Open Journal of Medical Microbiology
Vol.04 No.03(2014), Article ID:48958,7 pages
10.4236/ojmm.2014.43018
Detection of the Mex Efflux Pumps in Pseudomonas aeruginosa by Using a Combined Resistance-Phenotypic Markers and Multiplex RT-PCR
Kanchana Poonsuk, Rungtip Chuanchuen
Department of Veterinary Public Health, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand
Email: rchuanchuen@yahoo.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 16 June 2014; revised 14 July 2014; accepted 14 August 2014
ABSTRACT
The aim of this study was to detect the expression of 4 clinically-important efflux pumps in the Resistance-Nodulation-Cell Division (RND) family including MexAB-OprM, MexXY, MexCD-OprJ and MexEF-OprN in Pseudomonas aeruginosa using a combination of resistance-phenotypic markers and multiplex RT-PCR (mRT-PCR). The antibiotic substrates specific for each Mex systems were used as phenotypic markers including carbenicillin, MexAB-OprM, erythromycin, MexCD-OprJ, norfloxacin and imipenem, MexEF-OprN and gentamicin, MexXY-OprM. The methods were validated with reference strains with known genotypes of the Mex systems and the potential applicability in clinical practice was tested with clinical isolates. The results for the reference strains support that the combination of resistance phenotype and mRT-PCR is a potential-attractive method for diagnosis of efflux-mediated resistance in P. aeruginosa. Further development to make it more practical for clinical use and study in a larger number of clinical isolates is required.
Keywords:
Multidrug Efflux Pumps, Multiplex RT-PCR, Pseudomonas aeruginosa, Resistance-phenotypic marker

1. Introduction
Pseudomonas aeruginosa, a common cause of nosocomia infections is infamous for its resistance to multiple drugs [1] that is mainly attributed to the synergy between the low outer membrane permeability and the expression of multidrug efflux systems, particularly in the Resistance-Nodulation-Cell Division (RND) family [2] . Most RND-type drug efflux operons are chromosomally encoded. It is now generally accepted that the RND multidrug efflux systems function as tripartite systems consisting of a cytoplasmic membrane-associated RND transporter (e.g. MexB, MexD, MexF, MexY), periplasmic membrane fusion protein (MFP) e.g. MexA, MexC, MexE and MexX and an outer membrane protein (e.g. OprM, OprJ, and OprN) [3] . The P. aeruginosa genome contains at least 12 structural genes for the RND efflux systems, of which four are clinically-important (i.e. MexAB- OprM, MexCD-OprJ, MexEF-OprN and MexXY) [2] . Due to their constitutive expression, MexAB-OprM and MexXY contribute to intrinsic resistance to many antibiotics [2] . MexCD-OprJ and MexEF-OprN do not express wild-type cells, but can overexpress after the acquisition of regulatory mutations, resulting in acquired multidrug resistance [4] [5] . Coexpression of Mex systems has been reported in the P. aeruginosa clinical isolates where its variable impact on antibiotic susceptibility has been observed [6] - [8] .
Diagnosis of efflux-mediated resistance generates data that is helpful for both routine clinical analysis (e.g. rationalizing the antibiotic selection and dose) and epidemiological studies (e.g. monitoring the existing and prevalent resistance mechanisms) [9] . Efflux pump inhibitors (EPIs) have been under investigation as an alternative to the development of new antibiotics for treatment of P. aeruginosa infection [10] . As yet, no EPIs are approved for clinical use. However, detection methods for efflux-mediated resistance should be concurrently developed in preparation for the new treatment protocol. Phenotypic-based methods usually yield vague outcomes due to the possible existence of other resistance mechanisms, the simultaneous over expression of variable Mexs and the difficulty in assessing the pumps conferring low or moderate resistance level [9] . Detection of the Mex systems has long relied on western blot analysis using monoclonal or polyclonal antibodies. The technique is complicated and time consuming and the antibodies specific for the Mex protein are not commercially available. Quantitative realtime RT-PCR (realtime qRT-PCR) is a rapid method for measuring gene expression. However, many probes are required for simultaneous detection of many Mex genes, resulting in increased cost. In a previous study, realtime qRT-PCR was applied to measure the Mex expression level but limited to that of only 2 Mex systems (i.e. MexAB-OprM and MexXY) [11] . In contrast, quantitative RT-PCR (qRT-PCR) is less expensive but laborious. The disadvantage may be resolved by concomitant detecting of multiple Mex genes using multiplex-qRT-PCR (mRT-PCR). The latter allows us to easily observe the amplification products and is feasible in the laboratory where a realtime PCR machine is not available. Recently, the combined phenotypic and genotypic methods were used for detecting the expression of all 4 clinically-important Mex systems [9] . However, two different PCR-based methods were used for detecting each two systems in the same sample. So far, none of these new diagnostic methods have been commercially available.
