maldi or “laser desorption ionization” or spectrometry (all fields)] in Medline; and ((((“laryngeal or larynx or glottis or glottis or supraglottis or supraglottic or subglottis or subglottic”) and “squamous cell carcinoma” or SCC or cancer) and biomarker) and (proteomic or proteome or electrophoresis or Maldi or “laser desorption ionization” or spectrometry)). mp. [mp = title, abstract, subject headings, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword] in Embase.
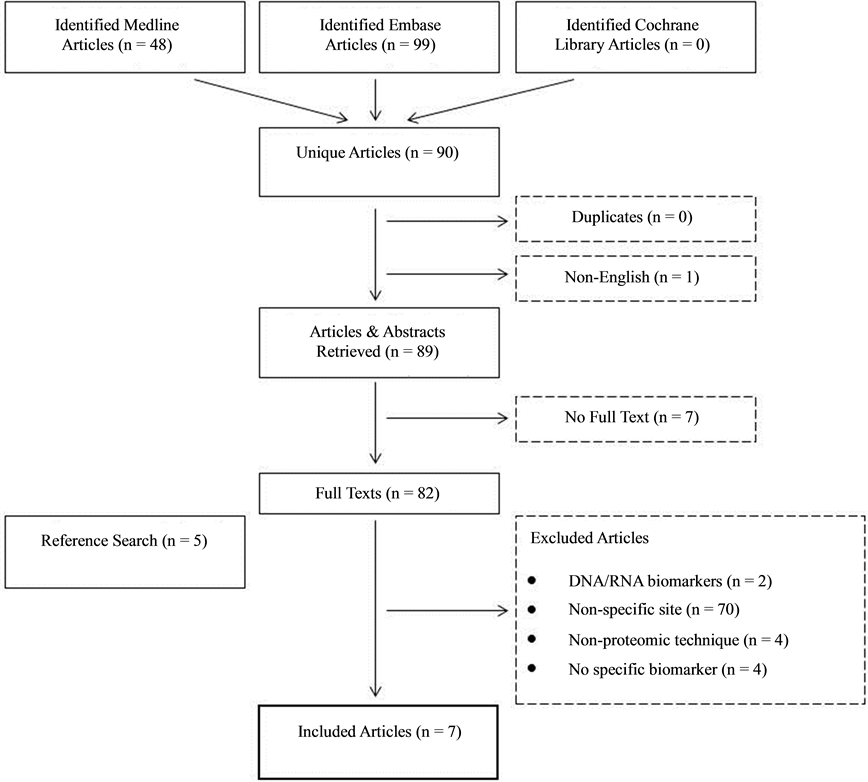
The search strategy is outlined in Figure 1 and follows the PRISMA statement. All articles published from 1950 to 1st March 2014 in the above databases were searched for relevant articles. Full text articles retrieved
Figure 1. Systematic search strategy for studies included in this review.
were reviewed individually for eligibility into this study. Additionally, a manual search was performed on all reference lists of included articles.
Inclusion criteria were defined as articles which identify using a proteomic approach and/or validate biomarkers in laryngeal squamous cell carcinoma using immunohistochemical techniques on larger samples. Non-English articles, case reports, opinions, reviews and news articles were excluded from this study. Articles assessing DNA/ RNA and those which did not identify specific biomarkers were also excluded.
Data retrieved from all unique articles identified include the date of publication, type of biomarker identified and its potential use in laryngeal SCC, proteomic technique used, source and tissue preparation, number of diseased and healthy subjects, and whether or not any validation of the biomarker had been performed. Due to the limited data available in the current literature, no statistical analysis could be performed.
3. Results
90 unique articles were identified in our search, with 7 meeting inclusion criteria for review in this study. The majority of excluded articles (n = 70) assessed head and neck SCC biomarkers on a non-specific anatomical site. Table 1 outlines a summary of information retrieved from included studies.
Studies were published between 1996 and 2012, with 5 studies published in the last 10 years [23] [30] - [33] . A variety of proteomic techniques were identified, including liquid chromatography, mass spectrometry, electrospray ionization, matrix-assisted laser desorption/ionization, sodium dodecyl sulfate polyacrylamide gel electrophoresis, enzyme-linked immunosorbent assay and 2-dimensional electrophoresis.
Table 1. Summary of information obtained from included studies.
LC: Liquid chromatography; MS: Mass spectrometry; ESI: Electrospray ionization; MALDI: Matrix-assisted laser desorption/ionization; TOF: Time of flight; SDS-PAGE: Sodium dodecyl sulfate polyacrylamide gel electrophoresis; ELISA: Enzyme-linked immunosorbent assay; 2D-DIGE: 2-di- mensional difference gel electrophoresis; 2DE: 2-dimensional electrophoresis.
