Advances in Microbiology
Vol.4 No.5(2014), Article ID:44793,9 pages DOI:10.4236/aim.2014.45030
Identification and Expression Analysis of Regulatory Genes Induced by Near-Ultraviolet Irradiation in Bipolaris oryzae
Junichi Kihara*, Nozomi Tanaka, Makoto Ueno, Sakae Arase
Faculty of Life and Environmental Science, Shimane University, Matsue, Japan
Email: *j-kihara@life.shimane-u.ac.jp
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 25 February 2014; revised 25 March 2014; accepted 1 April 2014
ABSTRACT
Bipolaris oryzae is the causal agent of brown leaf spot disease in rice, and its asexual spore (conidium) formation is known to be induced by near-ultraviolet (NUV) irradiation. In order to reveal the photomorphogenic response and to identify new genes upregulated by NUV irradiation, suppression subtractive hybridization (SSH) was carried out in B. oryzae. To confirm the differential gene expression in NUV-irradiated mycelia, quantitative real-time PCR (qRT-PCR) analysis was performed among 301 genes arbitrarily chosen from 1170 cDNA clones. The expression of 46 genes (named NUV01 to NUV46) was found to be significantly enhanced (>4-fold) by NUV irradiation. Sequence analysis revealed that 23 out of the 46 sequences (50%) showed significant matches to known fungal genes. The 46 genes were categorized as either BLR1-dependent or BLR1- independent expression groups using the BLR1-deficient mutant, which presumably lacks the blue/UVA-absorbing photoreceptor. This finding demonstrates that NUV irradiation can induce gene regulation, and that this response may be mediated by both a blue/UVA-absorbing photoreceptor and an as-yet-unidentified photoreceptor in B. oryzae.
Keywords
Bipolaris oryzae, Gene Expression, Near-Ultraviolet (NUV), Quantitative Real-Time PCR, Suppression Subtractive Hybridization, UVB

1. Introduction
Fungal life is greatly modulated by light, as observed in processes such as pigment biosynthesis, the formation of reproductive structures, phototropism, and so on [1] [2] . Blue light is most effective in fungal photomorphogenesis such as that in Neurospora crassa and Trichoderma atroviride, whereas red light and near-ultraviolet (NUV; 300 - 400 nm) can be effective for the induction of conidiation in Aspergillus nidulans [3] and Bipolarisoryzae [4] [5] , respectively. The molecular bases of fungal photoreceptors as well as of photomorphogenesis by blue and red light have been widely studied, as detailed in a number of published reviews [2] [6] [7] .
Bipolaris oryzae (Breda de Haan) Shoem. [anamorph of the ascomycetous fungus Cochliobolus miyabeanus (Ito et Kuribayashi) Drechsler ex Dastur] is the causal agent of brown leaf spot disease in rice, and NUV irradiation induces its asexual spore (conidium) formation. In previous studies, to reveal the mechanisms underlying the photomorphogenic response to NUV irradiation in B. oryzae, we focused on the regulatory genes induced by NUV irradiation, and reported that NUV irradiation specifically enhanced the expression of the novel NUV-inducible gene (UVI-1) [8] , three melanin biosynthesis genes [9] -[11] , the BMR1 gene encoding a transcriptional factor for these melanin biosynthesis genes [12] , and the photolyase gene [13] . On the other hand, in N. crassa and T. atroviride, many blue light-inducible genes have been identified [14] [15] . Thus, in the present study, we constructed subtractive cDNA libraries from cultures of B. oryzae grown under NUV irradiation in order to perform a more comprehensive search for the regulatory genes induced by NUV irradiation in B. oryzae. Suppression subtractive hybridization (SSH) is a method based on suppressive PCR that allows the creation of subtracted cDNA libraries for the identification of genes differentially expressed in response to different experimental conditions. It requires relatively few steps to identify cDNAs corresponding to differentially expressed genes. SSH has been widely used to identify differentially expressed genes, as shown in the appressorium development of Magnaporthe grisea [16] and in the conidiation of Exserohilum turcicum [17] in phytopathogenic fungi. In this paper, we report 46 newly identified genes that were upregulated by NUV irradiation in B. oryzae using SSH methods.