In this study, we have combined antibiotic-resistance-phenotypic markers and mRT-PCR for detecting the ex- pression of MexAB-OprM, MexXY, MexCD-OprJ and MexEF-OprN in P. aeruginosa. The methods were validated in reference strains with known genotypes of the Mex systems and the potential applicability in clinical practice was tested with clinical isolates.
2. Methods
2.1. Bacterial isolates and growth conditions
The P. aeruginosa reference strains including PAO1 [12] , PAO200 , PAO200-2, PAO238, PAO7H1A, PAO255 [13] , PAO267 and PAO280 [13] [14] and clinical isolates used in this study are listed in Table 2 and Table 3. The reference strains were selected as carrying known-RND efflux pumps expressed. The clinical isolates were randomly selected from our strain collection. Each of them was isolated from different patients admitted at Siriraj Hospital, Bangkok, Thailand in previous studies [15] . The clonality of the clinical isolates was examined by using ERIC PCR [16] and all were confirmed to be nonrepetitive strains (data not shown). All the P. aeruginosa strains were grown on Luria Bertani (LB) agar, LB broth (Difco, BD Diagnostic Systems, MD, USA) or in Mueller-Hinton broth (MHB; Difco). All the bacterial cultures in LB broth were incubated at 37˚C, with agitation at 120 rpm and under aerobic condition for 12 h. All the bacteria in LB agar and MHB were grown with aeration at the same temperature and period of time.
2.2. Antimicrobial susceptibility testing
MICs of antibiotics tested including carbapenem (Car), erythromycin (Ery), imipenem (Imi), norfloxacin (Nor) and gentamicin (Gen), were determined by using two-fold both microdilution method in the presence and absence of Phe-Arg-β-naphthylamide (PAβN), a broad spectrum EPI, at the concentration of 50 mg/ml (Sigma Aldrich, St. Louis, MO, USA) [17] . All antibiotics were purchased from Sigma Aldrich. P. aeruginosa ATCC 27853 [18] and wild-type PAO1 were used as control strains.
2.3. RNA extraction and cDNA synthesis
The P. aeruginosa cells grown in LB were harvested at 12 h of growth (A540~5.5) by centrifugation. The cells were immediately used for RNA extraction using Total RNA Extraction Mini Kit (RBC Bioscience, New Taipei City, Taiwan) and subsequently treated with RNase-free DNaseI (Fermentas®, Mainz, Germany) as suggested by the manufacture’s instruction. The absence of genomic DNA residuals was determined by PCR. Synthesis of cDNA was performed by reverse transcription using ImProm-IITM Reverse Transcriptase (Promega, WI, USA). Each 5 ml RNA-primer mixture contained 0.5 µg of free DNA-RNA, 10 pmol of each reverse primer (mexBMRT down, mexDRTdown, mexFRTdown and mexYMRTdown). The mixture was incubated at 70˚C for 5 min, quickly chilled at 4˚C for 5 min and hold on ice. The reverse transcription PCR reaction was performed in a 20 ml volume containing 6 µl of the RNA-primer mixture, 4 µl of Improm-IITM 5X Reaction Buffer, 2 µl of 25 mM MgCl2, 1 µl of dNTPs (10 mM each), 1 µl of Improm-IITM Reverse Transcriptase and nuclease free water added to 20 ml. The PCR cycles were as follows: annealing for 5 min at 25˚C, extension for 45 min at 45˚C and heat-inactivation for 15 min at 70˚C. The cDNA was stored at −20˚C until used. All the primers were designed by Primer3 software available at http://bioinfo.ut.ee/primer3-0.4.0/primer3/ (Table 1).
