Open Journal of Veterinary Medicine
Vol.04 No.12(2014), Article ID:52255,7 pages
10.4236/ojvm.2014.412035
Development of a drtqPCR Assay for Differentiation of PCV-2a and 2b in Shanghai Area
Jian Liu, Feifei Ge, Houbing Ju, Dequan Yang, Xin Li, Jian Wang, Peihong Liu, Jinping Zhou*
Shanghai Animal Disease Control Center, Shanghai, China
Email: jianjian115@sohu.com, qyfgff2369@aliyun.com, chengzinono@126.com, dequan_yang@126.com,
star880502@sina.com, jianwhlj@163.com, cathrinevet@163.com, *shzjpvet@163.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 10 October 2014; revised 25 November 2014; accepted 8 December 2014
ABSTRACT
A dual real-time quantitative polymerase chain reaction assay (drtqPCR) was established to detect and differentiate between Porcine circovirus-2a (PCV-2a) and Porcine circovirus-2b (PCV-2b). Genotype-specific primer sets and probes were designed by using sequence data published for different PCV-2 strains. Specificity and sensitivity of the drtqPCR were examined by using PCV-2 isolates with known genotype. Among 367 tissue samples, 44.69% (164/367) were PCV-2 positive. From 164 PCV-2 positive samples, 10.98% (18/164), 92.56% (137/164), and 3.31% (9/164) were positive for PCV-2a, PCV-2b, and both genotypes, respectively. These results suggest that the differential drtqPCR can be used to detect PCV-2 and to differentiate the 2 genotypes from field samples. The PCV-2 infection is quite common in swine of Shanghai area. Furthermore, the PCV-2b infective ratio is far higher than PCV-2a, and PCV-2a/2b mixed infections are also observed but at a lower prevalence in Shanghai area.
Keywords:
Genotype Differentiation, drtqPCR, PCV-2

1. Introduction
Porcine circovirus-2 (PCV-2) is a very small, circular, single stranded DNA virus about 1.76 kb in length [1] . Viral DNA possesses at least 3 functional open reading frames (ORFs): ORF1 encodes the Rep proteins involved in virus replication; ORF2 encodes the nucleocapsid (Cap) protein; and ORF3 encodes a protein that induces apoptosis and is involved in viral pathogenesis in vivo [2] . The virus has been associated with several dis-
ease syndromes in pigs, including postweaning multisystemic wasting syndrome (PMWS), porcine dermatitis and nephropathy syndrome, reproductive failure, and porcine respiratory disease complex. These syndromes together are considered porcine circovirus-associated disease (PCVAD) [1] [3] .
In general, PCV-2 strains from different geographical areas were also shown to be differentiated into genetic groups 1 and 2. PCV-2 groups 1 and 2 were found to contain 1767 and 1768 nucleotides, respectively [4] [5] . In phylogenetic trees, group 1 viruses were classified in the same cluster of PCV-2b, whereas group 2 viruses were classified in the cluster of PCV-2a. PCV-2 genotypes have been classified by several methods, including viral genomic sequencing, restriction fragment length polymorphism (RFLP) analysis, situ hybridization assay, nested polymerase chain reaction assay, and genotype-specific PCR [6] - [8] . Therefore, a dual real-time quantitative polymerase chain reaction (drtqPCR) assay was developed to detect with high sensitivity PCV-2 strains present in tissue samples of pigs, as well as this PCR assay was able to identify PCV-2 genotypes by targeting the ORF2 gene. Each of these methods has advantages and disadvantages in obtaining the results. It is generally accepted that drtqPCR is less time consuming and labor consuming than RFLP, nPCR, and sequencing methods. In this study, we will use the drtqPCR assay to show the PCV-2a and PCV-2b infection in pigs of Shanghai area.
2. Materials and Methods
2.1. Reference Strains
Download reference strains sequences from GenBank (Table 1). The 12 gene sequences of PCV-2 strains in GenBank were from different countries in North America, Europe, and Asia.