The number of samples in each study ranged between two to 70, 5 of the 6 studies having less than 50 samples. Laryngeal SCC samples were obtained from fresh tissue in four studies, saliva in one study and from cell lines in two studies. One study [30] validated biomarkers identified in oral cavity SCC using laryngeal SCC samples.
A number of laryngeal SCC biomarkers were identified in this review. These include ubiquitin cross-reactive protein (UCRP), C16-, C24-, C24:1-ceramides, urokinase plasminogen activator (uPA), membrane associated type 1 matrix metalloproteinase (MT1-MMP), stratifin, transferrin, albumin, tumor-associated calcium signal transducer, melanoma-associated antigen D3 (MAGE D3), S100 calcium-binding protein A9, p21-ARC, stathmin, enolase, plasminogen activator, urokinase (PLAU), insulin-like growth factor-binding protein 7 (IGFBP7), matrix metalloproteinase 14 (MMP14), Thrombospondin 1 (THBS1), beta fibrin, immunoglobulin heavy chain constant region gamma, cofilin, and transthyretin. A summary of the differential expression of laryngeal SCC biomarkers identified in this review are presented in Table 2. Transferrin was the only biomarker identified using proteomic techniques in more than one study. Only one study validated their identified biomarker using in a separate patient population [23] [31] .
4. Discussion
LSCC remains to be associated with significant morbidity and mortality despite extensive research recently, being one of only two cancer types to have a reduction in 5 year survival over the last decades [1] - [3] . The lack of specific molecular biomarkers in the management of LSCC may contribute to these statistics [23] . A better understanding of biomarkers may therefore provide new management options in the diagnosis, characterization, and targeted therapy for LSCC [5] . Recent developments in biomarker research include p16 and EGFR, which are implicated in the carcinogenesis and targeted therapy for LSCC, respectively [9] - [14] . A review of LSCC biomarkers in 2004 by Almadori et al. [5] suggested the possible association of protein biomarkers S100A2 calcium binding protein and galectin-3 with the molecular characterization of LSCC.
Table 2. Differential expression of biomarkers identified.
With the use of proteomic techniques such as 2DIGE and MALDI-TOF MS, it is now possible to sample large amount of proteins in the identification of potential cancer biomarkers [16] - [19] . This may aid in the process of identifying and validating biomarkers for LSCC. Although previous reviews had been conducted regarding the proteomic analysis of biomarkers associated with colorectal and endometrial cancer [29] [34] , this is the first systematic review evaluating the use of proteomic techniques in the analysis of LSCC biomarkers.
This review has identified 25 proteins reported as differentially expressed by 7 studies [23] [30] - [33] [35] [36] . 2 of these proteins―S100 calcium binding protein A9 and transferrin were reported as differentially expressed by more than one study, with transferrin [23] [31] reported to be up-regulated in 2 studies, while the differential expression of S100 calcium binding protein A9 was inconsistently reported in 2 studies [23] [31] . A selection of identified proteins will be discussed below.
The S100 family is a subtype of calcium binding proteins consisting of at least 25 different proteins [37] . There is current evidence to suggest the role of S100 proteins in cancer cell differentiation [38] , cell proliferation [39] [40] , cell apoptosis [41] [42] and tumor metastasis [43] [44] . Members of the S100 family have been implicated in various types of cancers. For example, non-small cell lung cancer (NSCLC) had been associated with over-expression of S100A2, S100A8 and S100A9 [45] - [47] , while S100A2 and S100A11 may be a prognostic marker for post-operative patients with pancreatic cancer [48] [49] . In HNSCC, patients with S100A2 positive LSCC had been shown to have better survival compared to S100A2 negative tumors [50] . S100A7 expression may also be implicated in the prognosis of HNSCC tumors [51] . Although serum S100 proteins may have potential implications in various types of cancers, there are still limited clinical applications for S100 proteins, with the only independent prognostic marker being serum S100B in melanoma [52] [53] . The differential reporting of S100 calcium binding protein by Dowling et al. and Sewell et al. as identified in this review may be secondary to the different sample sources (saliva and tumor tissue, respectively) and the use of control samples from the same patient by Dowling et al. [23] [31] .
A number of biomarkers identified in this review may not have significance in the carcinogenesis of LSCC due to their non-specific nature in multiple disease processes, notably albumin and transferrin, which may be differentially expressed due to various physiological or pathological processes [54] [55] . Transthyretin has been shown to be a potential biomarker in pancreatic cancer [56] [57] , but its association with multiple inflammatory disease processes may reduce its usefulness as a cancer biomarker [58] . Similarly, the use of fibrin and cofilin as LSCC biomarkers is limited by its non-specificity even though it is involved with various carcinogenic pro- cesses [59] [60] .