2. Materials and Methods
2.1. Fungal Strains and Growth Conditions
Bipolaris oryzae strain D9/F6-69 (stock culture at the Laboratory of Plant Pathology, Shimane University) was used as the WT strain. The experimental fungus was grown on potato dextrose agar (PDA) in 9-cm petri plates (Iwaki, Tokyo, Japan). To peel off the mycelia from the agar medium, the agar surface of a petri plate (15 mL PDA) was overlaid with a single layer of dialysis tubing (8 cm in flat width; Eidia, Tokyo, Japan) that had been autoclaved at 121˚C for 15 min. The plates were inoculated centrally with a mycelial plug. The cultures were incubated at 25˚C ± 1˚C for 4 days in continuous darkness and then subjected to light treatments. Black-light (BLB) lamps (FL-BLB; Toshiba Electric, Tokyo, Japan), which emitted wavelengths of 300 to 400 nm (mainly 352 nm), were used as the source of near-ultraviolet (NUV) radiation. The irradiance of NUV radiation was measured by an USB4000-UV-VIS miniature fiber optic spectrophotometer (Ocean Optics, Dunedin, FL). The average irradiance of NUV radiation was 280 mW∙m−2.
2.2. Isolation of Total RNA and mRNA
Colonies grown on a PDA medium for 4 days in darkness were exposed to BLB light as a source of NUV radiation for 1 h (light-treated mycelia). These mycelia were scratched from dialysis tubes by a sterilized stainless steel spatula, frozen in liquid nitrogen, and ground with a mortar and pestle to a fine frozen powder. Total RNA was isolated from the frozen mycelial powder by phenol/chloroform extraction, followed by LiCl precipitation according to the protocols [18] . Poly(A)+ RNA was purified using an OligotexTM-dT30
2.3. SSH
SSH was carried out between NUV-irradiated mycelia (tester) and dark-grown mycelia (driver) using a PCR Select cDNA subtraction kit (Clontech, Palo Alto, CA) according to the manufacturer’s instructions. The subtracted cDNA fragments were cloned into the cloning vector pT7Blue T-vector (TaKaRa Bio) and sequenced. Sequencing reactions were performed using a Big Dye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. DNA sequence analysis was performed on an ABI Prism 310 genetic analyzer (Applied Biosystems).
2.4. Quantitative Real-Time PCR Analysis
Total RNA was used as a template for cDNA synthesis. cDNA was prepared using Ready-To-Go You-prime First-Strand Beads (GE Healthcare UK, Ltd., Buckinghamshire, UK) with anoligo(dT)15 primer. DNA contamination was eliminated by treatment with RNase-free DNase I (TaKaRa Bio). Real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa Bio) and a Thermal Cycler Dice Real-Time System TP800 (TaKaRa Bio). The primers used in the reaction (Table S1) were designed by Primer3 at the website http://frodo.wi.mit.edu. Post-PCR melting curves confirmed the specificity of single-target amplification, and the fold expression of each gene relative to the glyceraldehyde-3-phosphate dehydrogenase gene (GPD) was determined.For reverse transcriptase polymerase chain reaction (RT-PCR) analysis, PCR amplification was carried out using 25 cycles of denaturation at 94˚C for 1 min, annealing at 50˚C for 30 s, and elongation at 72˚C for 45 s. Amplification fragments were electrophoresed on a 1% agarose gel (Agarose L03; TaKaRa). The gel was stained with ethidium bromide, destained in water, and photographed under UV illumination (302 nm).