2.2. Primer Sets and Probes Design for drtqPCR
Primer sets that could detect the 2 genotypes of PCV-2 were designed in the ORF2 region. By phylogenetic tree, PCV-2a and PCV-2b are obviously different in 1454-1488 regions; therefore the two probes combining PCV-2a and PCV-2b respectively were designed in the variant regions, and prime sets were designed in the common regions. For example PCV-2a, the forward primer sequences were C-F (1418) 5’-CCA, GAA, TTC, AAC, YTT, AAC, CTT, YCT, TAT-3’, and the reverse primer sequences were C-R (1535) 5’-GRC, RGT, GGA, CAT, GMT, GAG, A-3’. The probes were designed in their variant region 1454-1488, so the probe of PCV-2a was C-2a-Probe (1459) FAM-AGG, GTA, TAG, AGA, TTT, TGT, TGG, TCC, CCC, CTC-BHQ, and the probe of PCV-2b was C-2b-Probe (1478) HEX-CAA, ACC, CCC, kCw, CTG, TGC, CCT, TTG, A-BHQ (Y=C or T; M=A or C; R=A or G; K=G or T; W=A or T).
2.3. Porcine Circovirus-2 Recombinant Plasmids
Extraction of KF850469 and KF850463 isolates of DNA was performed using the Qiagen QIAamp DNA Mini Kit according to the manufacturer’s instructions. The PCR products encompassing the partial viral genome of both strains were produced using the primers: C-F (5’-CCA, GAA, TTC, AAC, YTT, AAC, CTT, YCT,
Table 1. PCV-2 strains for sequence comparison.
TAT-3’), C-R (5’-GRC, RGT, GGA, CAT, GMT, GAG, A-3’). The PCR products were subsequently cloned in the plasmid vector pMD18-T. Finally, the recombinant plasmids (pMD18-T/PCV-2a and pMD18-T/PCV-2b) were sequenced to verify the accuracy of the nucleotide sequences and were used as DNA templates and positive samples to validate the new drtqPCR diagnostic assay.
2.4. drtqPCR Assays
In the dual reaction, 2 µl of sample containing the DNA template was added to a 28-µl reaction mixture (Final concentration: dATP, dTTP, dGTP and dCTP 0.24 mM respectively; Mg2+ 2.4 mM; C-F and C-R 0.32 µM; C-2a-Prob and C-2b-Probe 5 µM; Taq1 U/µL; Taq protection 0.2 U/µL). The PCR reaction started with an initial denaturation and polymerase-activating step of 94˚C for 3 min, followed by 40 amplification cycles of a 3-step PCR (94˚C for 15 sec; 60˚C for 40 sec and detecting fluorescent light). Fluorescence channels are FAM and HEX.
2.5. Standard Curves
The standard curve were assayed for assessing the stability of the drtqPCR. The recombinant plasmids pMD18-T/PCV-2a and pMD18-T/PCV-2b were 10-fold serial diluted with the original concentrations 5.4 ng/µl and 7.2 ng/µl by Thermo NANODROP 2000 respectively.
2.6. Sensitivity and Specificity
To assess the sensitivity and specificity of the drtqPCR assay and to determine its ability to differentiate the 2 PCV-2 genotypes, 10-fold serial dilutions of pMD18-T/PCV-2a and pMD18-T/PCV-2b were assayed. The plas- mids were then quantified by Thermo NANODROP 2000. Furthermore, all 12 Shanghai PCV-2 isolates (PCV- 2a: KF850458, KF850466, and KF850468; PCV-2b: KF850459, KF850460, KF850461, KF850462, KF850463, KF850464, KF850465, KF850467, and KF850469) were tested by drtqPCR. The specificities of the primer- probe sets were determined by testing several specimens from different animals, bacterial DNA, and nucleic acids of swine viruses. The samples consisted of PCV-2-negative porcine tissues, semen, and sera, as well as bovine tissues and various pathogens isolated by the SCADC diagnostic laboratory, such as Escherichia coli, Haemophilus parasuis, Lawsonia intracellularis, Pasteurella multocida, Salmonella spp., Staphylococcus hyicus, Streptococcus suis, porcine reproductive and respiratory syndrome virus (PRRSV), classical swine fever virus (CSFV), swine influenza virus (SIV) H9N2, porcine pseudorabies virus (PRV), PCV-1, porcine parvovirus (PPV).
2.7. Application of drtqPCR Assay on Tissue Samples
A total of 367 tissue samples from pigs were randomly selected from Shanghai swine farms. 173 samples were from dead pigs tissue for disease, and 194 samples were healthy pigs tissue (Table 2).