Although Chi et al. identified the association of UCRP with LSCC, a protein previously shown to change tumor sensitivity to chemotherapy [61] [62] , the lack of validation using proteomic techniques in LSCC samples highlights the need for further classification of this biomarker [30] .
Stathmin over-expression is found in many types of cancers [63] , and is involved in processes such as the regulation of cell migration [64] [65] , which may have implications in the pathogenesis of LSCC. Likewise, IGFBP7 is differentially expressed in various cancers as a tumour suppressor protein [66] [67] . However, there is still insufficient evidence currently to demonstrate any significant associations between these proteins and LSCC.
Two biomarkers, uPA and MT1-MMP, were identified more than 10 years ago [32] [36] . uPA is a type of protease responsible for both proteolysis and fibrinolysis, processes which are required for tumor growth and metastasis [68] . Its expression is associated with various cancer types such as breast and colorectal tumors [69] [70] . It is the only biomarker identified in this review which had used a sample size of greater than 20. Moreover, recent studies have validated the over-expression of uPA in separate LSCC populations which suggests its potential as a biomarker in LSCC [71] [72] . MT1-MMP was also a novel biomarker and potential therapeutic target identified in the 1990s, but there had been limited studies regarding its use in LSCC since, with research focusing on other MMP subtypes such as MMP-2 and MMP-9 [73] [74] .
Ceramides, a well-studied group of molecules, are involved in apoptotic cellular pathways [75] . They have been shown to assist in the induction of cell death by chemotherapeutic agents both in vivo and in situ [76] [77] . Karahatay et al. reported a lower level of C18-ceramide in patients with LSCC as well as an inverse relationship between ceramide levels and the risk of nodal metastasis [32] . Ceramides may therefore be an important prognostic marker in LSCC.
The small number of studies identified by this review limits the use of quantitative methods to analyse current LSCC biomarkers discovered by proteomic techniques, and may be a source of bias in this review. Although 25 different proteins were identified, only two were reported by more than one study. This highlights some of the pitfalls of using proteomic techniques in cancer biomarker research. Firstly, identified biomarkers require validation in a larger population with reproducible results [78] . Only one study validated the identified biomarker in a separate population and the vast majority of studies had sample sizes of less than 20. Secondly, using different proteomic techniques and technologies as well as different tissue preparations may produce varying results. There was great variability in the type of tissue analyzed and the proteomic technique used between studies. There was no standardized method of identifying LSCC biomarkers using proteomic techniques across the seven studies identified in this review. This highlights the need for further research in evaluating the molecular mechanisms for LSCC biomarkers by proteomic techniques, with the aim of developing novel diagnostic and therapeutic options for this condition.
5. Conclusion
Despite advances in biomarker research, LSCC remains to be associated with significant morbidity and mortality. This systematic review assessing LSCC biomarkers identified using proteomic techniques has found various differentially expressed protein biomarkers associated with LSCC, with no specific marker of clinical significance. Future studies may aim to further characterize these biomarkers to better understand their mechanism of action in LSCC, and validate their use in a clinical setting.
Acknowledgements
The authors would like to acknowledge S. Chan and T. Jones for their contribution towards this manuscript.
References
- Ferlay, J.S.I., Ervik, M., Dikshit, R., et al. (2013) Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11; GLOBOCAN 2012 v1.0 2013. International Agency for Research on Cancer, Lyon. http://globocan.iarc.fr
- Siegel, R., Ma, J., Zou, Z. and Jemal, A. (2014) Cancer Statistics, 2014. CA: A Cancer Journal for Clinicians, 64, 9-29. http://dx.doi.org/10.3322/caac.21208
- Shah, J.P., Karnell, L.H., Hoffman, H.T., et al. (1997) Patterns of Care for Cancer of the Larynx in the United States. Archives of Otolaryngology―Head & Neck Surgery, 123, 475-483. http://dx.doi.org/10.1001/archotol.1997.01900050021002
- Mitri, Z., Constantine, T. and O’Regan, R. (2012) The HER2 Receptor in Breast Cancer: Pathophysiology, Clinical Use, and New Advances in Therapy. Chemotherapy Research and Practice, 2012, Article ID: 743193. http://dx.doi.org/10.1155/2012/743193
- Almadori, G., Bussu, F., Cadoni, G., Galli, J., Paludetti, G. and Maurizi, M. (2005) Molecular Markers in Laryngeal Squamous Cell Carcinoma: Towards an Integrated Clinicobiological Approach. European Journal of Cancer, 41, 683- 693.