3. Results
3.1. Identification of and Sequence Analysis of NUV-Regulated Genes
We previously reported that NUV irradiation specifically enhances the expression of a novel NUV-inducible gene (UVI-1) [8] , three melanin biosynthesis genes [9] -[11] , the BMR1 gene encoding a transcriptional factor for these melanin biosynthesis genes [12] , and a photolyase gene [13] . These transcripts accumulated to their maximum levels at 1 h after NUV irradiation in B. oryzae, suggesting that 1 h of NUV irradiation would be an appropriate condition under which to identify other genes upregulated by NUV irradiation in B. oryzae. Thus, total RNA was extracted from both dark-grown mycelia and mycelia irradiated with NUV for 1 h. SSH was carried out between dark-grown mycelia (driver) and NUV-irradiated mycelia (tester) in B. oryzae by using a PCR Select cDNA subtraction kit (Clontech). The PCR products of subtractive cDNA were then cloned separately into the cloning vector pT7Blue T-vector and stored at −20˚C before sequencing. We arbitrarily sequenced the inserts of 1170 randomly selected subtractive cDNA clones from the library and sorted these clones based on homology using the BLASTX program in the DNA Data Bank of Japan (DDBJ). These clones were candidates for transcripts specific to or upregulated in NUV-irradiated mycelia. In fact, the EST sequences of the three melanin biosynthesis genes and the photolyase gene mentioned above were included in these clones (data not shown). Furthermore, two opsin-like genes, OPS1 and OPS2, were identified from the subtractive cDNA library [19] . To confirm the differential gene expression in NUV-irradiated mycelia by the SSH strategy, quantitative real-time PCR (qRT-PCR) analysis was performed using each specific primer among 301 genes arbitrarily chosen from the 1170cDNA clones. The results showed that the expression levels of 46 (named NUV01 to NUV46) of the 301 genes were significantly enhanced (>4-fold) by NUV irradiation (Table 1). The average change among the 46 genes was 27.9-fold. NUV19 and NUV40 were particularly affected, showing 160- and 100-fold increases in mRNA levels after NUV irradiation, respectively. All 46 sequences (NUV01 to NUV46) have been submitted to the EST database of DDBJ andthe results of sequence matching with known genes are also summarized in Table 1. Sequence analysis revealed that 23 out of 46 sequences (50%) showed significant matches to known fungal genes. Putative photomorphogenesis-related genes were observed. NUV03 showed similarity to cryptochrome DASH as a putative blue light photoreceptor. NUV12 and NUV20 showed similarities to 1,3,6,8-tetrahydroxynaphthalene reductase and scytalone dehydratase, respectively, as melanin biosynthesis-related proteins. NUV36 showed similarity to deoxyribodipyrimidine photolyase. NUV19, NUV25, NUV39, and NUV46 showed similarity to several kinds of transporter/pump proteins. The remaining NUV-regulated genes showed similarity to cytochrome P450 (NUV01), acetyl-coA carboxylase (NUV08), monooxygenase (NUV10), linoleate diol synthase (NUV26), and so on. Sequence analysis also revealed that 21 out of 46 sequences (46%) were putative uncharacterized proteins, and the remaining 2 out of 46 sequences (4%) were novel sequences (NUV38 and NUV40).
Table 1. Genes upregulated >4-fold in NUV-irradiated mycelia compared to dark-grown mycelia.
*At, Aspergillus terreus; Ch, Cochliobolus heterostrophus; Cl, Cochliobolus lunatus; Gc, Grosmannia clavigera; Lm, Leptosphaeria maculans; Pn, Phaeosphaeria nodorum; Pter, Pyrenophora teres f. teres; Ptri, Pyrenophora tritici-repentis.