3. Results
3.1. Standard Curves
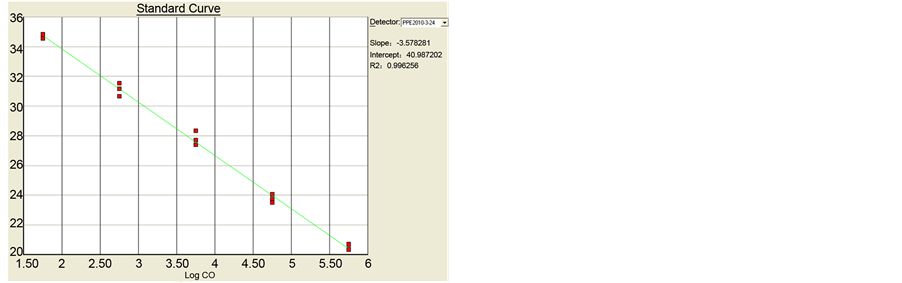
The standard curves of drtqPCR were performed, and R2 were 0.998 for PCV-2a and 0.984 for PCV-2b. The slope values were −3.578 and −3.477, and calculated E value were 90.3% and 93.9% respectively (Figure 1).
3.2. Sensitivity of drtqPCR
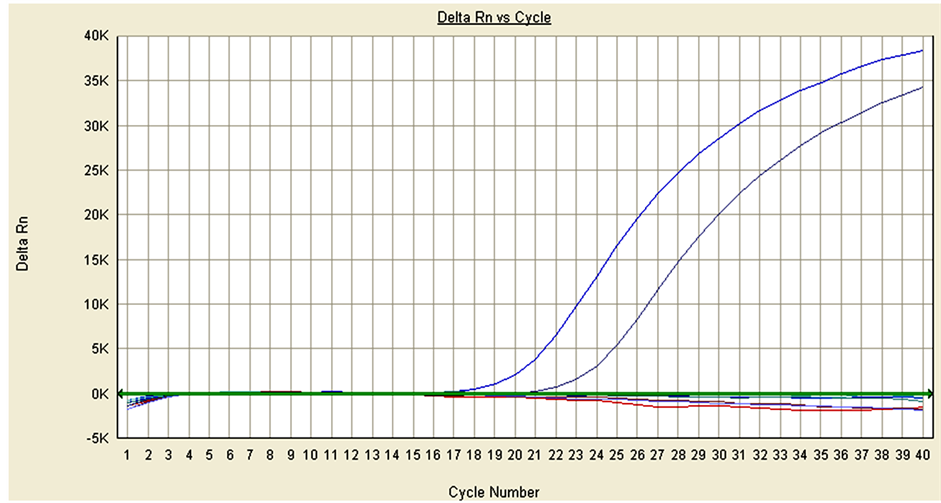
The analytical sensitivity of the drtqPCR was examined by 10-fold serially diluted plasmid of pMD18-T/PCV- 2a and pMD18-T/PCV-2b. The PCV-2a with an original concentrations 5.4 ng/µl was detected up to 10−6 dilution, and the PCV-2a sensitivity was 5.4 fg/µl. The PCV-2b with an original concentrations 7.2 ng/µl was up to 10−5 dilution, and the PCV-2b sensitivity was 0.72 fg/µl (Figure 2).
Table 2. Origin of samples.
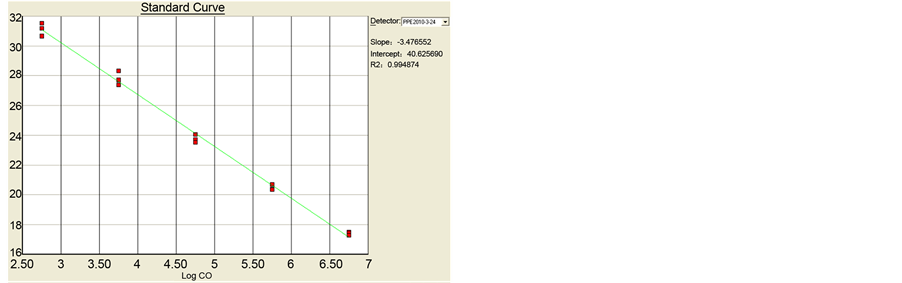
Figure 1. The standard curves of drtqPCR. (a) PCV-2a: R2 0.998, slope value −3.578, E value 90.3%; (b) PCV-2b: R2 0.984, slope value −3.477, E value 93.9%. (a) The standard curves of drtqPCR in PCV-2a; (b) The standard curves of drtqPCR in PCV-2b.