- Almadori, G., Bussu, F. and Paludettii, G. (2006) Predictive Factors of Neck Metastases in Laryngeal Squamous Cell Carcinoma. Towards an Integrated Clinico-Molecular Classification. Acta Otorhinolaryngologica Italica, 26, 326-334.
- Fletcher, A.M., Heaford, A.C. and Trask, D.K. (2008) Detection of Metastatic Head and Neck Squamous Cell Carcinoma Using the Relative Expression of Tissue-Specific Mir-205. Translational Oncology, 1, 202-208. http://dx.doi.org/10.1593/tlo.08163
- Patel, V., Martin, D., Malhotra, R., et al. (2013) DSG3 as a Biomarker for the Ultrasensitive Detection of Occult Lymph Node Metastasis in Oral Cancer Using Nanostructured Immunoarrays. Oral Oncology, 49, 93-101. http://dx.doi.org/10.1016/j.oraloncology.2012.08.001
- Grandis, J.R. and Tweardy, D.J. (1993) Elevated Levels of Transforming Growth Factor Alpha and Epidermal Growth Factor Receptor Messenger RNA Are Early Markers of Carcinogenesis in Head and Neck Cancer. Cancer Research, 53, 3579-3584.
- Reed, A.L., Califano, J., Cairns, P., et al. (1996) High Frequency of p16 (CDKN2/MTS-1/INK4A) Inactivation in Head and Neck Squamous Cell Carcinoma. Cancer Research, 56, 3630-3633.
- Combes, J.D. and Franceschi, S. (2014) Role of Human Papillomavirus in Non-Oropharyngeal Head and Neck Cancers. Oral Oncology, 50, 370-379. http://dx.doi.org/10.1016/j.oraloncology.2013.11.004
- Gheit, T., Abedi-Ardekani, B., Carreira, C., Missad, C.G., Tommasino, M. and Torrente, M.C. (2014) Comprehensive Analysis of HPV Expression in Laryngeal Squamous Cell Carcinoma. Journal of Medical Virology, 86, 642-646. http://dx.doi.org/10.1002/jmv.23866
- Bonner, J.A., Harari, P.M., Giralt, J., Azarnia, N., Shin, D.M., Cohen, R.B., et al. (2006) Radiotherapy plus Cetuximab for Squamous-Cell Carcinoma of the Head and Neck. The New England Journal of Medicine, 354, 567-578. http://dx.doi.org/10.1056/NEJMoa053422
- Bonner, J.A., Harari, P.M., Giralt, J., Cohen, R.B., Jones, C.U., Sur, R.K., et al. (2010) Radiotherapy plus Cetuximab for Locoregionally Advanced Head and Neck Cancer: 5-Year Survival Data from a Phase 3 Randomized Trial, and Relation between Cetuximab-Induced Rash and Survival. The Lancet Oncology, 11, 21-28. http://dx.doi.org/10.1016/S1470-2045(09)70311-0
- Rudert, F. (2000) Genomics and Proteomics Tools for the Clinic. Current Opinion in Molecular Therapeutics, 2, 633- 642.
- Celis, J.E. and Gromov, P. (1999) 2D Protein Electrophoresis: Can It Be Perfected? Current Opinion in Biotechnology, 10, 16-21. http://dx.doi.org/10.1016/S0958-1669(99)80004-4
- Unlu, M. (1999) Difference Gel Electrophoresis. Biochemical Society Transactions, 27, 547-549.
- Cheng, A.J., Chen, L.C., Chien, K.Y., Chen, Y.J., Chang, J.T.C., Wang, H.-M., et al. (2005) Oral Cancer Plasma Tumor Marker Identified with Bead-Based Affinity-Fractionated Proteomic Technology. Clinical Chemistry, 51, 2236- 2244. http://dx.doi.org/10.1373/clinchem.2005.052324
- Chapman, K. (2002) The Protein Chip Biomarker System from Ciphergen Biosystems: A Novel Proteomics Platform for Rapid Biomarker Discovery and Validation. Biochemical Society Transactions, 30, 82-87. http://dx.doi.org/10.1042/BST0300082
- Craven, R.A. and Banks, R.E. (2001) Laser Capture Microdissection and Proteomics: Possibilities and Limitation. Proteomics, 1, 1200-1204. http://dx.doi.org/10.1002/1615-9861(200110)1:10<1200::AID-PROT1200>3.0.CO;2-Q
- Qian, W.J., Jacobs, J.M., Liu, T., Camp II, D.G. and Smith, R.D. (2006) Advances and Challenges in Liquid Chromatography-Mass Spectrometry-Based Proteomics Profiling for Clinical Applications. Molecular & Cellular Proteomics: MCP, 5, 1727-1744.