3.2. BLR-Dependent NUV-Regulated Gene Expression
We previously isolated and characterized the Blue-light regulator 1 (BLR1) gene, which encodes a putative blue light photoreceptor similar to white collar 1 (WC-1) of N. crassa in B. oryzae [20] . In addition, it was suggested that the BLR1 protein is necessary not only for conidial formation but also for NUV radiation-enhanced photolyase gene expression in B. oryzae [20] . Thus, in the present work, we investigated whether or not the BLR1 gene is involved in NUV radiation-enhanced gene expression by qRT-PCR analysis using a BLR1-deficient mutant. Figure 1 shows the results of the expression analysis of 36 NUV-regulated genes (NUV1-NUV36) selected from the 46 NUV-regulated genes in the WT and the BLR1-deficient mutant (∆BLR1). Both genes with
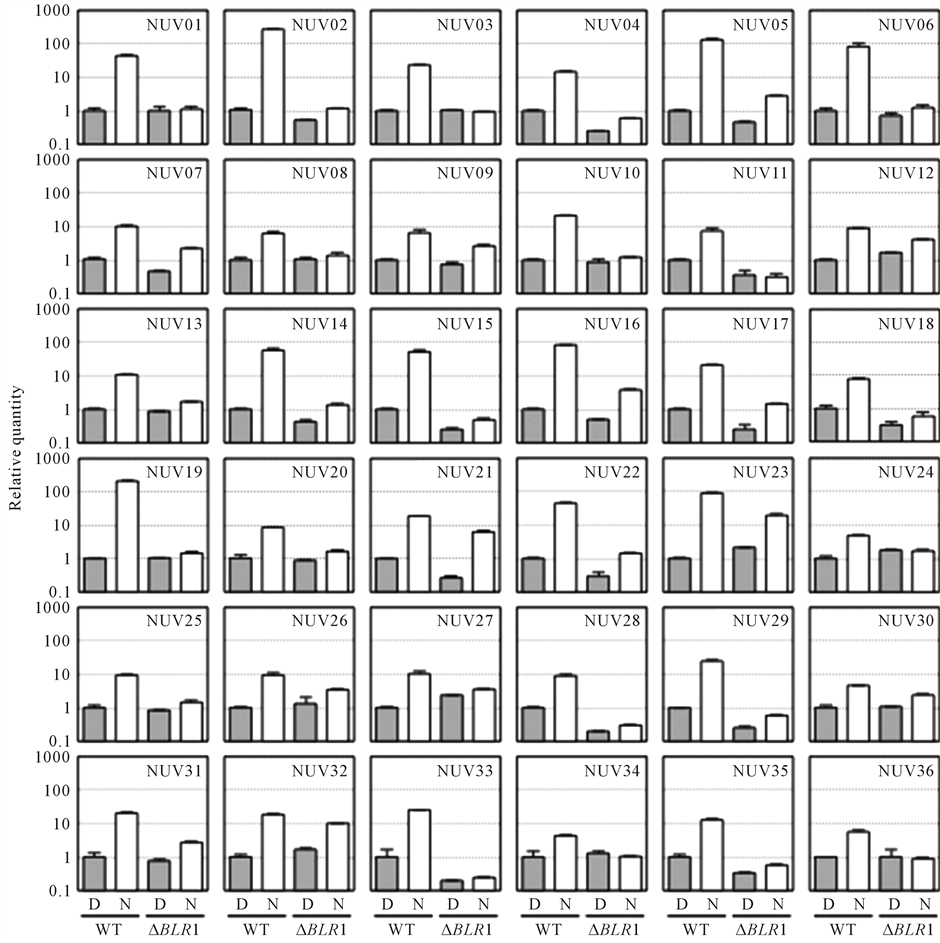
Figure 1. Expression analysis of 36 regulatory genes isolated from the NUV upregulated genes in the wild type (WT) and the BLR1-deficient mutant (ΔBLR1) [20] . Mycelia grown for 4 days on PDA medium in the dark (D, solid bar) were exposed to NUV radiation for 1 h (N, open bar), and total RNA was extracted. The expression was normalized by dividing the average value of the GPD as the internal control. The relative change in the expression of each mRNA was determined as a fold change relative to that of the WT control in the dark (=1). Vertical bars represent the SD. The primers used in this experiment are shown in Table S1.