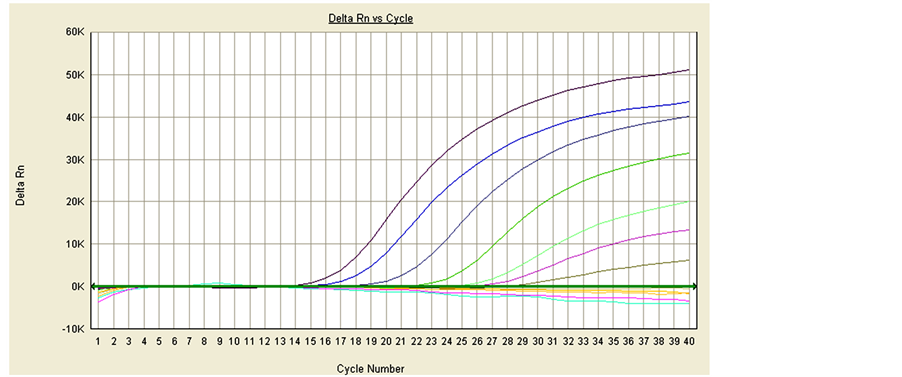
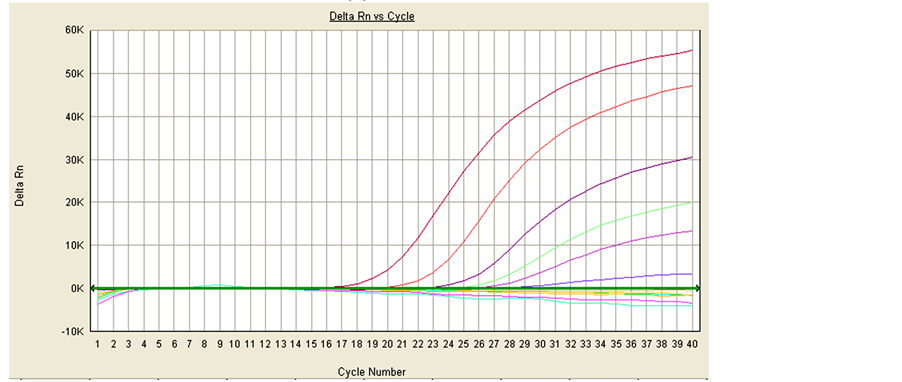
Figure 2. Detection of pMD18-T/PCV-2a and pMD18-T/PCV-2b with the original concentrations 5.4 ng/µl and 7.2 ng/µl respectively. (a) PCV-2a; form left to right: 10−1 - 10−12 dilution of pMD18-T/ PCV-2a; (b) PCV-2b; form left to right: 10−1 - 10−12 dilution of pMD18-T/PCV-2b. (a) The sensitivity of drtqPCR in PCV-2a; (b) The sensitivity of drtqPCR in PCV-2b.
3.3. Specificity of drtqPCR
The drtqPCR had not appeared positive curves in detecting samples except for PCV-2a/2b positive isolates (PCV-2a (KF850468), PCV-2b (KF850463)) (Figure 3). The samples consisted of PCV-2a/2b positive isolates, PCV-2-negative porcine tissues, semen, sera, and various pathogens isolated by the SCADC diagnostic laboratory, such as Escherichia coli, Haemophilus parasuis, Salmonella spp., Staphylococcus hyicus, Streptococcus suis, porcine reproductive and respiratory syndrome virus (PRRSV), classical swine fever virus (CSFV), porcine pseudorabies virus (PRV), PCV-1, porcine parvovirus (PPV), and uninfected porcine kidney (PK)-15 cells. The positive curves only occurred in PCV-2a/2b positive isolates, and no nonspecific curve appeared in other samples.
3.4. PCV-2 Results of Samples
164 samples were PCV-2 positive in 367 tissue samples. From 2009 to 2011, the positive ratio in PCVAD samples was 54.17%, 54.05%, 65.79% respectively, and the positive ratio in healthy samples was 20.00%, 22.64%, 39.69% (Table 3).
