Open Journal of Medical Imaging
Vol.07 No.04(2017), Article ID:79508,12 pages
10.4236/ojmi.2017.74014
Application of Sparse-Coding Super-Resolution to 16-Bit DICOM Images for Improving the Image Resolution in MRI
Junko Ota, Kensuke Umehara, Naoki Ishimaru, Takayuki Ishida
Department of Medical Physics and Engineering, Graduate School of Medicine, Osaka University, Suita, Japan

Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: September 12, 2017; Accepted: September 27, 2017; Published: September 30, 2017
ABSTRACT
Purpose: To improve the image resolution of magnetic resonance imaging (MRI), conventional interpolation methods are commonly used to magnify images via various image processing approaches; however, these methods tend to produce artifacts. While super-resolution (SR) schemes have been introduced as an alternative approach to apply medical imaging, previous studies applied SR only to medical images in 8-bit image format. This study aimed to evaluate the effectiveness of sparse-coding super-resolution (ScSR) for improving the image quality of reconstructed high-resolution MR images in 16-bit digital imaging and communications in medicine (DICOM) image format. Materials and Methods: Fifty-nine T1-weighted images (T1), 84 T2-weighted images (T2), 85 fluid attenuated inversion recovery (FLAIR) images, and 30 diffusion-weighted images (DWI) were sampled from The Repository of Molecular Brain Neoplasia Data as testing datasets, and 1307 non-medical images were sampled from the McGill Calibrated Color Image Database as a training dataset. We first trained the ScSR to prepare dictionaries, in which the relationship between low- and high-resolution images was learned. Using these dictionaries, a high-resolution image was reconstructed from a 16-bit DICOM low-resolution image downscaled from the original test image. We compared the image quality of ScSR and 4 interpolation methods (nearest neighbor, bilinear, bicubic, and Lanczos interpolations). For quantitative evaluation, we measured the peak signal-to-noise ratio (PSNR) and structural similarity (SSIM). Results: The PSNRs and SSIMs for the ScSR were significantly higher than those of the interpolation methods for all 4 MRI sequences (PSNR: p < 0.001, SSIM: p < 0.05, respectively). Conclusion: ScSR provides significantly higher image quality in terms of enhancing the resolution of MR images (T1, T2, FLAIR, and DWI) in 16-bit DICOM format compared to the interpolation methods.
Keywords:
Super-Resolution, Sparse-coding Super-Resolution (ScSR), MRI, DICOM, 16-Bit

1. Introduction
The spatial resolution of magnetic resonance imaging (MRI) is a crucial factor related to image quality, and affects the identification of anatomical features in medical imaging. However, as the image resolution is proportional to the scanning time, it is often difficult to obtain high-resolution images, especially in terms of MRI.
In order to improve the image resolution by image processing, conventional linear interpolation methods such as nearest neighbor, bilinear, and bicubic interpolations [1] can be used to magnify low-resolution images, although these interpolation methods can also cause artifacts (i.e., jagging, blurring, and ringing). As an alternative image processing, the super-resolution (SR) method was first proposed by Huang and Tsai [2] [3] , and, to date, many studies have demonstrated the usefulness of SR schemes in medical imaging [4] [5] . Our previous studies revealed the utility of sparse coding-based super-resolution (ScSR), a representative example-based SR scheme, when applied to CT imaging [4] and chest X-rays [6] .
Existing SR schemes operate only on 8-bit images, because the SR schemes were originally proposed in computer vision with RGB color images. Accordingly, most previous SR studies have applied to use 8-bit medical images [3] [7] . However, according to the international digital imaging and communications in medicine (DICOM) standard, which was published by the medical imaging and technology alliance [8] , MR images should be in 16-bit DICOM image format. Therefore, since the existing 8-bit-based SR methods are not suitable for MR images, the development of an SR scheme using 16-bit DICOM format is required.