BLR1-dependent and genes with BLR1-independent expression were recognized. In the BLR1-dependent expression group, which included NUV01, NUV06, and NUV35, the gene expression was significantly enhanced in mycelia exposed to NUV radiation in the WT, whereas the expression levels of these genes were the same as those under dark conditions in the BLR1-deficient mutant (Figure 1). On the other hand, in the BLR1-independent expression group, which included NUV07, NUV21, NUV23, and NUV32, the expression of genes was significantly enhanced in mycelia exposed to NUV radiation in both the WT and the BLR1-deficient mutant, suggesting that the expression of these genes could not be involved inBLR1 regulation. To confirm these differences in the expression profile between the two groups, RT-PCR analysis was performed using the same samples, and the amplified bands were visualized after electrophoresis (Figure 2). In the BLR1-dependent expression group, which included NUV01, NUV06, and NUV35, expression was significantly enhanced in mycelia exposed to NUV radiation, whereas NUV radiation did not enhance the expression levels of these genes in the BLR1-deficient mutant. On the other hand, in the BLR1-independent expression group, which included NUV07, NUV21, NUV23, and NUV32, gene expression was significantly enhanced in mycelia exposed to NUV radiation in both the WT and the BLR1-deficient mutant. In addition, the constitutively expressed GPD gene was observed in both the WT and the BLR1-deficient mutant.
4. Discussion
Generally, photomorphogenic responses in fungi are influenced by blue light, red light, and NUV radiation. The blue light responses have been best studied in Neurospora crassa, and White Collar-1 (WC-1) has been identified as a blue light photoreceptor protein [21] [22] . The red light responses have been best studied in Aspergillus nidulans, and phytochrome (FphA) has been identified as a red light-sensing photoreceptor protein [7] [23] . On the other hand, the effects of NUV irradiation on conidial induction have been studied in some phytopathogenic fungi, such as B. oryzae and Alternaria solani, and the action spectrum for conidial induction suggested that a putative UVB-absorbing photoreceptor would be involved in conidial induction [5] . A putative UVB-absorbing photoreceptor in fungi has not been identified thus far, although UVR8 as the UVB-absorbing photoreceptor in plantae was first identified and characterized in Arabidopsis thaliana [24] [25] . The possibility of a UVB-absorbing photoreceptor in B. oryzae has been suggested by the findings that the expression levels of UVI-1 [8] and of melanin biosynthesis genes [9] -[11] were enhanced by NUV irradiation but not by blue irradiation. In order to identify the putative UVB-absorbing photoreceptor in fungi, it is necessary to determine the global
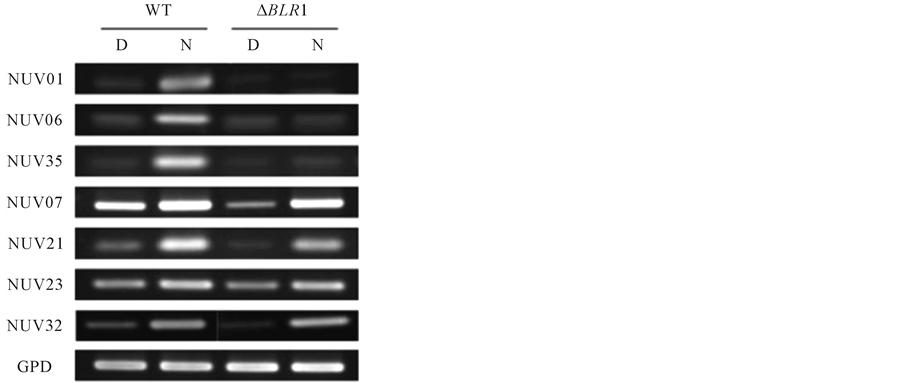
Figure 2. RT-PCR analysis of the NUV upregulated genes in the wild type (WT) and the BLR1-deficient mutant (ΔBLR1) [20] . Total RNA was extracted from mycelia growing in the dark (D) or exposed to NUV radiation for 1 h (N) from the WT and the BLR1-deficient mutant (∆BLR1). The GPD gene was used as an internal control. The primers used in this experiment are shown in Table S1.