Figure 3. The specificity of drtqPCR. Detecting samples including:PCV-2a (KF850468), PCV-2b (KF850463), PCV-2- negative porcine tissues, semen, sera, Escherichia coli, Haemophilus parasuis, Salmonella spp., Staphylococcus hyicus, Stre- ptococcus suis, PRRSV, CSFV, PRV, PCV-1, PPV, uninfected porcine kidney (PK)-15 cell. Positive curves were only PCV- 2a (right) and PCV-2b (left).
Table 3. PCV-2 results of samples.
3.5. PCV-2a/2b in PCV-2 Positive Samples
From 2009 to 2011, PCV-2a single positive ratio was 13.33%, 4.17% and 16.88% respectively, PCV-2b single positive ratio was 86.67%, 91.67% and 75.32%, and PCV-2a/2b co-infection ratio was 0%, 4.17% and 7.79% (Table 4).
3.6. PCV-2a/2b in PCV-2 Positive Samples from Dead for Disease
In PCV-2 positive samples from dead for disease, the PCV-2a single positive ratio was 7.69%, 3.57% and 0 in 2009, 2010 and 2011 respectively, the PCV-2b single positive ratio was 92.31%, 91.07% and 88.00%, and the PCV-2a/2b co-infected ratio was 0, 5.36% and 12.00% (Table 5).
3.7. PCV-2a/2b in PCV-2 Positive Samples from Healthy Swine Farms
In healthy swine farms samples, the PCV-2a single positive ratio was 50.00%, 12.50% and 23.08% in 2009, 2010 and 2011 respectively, the PCV-2b single positive ratio was 50.00%, 87.50% and 71.15%, and the PCV-2a/2b co-infected ratio was 0, 0 and 5.77% (Table 6).
4. Discussion
Porcine circovirus-2 is now considered a global disease and the cause of significant economic losses in the swine industry [9] . Nonetheless, it strongly suggests that PCV-2b virus could be more virulent than PCV-2a virus. Moreover, others have reported that genetic variations may have a great impact on the virulence of PCV-2 [10] . One of the main objectives of the swine industry and health scientists is to establish if there is virulence variation between PCV-2a and PCV-2b viruses. Consequently, it has become critical for swine producers and veterinarians to establish which PCV-2 genotypes are present in their herds [11] . In the present study, the established drtqPCR was believed that it combines specificity and sensitivity, and reduces the labor and time
Table 4. PCV-2a/2b differential results in PCV-2 positive samples.
Table 5. PCV-2a/2b in PCV-2 positive samples from dead for disease.
Table 6. PCV-2a/2b in PCV-2 positive samples from healthy swine farms.
intensiveness of diagnosing PCV-2a and PCV-2b. The drtqPCR was verified by various samples in this study.
In the present study, the ratio of PCV-2 in dead pigs was 54.34%, as 34.02% in healthy. The ratio in dead pigs was higher than in healthy, and both of them raised in recent years of Shanghai area. This suggests that dead pigs for infectious diseases was related of PCV-2, but not all pigs infected with PCV-2 virus may lead to death, and pigs infected PCV-2a/2b could be no any clinics. 9 PCV-2a/2b co-infections were also observed but at a lower prevalence in Shanghai area (2.45%). It was remarkable that the co-infection ratio was rising in recent years. The ratio of PCV-2b was much higher than PCV-2a. The ratio of PCV-2a/2b co-infected rose from 2009 to 2011, whereas PCV-2a was low. This suggests PCV-2b was the most viruses in Shanghai area, and PCV- 2a/2b co-infected would impact the farms of Shanghai in the future.
5. Conclusion
In conclusion, the new drtqPCR diagnostic assay is suitable for the sensitive identification and differentiation of PCV-2 and is a more convenient approach than RFLP [12] , partial or entire viral genome sequencing, and conventional PCR techniques [13] . Furthermore, it could efficiently quantify PCV-2 in submitted samples and provide a risk assessment in regard to the odds of developing death.
Acknowledgements
This work was supported by a grant from Shanghai Scientific Agricultural Key Program (No. 2010(5-6)).