- Evans, C., Noirel, J., Ow, S.Y., et al. (2012) An Insight into iTRAQ: Where Do We Stand Now? Analytical and Bioanalytical Chemistry, 404, 1011-1027. http://dx.doi.org/10.1007/s00216-012-5918-6
- Sewell, D.A., Yuan, C.X. and Robertson, E. (2007) Proteomic Signatures in Laryngeal Squamous Cell Carcinoma. ORL: Journal for Oto-Rhino-Laryngology and Its Related Specialties, 69, 77-84. http://dx.doi.org/10.1159/000097406
- Conrads, T.P., Zhou, M., Petricoin 3rd, E.F., Liotta, L. and Veenstra, T.D. (2003) Cancer Diagnosis Using Proteomic Patterns. Expert Review of Molecular Diagnostics, 3, 411-420. http://dx.doi.org/10.1586/14737159.3.4.411
- Schirle, M., Heurtier, M.A. and Kuster, B. (2013) Profiling Core Proteomes of Human Cell Lines by One-Dimensional PAGE and Liquid Chromatography-Tandem Mass Spectrometry. Molecular & Cellular Proteomics: MCP, 2, 1297- 1305.
- Tanca, A., Pagnozzi, D., Burrai, G.P., Polinas, M., Uzzau, S., Antuofermo, E. and Addis, M.F. (2012) Comparability of Differential Proteomics Data Generated from Paired Archival Fresh-Frozen and Formalin-Fixed Samples by GeLC-MS/MS and Spectral Counting. Journal of Proteomics, 77, 561-576. http://dx.doi.org/10.1016/j.jprot.2012.09.033
- Streckfus, C.F. and Dubinsky, W.P. (2007) Proteomic Analysis of Saliva for Cancer Diagnosis. Expert Review of Proteomics, 4, 329-332. http://dx.doi.org/10.1586/14789450.4.3.329
- Cheng, Y.S., Rees, T. and Wright, J. (2014) A Review of Research on Salivary Biomarkers for Oral Cancer Detection. Clinical and Translational Medicine, 3, 3. http://dx.doi.org/10.1186/2001-1326-3-3
- Ma, Y., Zhang, P., Wang, F. and Qin, H. (2012) Searching for Consistently Reported Up- and Down-Regulated Biomarkers in Colorectal Cancer: A Systematic Review of Proteomic Studies. Molecular Biology Reports, 39, 8483-8490. http://dx.doi.org/10.1007/s11033-012-1702-0
- Chi, L.M., Lee, C.W., Chang, K.P., Hao, S.-P., Lee, H.-M., Liang, Y., et al. (2009) Enhanced Interferon Signaling Pathway in Oral Cancer Revealed by Quantitative Proteome Analysis of Microdissected Specimens Using 16O/18O Labeling and Integrated Two-Dimensional LC-ESI-MALDI Tandem MS. Molecular & Cellular Proteomics: MCP, 8, 1453-1474.
- Dowling, P., Wormald, R., Meleady, P., Henry, M., Curran, A. and Clynes, M. (2008) Analysis of the Saliva Proteome from Patients with Head and Neck Squamous Cell Carcinoma Reveals Differences in Abundance Levels of Proteins Associated with Tumour Progression and Metastasis. Journal of Proteomics, 71, 168-175. http://dx.doi.org/10.1016/j.jprot.2008.04.004
- Karahatay, S., Thomas, K., Koybasi, S., Senkal, C.E., ElOjeimy, S., Liu, X., et al. (2007) Clinical Relevance of Ceramide Metabolism in the Pathogenesis of Human Head and Neck Squamous Cell Carcinoma (HNSCC): Attenuation of C18-Ceramide in HNSCC Tumors Correlates with Lymphovascular Invasion and Nodal Metastasis. Cancer Letters, 256, 101-111. http://dx.doi.org/10.1016/j.canlet.2007.06.003
- Sepiashvili, L., Hui, A., Ignatchenko, V., Shi, W., Su, S., Xu, W., et al. (2012) Potentially Novel Candidate Biomarkers for Head and Neck Squamous Cell Carcinoma Identified Using an Integrated Cell Line-Based Discovery Strategy. Molecular & Cellular Proteomics: MCP, 11, 1404-1415.
- Galazis, N., Pang, Y.L., Galazi, M., Haoula, Z., Layfield, R. and Atiomo, W. (2013) Proteomic Biomarkers of Endometrial Cancer Risk in Women with Polycystic Ovary Syndrome: A Systematic Review and Biomarker Database Integration. Gynecological Endocrinology: The Official Journal of the International Society of Gynecological Endocrinology, 29, 638-644.