changes in gene expression induced by UVB irradiation. In this paper, we found that the expression of 46 newly identified genes was upregulated by NUV irradiation, based on the SSH procedure and quantitative real-time PCR analysis in B. oryzae (Table 1). NUV irradiation induced gene regulation mediated through both a blue/ UVA-absorbing photoreceptor and a putative UVB-absorbing photoreceptor, because NUV radiation (300 - 400 nm) contains UVA radiation (320 - 400 nm) and little UVB radiation (280 - 320 nm). Thus, we tried to categorize the 46 genes as either blue/UVA-regulated genes or UVB-regulated genes using the BLR1-deficient mutant, which presumably lacks the blue/UVA-absorbing photoreceptor, similar to the blue light photoreceptor WC-1 of N. crassa [20] . As a result, BLR1-dependent and BLR1-independent expression groups were recognized (Figure 1 and Figure 2). In the BLR1-dependent expression group, which included NUV01, NUV06, and NUV35, enhanced gene expression by NUV irradiation was not observed in the BLR1-deficient mutant, suggesting that the expression of these genes could be regulated by BLR1 as the blue/UVA-absorbing photoreceptor (Figure 2). On the other hand, in the BLR1-independent expression group, which included NUV07, NUV21, NUV23, and NUV32, enhanced gene expression by NUV irradiation occurred in both the WT and the BLR1-deficient mutant, suggesting that the expression of these genes could be regulated by a putative UVB-absorbing photoreceptor, which was distinct from the regulation by BLR1 as a blue/UVA-absorbing photoreceptor (Figure 2). Interestingly, NUV23 and NUV32 showed similarities to the zinc finger protein and the FAD-binding domain protein, respectively, suggesting that NUV23 and NUV32 could be regulatory elements controlled by a putative UVBabsorbing photoreceptor. The remaining products of the newly identified genes, NUV07 and NUV21, displayed similarities to hypothetical/putative uncharacterized proteins. BLR1-independent blue light-regulated gene expression was also reported in Trichoderma atroviride, indicating the existence of a blue light perception pathway independent of the blue/UVA-absorbing photoreceptor, BLR-1 [15] .
It has previously been considered that NUV radiation emitted by black-light lamps is not a suitable radiation source for research into the photomorphogenic response in B. oryzae, because two photosensory pathways—a blue/UVA-absorbing photoreceptor and a putative UVB-absorbing photoreceptor—would be involved in the photomorphogenic response in B. oryzae, and because NUV radiation (300 - 400 nm) contains UVA radiation (320 - 400 nm) and little UVB radiation (280 - 320 nm). Thus, light-emitting diode (LED) emitting sharp UVB wavelengths would further characterize NUV-regulated genes in B. oryzae. The functions of NUV-regulated genes remain unclear. Further research, such as experiments investigating overexpression, gene disruption, or the introduction of antisense RNA, would help to reveal these roles in the NUV response of B. oryzae. Several kinds of photoreceptor proteins in fungi have been identified, including the White Collar protein complex (WC-1/WC-2), phytochromes, cryptochromes, and opsins [2] [26] . On the other hand, only BLR1 and BLR2 were thought to be blue light-absorbing photoreceptors in B. oryzae [20] [27] , whereas a putative UVB-absorbing photoreceptor has not been identified. Thus, the NUV-regulated genes, especially BLR1-independent genes, will be helpful targets for the identification of unknown UVB-absorbing photoreceptors in B. oryzae.
Acknowledgements
This work was supported by JSPS KAKENHI Grants, Numbers 22780067 and 25450101.