References
- Opriessnig, T., Meng, X.J. and Halbur, P.G. (2007) Porcine Circovirus Type 2 Associated Disease: Update on Current Terminology, Clinical Manifestations, Pathogenesis, Diagnosis, and Intervention Strategies. Journal of Veterinary Diagnostic Investigation, 19, 591-615. http://dx.doi.org/10.1177/104063870701900601
- Carl, A.G., Jerome, R.E., Nedzad, M., Guy, F., Josee, H. and Donald, T. (2008) Development and Use of a Multiplex Real-Time Quantitative Polymerase Chain Reaction Assay for Detection and Differentiation of Porcine Circovirus-2 Genotypes 2a and 2b in an Epidemiological Survey. Journal of Veterinary Diagnostic Investigation, 20, 545-558. http://dx.doi.org/10.1177/104063870802000503
- Carl, A.G., Donald, T., Peter, T., Venne, M.H., Alain, H. and Seyyed, M.E. (2007) The Emergence of Porcine Circovirus 2b Genotype (PCV-2b) in Swine in Canada. Canadian Veterinary Journal, 48, 811-819.
- Cheung, A.K., Lager, K.M., Kohutyuk, O.I., Vincent, A.L., Henry, S.C., Baker, R.B., Rowland, R.R. and Dunham, A.G. (2007) Detection of Two Porcine Circovirus Type 2 Genotypic Groups in United States Swine Herds. Archives of Virology, 152, 1035-1044. http://dx.doi.org/10.1007/s00705-006-0909-6
- Olvera, A., Cortey, M. and Segales, J. (2007) Molecular Evolution of Porcine Circovirus Type 2 Genomes: Phylogeny and Clonality. Virology, 357, 175-185. http://dx.doi.org/10.1016/j.virol.2006.07.047
- Duyeol, K., Yooncheol, H., Yeonsu, O., Kiwon, H., Seung, H.N., Cheolwoo, K., Kim, S.H. and Chanhee, C. (2010) Development of in Situ Hybridization Assay That Differentiates between Two Genotypes of Porcine Circovirus-2 in Formalin-Fixed, Paraffin-Embedded Tissues. Journal of Veterinary Diagnostic Investigation, 22, 231-233. http://dx.doi.org/10.1177/104063871002200209
- Kwang, S.L., Hyeun, B.K. and Han, S.J. (2008) Evaluation of a Nested Polymerase Chain Reaction Assay to Differentiate between Two Genotypes of Porcine Circovirus-2. Journal of Veterinary Diagnostic Investigation, 20, 283-288. http://dx.doi.org/10.1177/104063870802000304
- Guo, L.J., Lu, Y.H., Wei, Y.W., Huang, L.P. and Liu, C.M. (2010) Porcine Circovirus Type 2 (PCV2): Genetic Variation and Newly Emerging Genotypes in China. Virology Journal, 7, 273. http://dx.doi.org/10.1186/1743-422X-7-273
- Shibata, I., Okuda, Y., Kitajima, K. and Asai, T. (2006) Shedding of Porcine Circovirus into Colostrum of Sows. Journal of Veterinary Medicine, 53, 278-280. http://dx.doi.org/10.1111/j.1439-0450.2006.00953.x
- Fenaux, M., Opriessnig, T., Halbur, P.G., Elvinger, F. and Meng, X.J. (2004) Two Amino Acid Mutations in the Capsid Protein of Type 2 Porcine Circovirus (PCV2) Enhanced PCV2 Replication in Vitro and Attenuated the Virus in Vivo. Journal of Virology, 78, 13440-13446. http://dx.doi.org/10.1128/JVI.78.24.13440-13446.2004
- Kwang, S.L., Hyeun, B.K. and Han, S.J. (2008) Evaluation of a Nested Polymerase Chain Reaction Assay to Differentiate between Two Genotypes of Porcine Circovirus-2. Journal of Veterinary Diagnostic Investigation, 20, 283-288. http://dx.doi.org/10.1177/104063870802000304
- Grau-Roma, L., Crisci, E., Sibila, M., Lopez-Soria, S., Nofrarias, M., Cortey, M., Fraile, L., Olvera, A. and Segales, J. (2008) A Proposal on Porcine Circovirus Type 2 (PCV2) Genotype Definition and Their Relation with Postweaning Multisystemic Wasting Syndrome (PMWS) Occurrence. Veterinary Microbiology, 128, 23-35. http://dx.doi.org/10.1016/j.vetmic.2007.09.007
- Larochelle, R., Morin, M., Antaya, M. and Magar, R. (1999) Identification and Incidence of Porcine Circovirus in Routine Field Cases in Quebec as Determined by PCR. Veterinary Record, 145, 140-142. http://dx.doi.org/10.1136/vr.145.5.140
NOTES
*Corresponding author.