2.4. Multiplex RT-PCR
cDNA from each bacterial strain was individually used as DNA template in mRT-PCR, as well as the mixture of cDNA from all reference strains. All the mRT-PCR reactions were carried out in a 30 ml volume containing 5 µl cDNA (100 - 2000 ng/ml), 10 pmol of each primer, 15 µl of KAPATaq ReadyMix DNA polymerase (Kapabiosystems, MA, USA) and nuclease free water added to 30 µl. The PCR amplication was performed according to the following cycles: one predenaturation for 5 min at 95˚C and 30 cycles of denaturation for 45 s at 95˚C, annealing for 45 s at 54˚C and extension for 30 s at 72˚C, followed by final extension for 10 min at 72˚C.
2.5. PCR amplification and DNA sequencing
All the conventional PCR amplifications were conducted using KAPA Taq ReadyMix (KAPAbiosystem) according to the manufacturer’s instruction. The PCR amplicons from either conventional PCR or mRT-PCR were gel- purified using Nucleospin® ExtractII (Mccherey-Nagel, Düren, Germany) and submitted for sequencing with the PCR primers at 1st BASE Pte Ltd. (Gemini Singapore Science ParkII, Singapore). The DNA sequences obtained were compared with the corresponding sequences of PAO1 available at the Pseudomonas Genome Project [19] .
3. Results
3.1. Resistance phenotypes associated with the Mex efflux pumps
Five antibiotics were used as resistance-phenotypic markers for 4 clinically-important Mex systems tested in this study: carbenicillin, MexAB-OprM; erythromycin, MexCD-OprJ; norfloxacin and imipenem, MexEF-OprN and gentamicin, MexXY-OprM. The MIC values for these antibiotic markers in the presence and absence of PAβN are shown in Table 2 and Table 3. For the reference strains overexpressing a Mex pump, the addition of Phenylalanine-Arginine β-Naphthylamide (PAβN) reduced the MIC values for the corresponding antibiotic markers 2 to 128 folds. The lowest-reduction (2 fold) was observed for imipenem MIC in PAO7H1A overexpressing MexEF-OprN. Most of the clinical isolates exhibited higher resistance level to all antibiotics tested than PAO1 did. PAβN, also called MC-207,110, is a peptidomimetic compound that functions as a broad-spectrum EPI. At the concentration above 16 µg/mL, PAβN additionally permeabilizes membranes in MexAB-OprM deficient P. aeruginosa mutants [20] .
3.2. Expression of the Mex efflux pumps determined by mRT-PCR
For the reference strains, the expression of the Mex efflux pumps is shown in Table 2. When the mixture of cDNA
Table 1. Primers used in this study.
Table 2. Phenotypic and genotypic properties of the P. aeruginosa reference strains.
aPAO7H, overproduced Mex EF-OprN [21] , bPAO3579, PAO1D(amr) [22] .
Table 3. Antimicrobial susceptibility and expression of the RND efflux determined by Multiplex RT-PCR in the P. aeruginosa Clinical isolates.
Not known. AB, MexAB-OprM; CD, MexCD-OprJ; EF, MexEF-OprN, XY, MexXY-OprM.
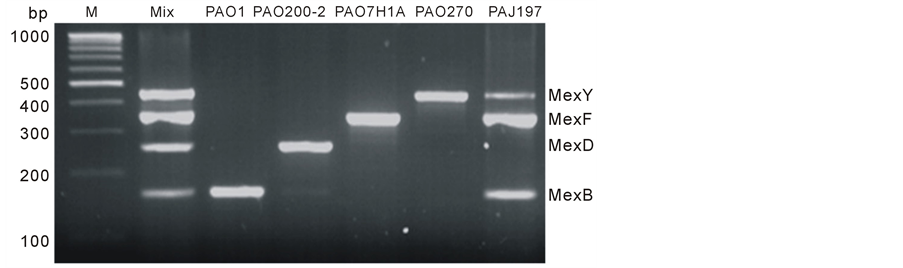
from all the reference strains was used for template, PCR amplicons of all 4 Mex systems was obtained (Figure 1) and DNA sequencing analyses confirmed their specificity. By using mRT-PCR, MexB, MexD, MexF and MexY expression was detected in PAO1, PAO200-2, PAO7H1A and PAO267, respectively. None of the Mex expression was observed in PAO200, PAO238, PAO255 and PAO280 that are null mutant derivatives.