- Parolini, S., Flagiello, D., Cinquetti, A., Gozzi, R., Cristini, S., Cappiello, J., et al. (1996) Up-Regulation of Urokinase-Type Plasminogen Activator in Squamous Cell Carcinoma of Human Larynx. British Journal of Cancer, 74, 1168-1174. http://dx.doi.org/10.1038/bjc.1996.512
- Yoshizaki, T., Sato, H., Maruyama, Y., Murono, S., Furukawa, M., Park, C.-S. and Seiki, M. (1997) Increased Expression of Membrane Type 1-Matrix Metalloproteinase in Head and Neck Carcinoma. Cancer, 79, 139-144. http://dx.doi.org/10.1002/(SICI)1097-0142(19970101)79:1<139::AID-CNCR20>3.0.CO;2-4
- Donato, R. (2001) S100: A Multigenic Family of Calcium-Modulated Proteins of the EF-Hand Type with Intracellular and Extracellular Functional Roles. The International Journal of Biochemistry & Cell Biology, 33, 637-668. http://dx.doi.org/10.1016/S1357-2725(01)00046-2
- Cao, L.Y., Yin, Y., Li, H., Jiang, Y. and Zhang, H.F. (2009) Expression and Clinical Significance of S100A2 and p63 in Esophageal Carcinoma. World Journal of Gastroenterology: WJG, 15, 4183-4188. http://dx.doi.org/10.3748/wjg.15.4183
- Fuentes, M.K., Nigavekar, S.S., Arumugam, T., Logsdon, C.D., Schmidt, A.M., Park, J.C. and Huang, E.H. (2007) RAGE Activation by S100P in Colon Cancer Stimulates Growth, Migration, and Cell Signaling Pathways. Diseases of the Colon and Rectum, 50, 1230-1240. http://dx.doi.org/10.1007/s10350-006-0850-5
- Arumugam, T., Simeone, D.M., Van Golen, K. and Logsdon, C.D. (2005) S100P Promotes Pancreatic Cancer Growth, Survival, and Invasion. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 11, 5356-5364.
- Mahon, P.C., Baril, P., Bhakta, V., Chelala, C., Caulee, K., Harada, T. and Lemoine, N.R. (2007) S100A4 Contributes to the Suppression of BNIP3 Expression, Chemoresistance, and Inhibition of Apoptosis in Pancreatic Cancer. Cancer Research, 67, 6786-6795. http://dx.doi.org/10.1158/0008-5472.CAN-07-0440
- Pedersen, K.B., Andersen, K., Fodstad, O. and Maelandsmo, G.M. (2004) Sensitization of Interferon-Gamma Induced Apoptosis in Human Osteosarcoma Cells by Extracellular S100A4. BMC Cancer, 4, 52. http://dx.doi.org/10.1186/1471-2407-4-52
- Jenkinson, S.R., Barraclough, R., West, C.R. and Rudland, P.S. (2004) S100A4 Regulates Cell Motility and Invasion in an in Vitro Model for Breast Cancer Metastasis. British Journal of Cancer, 90, 253-262.z http://dx.doi.org/10.1038/sj.bjc.6601483
- Lo, J.F., Yu, C.C., Chiou, S.H., Huang, C.Y., Jan, C.I., Lin, S.C., et al. (2011) The Epithelial-Mesenchymal Transition Mediator S100A4 Maintains Cancer-Initiating Cells in Head and Neck Cancers. Cancer Research, 71, 1912-1923. http://dx.doi.org/10.1158/0008-5472.CAN-10-2350
- Kawai, H., Minamiya, Y. and Takahashi, N. (2011) Prognostic Impact of S100A9 Overexpression in Non-Small Cell Lung Cancer. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine, 32, 641-646.
- Smith, S.L., Gugger, M., Hoban, P., Ratschiller, D., Watson, S.G., Field, J.K., et al. (2004) S100A2 Is Strongly Expressed in Airway Basal Cells, Preneoplastic Bronchial Lesions and Primary Non-Small Cell Lung Carcinomas. British Journal of Cancer, 91, 1515-1524. http://dx.doi.org/10.1038/sj.bjc.6602188
- Wang, H., Zhang, Z., Li, R., Ang, K.K., Zhang, H., Caraway, N.P., et al. (2005) Overexpression of S100A2 Protein as a Prognostic Marker for Patients with Stage I Non Small Cell Lung Cancer. International Journal of Cancer, 116, 285- 290.