References
- Herrera-Estrella, A. and Horwitz, B.A. (2006) Looking through the Eyes of Fungi: Molecular Genetics of Photoreception. Molecular Microbiology, 64, 5-15. http://dx.doi.org/10.1111/j.1365-2958.2007.05632.x
- Corrochano, L.M. (2007) Fungal Photoreceptors: Sensory Molecules for Fungal Development and Behaviour. Photochemical & Photobiological Sciences, 6, 725-736. http://dx.doi.org/10.1039/b702155k
- Moony, J.J. and Yager, L.N. (1990) Light Is Required for Conidiationin Aspergillus nidulans. Genes & Development, 4, 1473-1482. http://dx.doi.org/10.1101/gad.4.9.1473
- Leach, C.M. (1961) The Sporulation of Helminthosporium oryzae as Affected by Exposure to Near Ultraviolet Radiation and Dark Periods. Canadian Journal of Botany, 39, 705-715. http://dx.doi.org/10.1139/b61-057
- Kumagai, T. (1983) Action Spectra for the Blue and Near-Ultraviolet Reversible Photoreaction in the Induction of Fungal Conidiation. Physiologia Plantarum, 57, 468-471. http://dx.doi.org/10.1111/j.1399-3054.1983.tb02770.x
- Purschwits, J., Müller, S., Kastner, C. and Fischer, R. (2006) Seeing the Rainbow: Light Sensing in Fungi. Current Opinion in Microbiology, 9, 566-571. http://dx.doi.org/10.1016/j.mib.2006.10.011
- Rodriguez-Romero, J., Hedtke, M., Kastner, C., Müller, S. and Fischer, R. (2010) Fungi, Hidden in Soil or Up in the Air: Light Makes a Difference. Annual Review of Microbiology, 64, 585-610. http://dx.doi.org/10.1146/annurev.micro.112408.134000
- Kihara, J., Sato, A., Okajima, S. and Kumagai, T. (2001) Molecular Cloning, Sequence Analysis and Expression of Novel Gene Induced by Near-UV Light in Bipolaris oryzae. Molecular Genetics and Genomics, 266, 64-71. http://dx.doi.org/10.1007/s004380100519
- Kihara, J., Moriwaki, A., Ito, M., Arase, S. and Honda, Y. (2004) Expression of THR1, a 1,3,8-THN Reductase Gene Involved in Melanin Biosynthesis in the Phytopathogenic Fungus Bipolaris oryzae, Is Enhanced by Near-Ultraviolet Radiation. Pigment Cell Research, 17, 15-23. http://dx.doi.org/10.1046/j.1600-0749.2003.00102.x
- Kihara, J., Moriwaki, A., Ueno, M., Tokunaga, T., Arase, S. and Honda, Y. (2004) Cloning, Functional Analysis and Expression of a Scytalonedehydratase Gene (SCD1) Involved in Melanin Biosynthesis of the Phytopathogenic Fungus Bipolaris oryzae. Current Genetics, 45, 197-204. http://dx.doi.org/10.1007/s00294-003-0477-1
- Moriwaki, A., Kihara, J., Kobayashi, T., Tokunaga, T., Arase, S. and Honda, Y. (2004) Insertional Mutagenesis and Characterization of a Polyketide Synthase Gene (PKS1) Required for Melanin Biosynthesis in Bipolaris oryzae. FEMS Microbiology Letters, 238, 1-8. http://dx.doi.org/10.1111/j.1574-6968.2004.tb09729.x
- Kihara, J., Moriwaki, A., Tanaka, N., Tanaka, C., Ueno, M. and Arase, S. (2008) Characterization of the BMR1 Gene Encoding a Transcriptional Factor for Melanin Biosynthesis Genes in the Phytopathogenic Fungus Bipolaris oryzae. FEMS Microbiology Letters, 281, 221-227. http://dx.doi.org/10.1111/j.1574-6968.2008.01101.x
- Kihara, J., Moriwaki, A., Matsuo, N., Arase, S. and Honda, Y. (2004) Cloning, Functional Characterization, and NearUltraviolet Radiation-Enhanced Expression of a Photolyase Gene (PHR1) from the Phytopathogenic Fungus Bipolaris oryzae. Current Genetics, 46, 37-46. http://dx.doi.org/10.1007/s00294-004-0507-7