All the clinical isolates produced MexAB-OprM and MexXY as determined by mRT-PCR. Four isolates expressed 3 Mex systems including PAJ128 and PAJ147 (overexpressing MexAB-OprM, MexCD-OprJ and MexXY) and PAJ197 and PAJ207 (overexpressing MexAB-OprM, MexEF-OprN and MexXY). All the isolates were resistant to imipenem and norfloxacin but MexEF-OprN expression was observed in 2 isolates (PAJ197 and
Figure 1. PCR amplicons of 4 clinically important Mex systems generated by RT-PCR. Lane 1) M, molecular weight marker; Lane 2) Mix, The mixture of cDNA from PAO1, PAO200-2, PAO7H1A and PAO27 was used as template. The size of mexB (155 bp), mexD (250 bp), mexF (350 bp) and mexY (445 bp). Lane 3 - 6) band for mexB (155 bp), mexD (250 bp), mexF (350 bp) and mexY (445 bp) PAO1 overexpressing MexB; PAO200-2 overexpressing MexD; PAO7H1A overexpressing MexF and PAO270 overexpressing MexY respectively. Lane 7, PAJ197 a clinical isolate overexpressing MexB/MexF and MexY.
PAJ207). The similar result was observed for erythromycin resistance.
4. Discussion
For phenotype detection, the antibiotics used for phenotypic markers are specific substrates for each of 4 clinically-important Mex systems based on previous studies [23] - [26] . For MexEF-OprN, imipenem was also included as an indirect indicator of pump as previously suggested [9] . In the up-regulated MexEF-OprN mutant strain, resistance to carbapenems is a result of down-regulated OprD that happens concomitantly with the elevated-expression of MexEF-OprN [21] [27] . In the present study, the addition of PAβN caused a 2 fold-reduc- tion of imipenem MIC in both PAO7H1A overexpressing only MexEF-OprN and its isogenic-null mutant PAO255. Similarly, PAβN also reduced the imipenem MIC in all clinical isolates either with or without MexEF-OprN expression. These observations suggest the possible existence of uncharacterized Mex systems that are also inhibited by PAβN in these strains. However, the OprD expression and its actual involvement of MexEF-OprN in the MexEF-OprN-overespressing isolates were not examined. Several studies showed that the synchronized expression and carbapenem resistance was not always observed in clinical isolates [28] . It is evident by our observation that some isolates with high imipenem MICs (e.g. PAJ128 and PAJ147) did not produce MexEF-OprN. Taken together, these data confirmed that imipenem is not a good-indirect phenotypic marker for MexEF-OprN.
In the laboratory reference strains, a good agreement was observed between the RND-efflux genotype and the Mex expression determined by mRT-PCR in all the reference strains (Table 2). For example, only expression of MexD was detected in PAO200-2 that is a spontaneous nfxB derivative of PAO200. The good correlation was also identified between resistance phenotype and the Mex expression. For the instance, PAO1 constitutively producing MexAB-OprM exhibited high carbenicillin MIC. PAO200-2 overexpressing MexCD-OprJ was highly resistant to erythromycin.