- Ohuchida, K., Mizumoto, K., Miyasaka, Y., Yu, J., Cui, L., Yamaguchi, H., et al. (2007) Over-Expression of S100A2 in Pancreatic Cancer Correlates with Progression and Poor Prognosis. The Journal of Pathology, 213, 275-282. http://dx.doi.org/10.1002/path.2250
- Xiao, M.B., Jiang, F., Ni, W.K., Chen, B.-Y., Lu, C.-H., Li, X.-Y. and Ni, R.-Z. (2012) High Expression of S100A11 in Pancreatic Adenocarcinoma Is an Unfavorable Prognostic Marker. Medical Oncology, 29, 1886-1891. http://dx.doi.org/10.1007/s12032-011-0058-y
- Almadori, G., Bussu, F., Galli, J., Rigante, M., Lauriola, L., Michetti, F., et al. (2009) Diminished Expression of S100A2, a Putative Tumour Suppressor, Is an Independent Predictive Factor of Neck Node Relapse in Laryngeal Squamous Cell Carcinoma. Journal of Otolaryngology―Head & Neck Surgery, 38, 16-22.
- Tripathi, S.C., Matta, A., Kaur, J., Grigull, J., Chauhan, S.S., Thakar, A., et al. (2010) Nuclear S100A7 Is Associated with Poor Prognosis in Head and Neck Cancer. PloS ONE, 5, e11939. http://dx.doi.org/10.1371/journal.pone.0011939
- Astrand, R., Unden, J. and Romner, B. (2013) Clinical Use of the Calcium-Binding S100B Protein. Methods in Molecular Biology, 963, 373-384. http://dx.doi.org/10.1007/978-1-62703-230-8_23
- Hartman, K.G., McKnight, L.E., Liriano, M.A. and Weber, D.J. (2013) The Evolution of S100B Inhibitors for the Treatment of Malignant Melanoma. Future Medicinal Chemistry, 5, 97-109. http://dx.doi.org/10.4155/fmc.12.191
- Szoke, D. and Panteghini, M. (2012) Diagnostic Value of Transferrin. Clinica Chimica Acta: International Journal of Clinical Chemistry, 413, 1184-1189. http://dx.doi.org/10.1016/j.cca.2012.04.021
- Sethi, A., Sher, M., Akram, M.R., Karim, S., Khiljee, S., Sajjad, A., et al. (2013) Albumin as a Drug Delivery and Diagnostic Tool and Its Market Approved Products. Acta Poloniae Pharmaceutica, 70, 597-600.
- Chen, J., Chen, L.J., Xia, Y.L., Zhou, H.-C., Yang, R.-B., Wu, W., et al. (2013) Identification and Verification of Transthyretin as a Potential Biomarker for Pancreatic Ductal Adenocarcinoma. Journal of Cancer Research and Clinical Oncology, 139, 1117-1127. http://dx.doi.org/10.1007/s00432-013-1422-4
- Lv, S., Gao, J., Zhu, F., Li, Z.S., Gong, Y.F., Xu, G.M. and Ma, L. (2011) Transthyretin, Identified by Proteomics, Is Overabundant in Pancreatic Juice from Pancreatic Carcinoma and Originates from Pancreatic Islets. Diagnostic Cytopathology, 39, 875-881. http://dx.doi.org/10.1002/dc.21484
- Fleming, C.E., Nunes, A.F. and Sousa, M.M. (2009) Transthyretin: More than Meets the Eye. Progress in Neurobiology, 89, 266-276. http://dx.doi.org/10.1016/j.pneurobio.2009.07.007
- Mizuno, K. (2013) Signaling Mechanisms and Functional Roles of Cofilin Phosphorylation and Dephosphorylation. Cellular Signalling, 25, 457-469. http://dx.doi.org/10.1016/j.cellsig.2012.11.001
- Fernandez, P.M., Patierno, S.R. and Rickles, F.R. (2004) Tissue Factor and Fibrin in Tumor Angiogenesis. Seminars in Thrombosis and Hemostasis, 30, 31-44. http://dx.doi.org/10.1055/s-2004-822969
- Desai, S.D., Mao, Y., Sun, M., Li, T.K., Wu, J. and Liu, L.F. (2000) Ubiquitin, SUMO-1, and UCRP in Camptothecin Sensitivity and Resistance. Annals of the New York Academy of Sciences, 922, 306-308. http://dx.doi.org/10.1111/j.1749-6632.2000.tb07050.x
- Desai, S.D., Wood, L.M., Tsai, Y.C., Hsieh, T.-S., Marks, J.R., Scott, G.L., et al. (2008) ISG15 as a Novel Tumor Biomarker for Drug Sensitivity. Molecular Cancer Therapeutics, 7, 1430-1439. http://dx.doi.org/10.1158/1535-7163.MCT-07-2345
- Belletti, B. and Baldassarre, G. (2011) Stathmin: A Protein with Many Tasks. New Biomarker and Potential Target in Cancer. Expert Opinion on Therapeutic Targets, 15, 1249-1266. http://dx.doi.org/10.1517/14728222.2011.620951
- Giampietro, C., Luzzati, F., Gambarotta, G., Giacobini, P., Boda, E., Fasolo, A. and Perroteau, I. (2005) Stathmin Expression Modulates Migratory Properties of GN-11 Neurons in Vitro. Endocrinology, 146, 1825-1834. http://dx.doi.org/10.1210/en.2004-0972
- Jin, K., Mao, X.O., Cottrell, B., et al. (2004) Proteomic and Immunochemical Characterization of a Role for Stathmin in Adult Neurogenesis. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 18, 287-299.