- Berlin, V. and Yanofsky, C. (1985) Isolation and Characterization of Genes Differentially Expressed during Conidiation of Neurospora crassa. Molecular and Cellular Biology, 5, 849-855. http://mcb.asm.org/content/5/4/849
- Rosales-Saavedra, T., Esquivel-Naranjo, E.U., Casas-Flores, S., Martínez-Hernández, P., Ibarra-Laclette, E., Cortes- Penagos, C. and Herrera-Estrella, A. (2006) Novel Light-Regulated Genes in Trichodera atroviride: A Dissection by cDNA Microarrays. Microbiology, 152, 3305-3317. http://dx.doi.org/10.1099/mic.0.29000-0
- Lu, J.P., Liu, T.B. and Lin, F.C. (2005) Identification of Mature Appressorium-Enriched Transcripts in Magnaporthe grisea, the Rice Blast Fungus, Using Suppression Subtractive Hybridization. FEMS Microbiology Letters, 245, 131- 137. http://dx.doi.org/10.1016/j.femsle.2005.02.032
- Flaherty, J.E. and Dunkle, L.D. (2005) Identification and Expression Analysis of Regulatory Genes Induced during Conidiation in Exserohilum turcicum. Fungal Genetics and Biology, 42, 471-481. http://dx.doi.org/10.1016/j.fgb.2005.02.001
- Verwoerd, T.C., Dekker, B.M.M. and Hoekema, A. (1989) A Small-Scale Procedure for the Rapid Isolation of Plant RNAs. Nucleic Acid Research, 17, 2362. http://dx.doi.org/10.1093/nar/17.6.2362
- Kihara, J., Tanaka, N., Ueno, M. and Arase, S. (2009) Cloning and Expression Analysis of Two Opsin-Like Genes in the Phytopathogenic Fungus Bipolaris oryzae. FEMS Microbiology Letters, 295, 289-294. http://dx.doi.org/10.1111/j.1574-6968.2009.01609.x
- Kihara, J., Moriwaki, A., Tanaka, N., Ueno, M. and Arase, S. (2007) Characterization of the BLR1 Gene Encoding a Putative Blue-Light Regulator in the Phytopathogenic Fungus Bipolarisoryzae. FEMS Microbiology Letters, 266, 110- 118. http://dx.doi.org/10.1111/j.1574-6968.2006.00514.x
- Linden, H., Ballario, P. and Macino, G. (1997) Blue Light Regulation in Neurosporacrassa. Fungal Genetics and Biology, 22, 141-150. http://dx.doi.org/10.1006/fgbi.1997.1013
- Chen, C.H., Dunlap, J.C. and Loros, J.J. (2010) Neurospora Illuminates Fungal Photoreception. Fungal Genetics and Biology, 47, 922-929. http://dx.doi.org/10.1016/j.fgb.2010.07.005
- Bayram, Ö., Braus, G.H., Fischer, R. and Rodriguez-Romero, J. (2010) Spotlight on Aspergillus nidulans Photosensory Systems. Fungal Genetics and Biology, 47, 900-908. http://dx.doi.org/10.1016/j.fgb.2010.05.008
- Kliebenstein, D.J., Lim, J.E., Landry, L.G. and Last, R.L. (2002) Arabidopsis UVR8 Regulates Ultraviolet-B Signaling Transduction and Tolerance and Contains Sequence Similarity to Human Regulator of Chromatin Condensation 1. Plant Physiology, 130, 234-243. http://dx.doi.org/10.1104/pp.005041
- Heijde, M. and Ulm, R. (2012) UV-B Photoreceptor-Mediated Signaling in Plants. Trends in Plant Science, 17, 230- 237. http://dx.doi.org/10.1016/j.tplants.2012.01.007
- Idnurm, A., Verma, S. and Corrochano, L.M. (2010) A Glimpse into the Basis of Vision in the Kingdom Mycota. Fungal Genetics and Biology, 47, 881-892. http://dx.doi.org/10.1016/j.fgb.2010.04.009
- Moriwaki, A., Katsube, H., Ueno, M., Arase, S. and Kihara, J. (2008) Cloning and Characterization of the BLR2, the Homologue of the Blue-Light Regulator of Neurospora crassa WC-2, in the Phytopathogen in Fungus Bipolaris oryzae. Current Microbiology, l56, 115-121. http://dx.doi.org/10.1007/s00284-007-9080-x
Supplement
Table S1 . Nucleotide sequences of the primers used in quantitative real-time PCR.
NOTES

*Corresponding author.