Unlike the laboratory reference strains, clinical isolates have diverse genetic backgrounds, resulting in diverse phenotypes. Therefore, it cannot simply use resistance phenotype to predict the expressed-Mex efflux pumps. The discrepancy may be overcome by increasing the number of antibiotic markers to cover all the possible Mex systems. This will markedly cause increasing cost and thus, decreasing the attraction of the method. The addition of a specific Mex inhibitor is of interest but such inhibitors are still not commercially available [9] . Transcription of MexB and MexY was detected in all the clinical isolates, is in agreement with the fact that these efflux systems are always expressed at basal level. The good correlation of resistance phenotype and MexCD- OprJ was observed in two isolates (PAJ128 and PAJ147) and that of MexEF-OprN was observed in PAJ197 and PAJ207. This may not be surprising because MexCD-OprJ and MexEF-OprN are expressed in regulatory mutants and their contribution to antibiotic markers may vary. The addition of PAβN revealed that the contribution of the Mex systems in resistance level varied in clinical isolates and suggested the existence of other enzymatic or non-enzymatic resistance mechanisms. The marginal effect (i.e. 2 fold reduction) of the Mex systems on antibiotic susceptibility was observed in some isolates. It could prevent antibiotics from reaching their optimal concentrations in target organs, especially where the antibiotic concentrations are hindered (e.g. in pus, biofilms, lung tissues), and therefore is still of clinical importance [15] .
A comment could be made that the usefulness of mRT-PCR may be less in comparison to realtime qRT-PCR because the Mex expression level was not quantified. Several studies showed no correlation between the level of transcription and resistance in P. aeruginosa clinical isolates from either animals or humans [29] [30] . Therefore, the measurement of expression level is not always essential for routine diagnosis. Importantly, mRT-PCR could be easily performed in most clinical laboratories, especially those without a sophisticated realtime PCR machine. Still, it cannot be disputed that MIC determination is a gold standard method for assessing susceptibility of P. aeruginosa before choosing antibiotic treatment. For better treatment regimen, mRT-PCR will allow optimal antibiotic choices, especially antibiotics available for use in combination with EPIs.
5. Conclusion
The results in this study support that the combination of resistance phenotype and mRT-PCR is a potential-at- tractive method for diagnosis of efflux-mediated resistance in P. aeruginosa. mRT-PCR is rapid and specific for detection of the Mex systems. However, further development to make it more practical for clinical use and study in a larger number of clinical isolates is still required. The appropriate antibiotics that can be specifically used for the Mex pumps need to be explored. Although the knowledge from this study requires more research before the application in clinical analysis, it is a useful tool for epidemiological studies of the prevalent Mex systems without delay.
Acknowledgements
References
- Aksamit, T.R. (1993) Pseudomonas Pneumonia and Bacteremia in the Immunocompromised Patient. In: Fick, R.B.J., Ed., Pseudomonas aeruginosa: The Opportunist―Pathogenesis and Disease, CRC Press, Boca Raton, 177-188.
- Lister, P.D., Wolter, D.J. and Hanson, N.D. (2009) Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clinical Microbiology Reviews, 22, 582-610. http://dx.doi.org/10.1128/CMR.00040-09
- Poole, K. and Srikumar, R. (2001) Multidrug Efflux in Pseudomonas aeruginosa: Components, Mechanisms and Clinical Significance. Current Topics in Medicinal Chemistry, 1, 59-71. http://dx.doi.org/10.2174/1568026013395605
- Morita, Y., Komori, Y., Mima, T., Kuroda, T., Mizushima, T. and Tsuchiya, T. (2001) Construction of a Series of Mutants Lacking All of the Four Major mex Operons for Multidrug Efflux Pumps or Possessing Each One of the Operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ Is an Inducible Pump. FEMS Microbiology Letters, 202, 139-143. http://dx.doi.org/10.1111/j.1574-6968.2001.tb10794.x
- Kohler, T., Michea-Hamzehpour, M., Henze, U., Gotoh, N., Curty, L.K. and Pechere, J.C. (1997) Characterization of MexE-MexF-OprN, a Positively Regulated Multidrug Efflux System of Pseudomonas aeruginosa. Molecular Microbiology, 23, 345-354. http://dx.doi.org/10.1046/j.1365-2958.1997.2281594.x
- Aendekerk, S., Ghysels, B., Cornelis, P. and Baysse, C. (2002) Characterization of a New Efflux Pump, MexGHI- OpmD, from Pseudomonas aeruginosa That Confers Resistance to Vanadium. Microbiology, 148, 2371-2381.