- Chen, D., Yoo, B.K., Santhekadur, P.K., et al. (2011) Insulin-Like Growth Factor-Binding Protein-7 Functions as a Potential Tumor Suppressor in Hepatocellular Carcinoma. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 17, 6693-6701.
- Shersher, D.D., Vercillo, M.S., Fhied, C., Basu, S., Rouhi, O., Mahon, B., et al. (2011) Biomarkers of the Insulin-Like Growth Factor Pathway Predict Progression and Outcome in Lung Cancer. The Annals of Thoracic Surgery, 92, 1805- 1811; Discussion 1811. http://dx.doi.org/10.1016/j.athoracsur.2011.06.058
- Mignatti, P. and Rifkin, D.B. (1993) Biology and Biochemistry of Proteinases in Tumor Invasion. Physiological Reviews, 73, 161-195.
- Duffy, M.J., Duggan, C., Mulcahy, H.E., McDermott, E.W. and O’Higgins, N.J. (1998) Urokinase Plasminogen Activator: A Prognostic Marker in Breast Cancer Including Patients with Axillary Node-Negative Disease. Clinical Chemistry, 44, 1177-1183.
- Kim, T.D., Song, K.S., Li, G., Choi, H., Park, H.-D., Lim, K., et al. (2006) Activity and Expression of Urokinase-Type Plasminogen Activator and Matrix Metalloproteinases in Human Colorectal Cancer. BMC Cancer, 6, 211. http://dx.doi.org/10.1186/1471-2407-6-211
- Wang, D. and Wang, T. (2005) Expressions and Clinical Significance of Urokinase-Type Activator (uPA) and uPA Receptor (uPAR) in Laryngeal Squamous Cell Carcinoma. Journal of Clinical Otorhinolaryngology, 19, 529-531.
- Wu, H.Y., Shen, X.H., Ni, R.S., Qian, X.Y. and Gao, X. (2009) Expression of E-Cadherin and uPA and Their Prognostic Value in Carcinoma of Human Larynx. Chinese Journal of Otorhinolaryngology Head and Neck Surgery, 44, 1024- 1028.
- Gou, X., Chen, H., Jin, F., Wu, W., Li, Y., Long, J., et al. (2013) Expressions of CD147, MMP-2 and MMP-9 in Laryngeal Carcinoma and Its Correlation with Poor Prognosis. Pathology Oncology Research: POR, Published Online.
- Yang, L., Shang, X., Zhao, X., Lin, Y. and Liu, J. (2012) Correlation Study between OPN, CD44v6, MMP-9 and Distant Metastasis in Laryngeal Squamous Cell Carcinoma. Journal of Clinical Otorhinolaryngology, Head, and Neck Surgery, 26, 989-992.
- Ogretmen, B. and Hannun, Y.A. (2004) Biologically Active Sphingolipids in Cancer Pathogenesis and Treatment. Nature Reviews Cancer, 4, 604-616. http://dx.doi.org/10.1038/nrc1411
- Koybasi, S., Senkal, C.E., Sundararaj, K., Spassieva, S., Bielawski, J., Osta, W., et al. (2004) Defects in Cell Growth Regulation by C18:0-Ceramide and Longevity Assurance Gene 1 in Human Head and Neck Squamous Cell Carcinomas. The Journal of Biological Chemistry, 279, 44311-44319. http://dx.doi.org/10.1074/jbc.M406920200
- Senkal, C.E., Ponnusamy, S., Rossi, M.J., Bialewski, J., Sinha, D., Jiang, J.C., et al. (2007) Role of Human Longevity Assurance Gene 1 and C18-Ceramide in Chemotherapy-Induced Cell Death in Human Head and Neck Squamous Cell Carcinomas. Molecular Cancer Therapeutics, 6, 712-722. http://dx.doi.org/10.1158/1535-7163.MCT-06-0558
- Issaq, H.J., Waybright, T.J. and Veenstra, T.D. (2011) Cancer Biomarker Discovery: Opportunities and Pitfalls in Analytical Methods. Electrophoresis, 32, 967-975. http://dx.doi.org/10.1002/elps.201000588