- Wolter, D.J., Smith-Moland, E., Goering, R.V., Hanson, N.D. and Lister, P.D. (2004) Multidrug Resistance Associated with mexXY Expression in Clinical Isolates of Pseudomonas aeruginosa from a Texas Hospital. Diagn. Microbiol. Diagnostic Microbiology and Infectious Disease, 50, 43-50. http://dx.doi.org/10.1016/j.diagmicrobio.2004.05.004
- Sevillano, E., Valderrey, C., Canduela, M.J., Umaran, A., Calvo, F. and Gallego, L. (2006) Resistance to Antibiotics in Clinical Isolates of Pseudomonas aeruginosa. Pathologie Biologie (Paris), 54, 493-497. http://dx.doi.org/10.1016/j.patbio.2006.07.030
- Mesaros, N., Glupczynski, Y., Avrain, L., Caceres, N.E., Tulkens, P.M. and Van Bambeke, F. (2007) A Combined Phenotypic and Genotypic Method for the Detection of Mex Efflux Pumps in Pseudomonas aeruginosa. Journal of Antimicrobial Chemotherapy, 59, 378-386. http://dx.doi.org/10.1093/jac/dkl504
- Lomovskaya, O., Warren, M.S., Lee, A., Galazzo, J., Fronko, R., Lee, M., et al. (2001) Identification and Characterization of Inhibitors of Multidrug Resistance Efflux Pumps in Pseudomonas aeruginosa: Novel Agents for Combination Therapy. Antimicrobial Agents and Chemotherape, 45, 105-116. http://dx.doi.org/10.1128/AAC.45.1.105-116.2001
- Yoneda, K., Chikumi, H., Murata, T., Gotoh, N., Yamamoto, H., Fujiwara, H., et al. (2005) Measurement of Pseudomonas aeruginosa Multidrug Efflux Pumps by Quantitative Real-Time Polymerase Chain Reaction. FEMS Microbiology Letters, 243, 125-131. http://dx.doi.org/10.1016/j.femsle.2004.11.048
- Watson, J.M. and Holloway, B.W. (1978) Chromosome Mapping in Pseudomonas aeruginosa. Journal of Bacteriology, 133, 1113-1125. http://dx.doi.org/10.103/00006450-01000-00007
- Chuanchuen, R., Beinlich, K., Hoang, T.T., Becher, A., Karkhoff-Schweizer, R.R. and Schweizer, H.P. (2001) Cross- Resistance between Triclosan and Antibiotics in Pseudomonas aeruginosa Is Mediated by Multidrug Efflux Pumps: Exposure of a Susceptible Strain to Triclosan Selects nfxB Mutants Overexpressing MexCD-OprJ. Antimicrobial Agents and Chemotherapy, 45, 428-432. http://dx.doi.org/10.1128/AAC.45.2.428-432.2001
- Chuanchuen, R., Narasaki, C.T. and Schweizer, H.P. (2002) The MexJK Efflux Pump of Pseudomonas aeruginosa Requires OprM for Antibiotic Efflux but Not for Efflux of Triclosan. Journal of Bacteriology, 184, 5036-5044. http://dx.doi.org/10.1128/JB.184.18.5036-5044.2002
- Poonsuk, K., Tribuddharat, C. and Chuanchuen, R. (2013) Aminoglycoside Resistance Mechanisms in Pseudomonas aeruginosa Isolates from Non-Cystic Fibrosis Patients in Thailand. Canadian Journal of Microbiology, 59, 51-56. http://dx.doi.org/10.1139/cjm-2012-0465
- Woods, C.R., Versalovic, J., Koeuth, T. and Lupski, J.R. (1993) Whole-Cell Repetitive Element Sequence-Based Polymerase Chain Reaction Allows Rapid Assessment of Clonal Relationships of Bacterial Isolates. Journal of Clinical Microbiology, 31, 1927-1931.
- CLSI. (2013) VET01-A4: Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard. 4th Edition. Clinical and Laboratory Standards Institute, Wayne, PA.
- Fang, X., Fang, Z., Zhao, J., Zou, Y., Li, T., Wang, J., et al. (2012) Draft Genome Sequence of Pseudomonas aeruginosa Strain ATCC 27853. Journal of Bacteriology, 194, 3755. http://dx.doi.org/10.1128/JB.00690-12
- Winsor, G.L., Lam, D.K., Fleming, L., Lo, R., Whiteside, M.D., Yu, N.Y., et al. (2011) Pseudomonas Genome Database: Improved Comparative Analysis and Population Genomics Capability for Pseudomonas genomes. Nucleic Acids Research, 39, D596-D600. http://dx.doi.org/10.1093/nar/gkq869
- Lamers, R.P., Cavallari, J.F. and Burrows, L.L. (2013) The Efflux Inhibitor Phenylalanine-Arginine Beta-Naphthy- lamide (PABetaN) Permeabilizes the Outer Membrane of Gram-Negative Bacteria. PLoS ONE, 8, e60666. http://dx.doi.org/10.1371/journal.pone.0060666
- Kohler, T., Michea-Hamzehpour, M., Henze, U., Gotoh, N., Curty, L.K. and Pechere, J.C. (1997) Characterization of MexE-MexF-OprN, a Positively Regulated Multidrug Efflux System of Pseudomonas aeruginosa. Molecular Microbiology, 23, 345-354. http://dx.doi.org/10.1046/j.1365-2958.1997.2281594.x
- Westbrock-Wadman, S., Sherman, D.R., Hickey, M.J., Coulter, S.N., Zhu, Y.Q., Warrener, P., et al. (1999) Characterization of a Pseudomonas aeruginosa Efflux Pump Contributing to Aminoglycoside Impermeability. Antimicrobial Agents and Chemotherapy, 43, 2975-2983.
- Masuda, N., Sakagawa, E., Ohya, S., Gotoh, N., Tsujimoto, H. and Nishino, T. (2000) Substrate Specificities of MexAB-OprM, MexCD-OprJ and MexXY-OprM Efflux Pumps in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy, 44, 3322-3327. http://dx.doi.org/10.1128/AAC.44.12.3322-3327.2000
- Murata, T., Kuwagaki, M., Shin, T., Gotoh, N. and Nishino, T. (2002) The Substrate Specificity of Tripartite Efflux Systems of Pseudomonas aeruginosa Is Determined by the RND Component. Biochemical and Biophysical Research Communications, 299, 247-251. http://dx.doi.org/10.1016/S0006-291X(02)02626-8
- Hocquet, D., Vogne, C., El Garch, F., Vejux, A., Gotoh, N., Lee, A., et al. (2003) MexXY-OprM Efflux Pump Is Necessary for Adaptive Resistance of Pseudomonas aeruginosa to Aminoglycosides. Antimicrobial Agents and Chemotherapy, 47, 1371-1375. http://dx.doi.org/10.1128/AAC.47.4.1371-1375.2003
- Kohler, T., Michea-Hamzehpour, M., Plesiat, P., Kahr, A.L. and Pechere, J.C. (1997) Differential Selection of Multidrug Efflux Systems by Quinolones in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy, 41, 2540-2543.
- Ochs, M.M., McCusker, M.P., Bains, M. and Hancock, R.E. (1999) Negative Regulation of the Pseudomonas aeruginosa Outer Membrane Porin OprD Selective for Imipenem and Basic Amino Acids. Antimicrobial Agents and Chemotherapy, 43, 1085-1090.
- Wolter, D.J., Hanson, N.D. and Lister, P.D. (2008) Novel Mechanism of mexEF-oprN Efflux Pump Overexpression in Pseudomonas aeruginosa without Co-Regulation of oprD Expression. Abstracts. 48th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington DC, October 26 2008.
- Islam, S., Jalal, S. and Wretlind, B. (2004) Expression of the MexXY Efflux Pump in Amikacin-Resistant Isolates of Pseudomonas aeruginosa. Clinical Microbiology and Infection, 10, 877-883. http://dx.doi.org/10.1111/j.1469-0691.2004.00991.x
- Poonsuk, K. and Chuanchuen, R. (2012) Contribution of the MexXY Multidrug Efflux Pump and Other Chromosomal Mechanisms on Aminoglycoside Resistance in Pseudomonas aeruginosa Isolates from Canine and Feline Infections. The Journal of Veterinary Medical Science, 74, 1575-1582. http://dx.doi.org/10.1292/jvms.12-0239