In this study, we applied ScSR to four types of MR images, i.e., T1-weighted imaging (T1), T2-weighted imaging (T2), fluid attenuated inversion recovery (FLAIR) imaging, and diffusion-weighted imaging (DWI) in 16-bit DICOM format and evaluated its performance on improving image quality while magnifying the images.
2. Methods
2.1. Sparse-Coding-Based Super-Resolution [9] [10]
In Equation (1), X is a high-resolution image and Y is a low-resolution image based on the known degradation model.
(1)
Further, S is a downscaling function and H is the blurring factor. To restore high-resolution images remains an ill-posed problem because of the above reconstruction constraints.
On the other hand, sparse coding contributes to finding a concise expression of the signal. For a given unlabeled image, the input image is represented by a weighted linear combination of a small number of bases with more meaningful features.
The ScSR method is an algorithm used for solving the ill-posed problem of recovering a high-resolution image from a low-resolution image using the above sparse coding algorithm. Figure 1 shows an overview of the ScSR scheme.
In the ScSR method, Dl (low-resolution dictionary) and Dh (high-resolution dictionary), selected bases with highly meaningful features from training images learned to efficiently represent input images, are prepared before reconstruction of a high-resolution image (output image) from a low-resolution image (input image). The problem of finding the sparsest representation of a patch y (a low-resolution image patch) can be formulated as follows:
(2)
where F is four 1-D high-pass filters used to find representation more efficiently, as summarized below:
(3)
and is a vector of coefficients of a sparse linear combination (w1-n in Figure 1). However, Equation (2) is non-deterministic polynomial-hard; when coefficients are adequately sparse, they can be effectively restored by minimizing the l1-norm:
(4)
After the training, each patch of the input image (low-resolution) is searched for a sparse representation of Dl, represented as downscaled image patches of the
Figure 1. Overview of the sparse-coding super-resolution (ScSR) scheme. Abbreviations: Dl, low-resolution dictionary; Dh, high-resolution dictionary; w1/2/3∙∙∙, coefficients of the sparse representation.
high-resolution image patches. Subsequently, , which is the corresponding vector of , is used to reconstruct the high-resolution output image.
(5)
2.2. Experiments
2.2.1. Dataset
In this study, we used the brain MRI dataset with brain cancers provided by The Repository of Molecular Brain Neoplasia Data [11] . This dataset is publicly available from the cancer imaging archive [12] . We arbitrarily sampled MR images from The Repository of Molecular Brain Neoplasia Data, only in cases which MRI sequences can be identified according to DICOM header information. The sampled cases comprised 59 T1-weighted, 84 T2-weighted, 85 FLAIR, and 30 DWI images. For training, 1307 non-medical images were sampled from the McGill Calibrated Color Image Database [13] .
2.2.2. Applying 16-Bit DICOM to ScSR
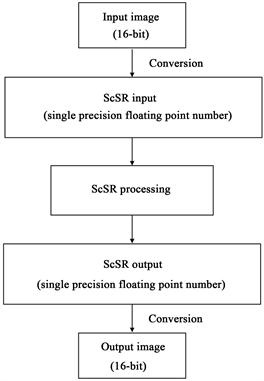
In our previous studies, we first transformed 8-bit bitmap images to a single precision floating point number to efficiently solve the optimization problem in ScSR, after which a single precision floating point number, as a result of the optimization problem, was transformed to 8-bit bitmap image format. In the present study, we inputted a 16-bit DICOM image instead of an 8-bit bitmap image and outputted a 16-bit DICOM image after the optimization problem using a single precision floating point number (Figure 2).
Figure 2. How to apply 16-bit DICOM images to sparse-coding super-resolution (ScSR).
2.2.3. Comparison of the ScSR Scheme with Conventional Interpolation Methods
First, we prepared low-resolution images by downscaling (128 × 128 pixels). Next, we reconstructed the high-resolution images from the prepared low-resolution images using ScSR to magnify by 2× to obtain the resulting image at the same matrix size as the original image (256 × 256 pixels). The same experiment was also performed using nearest neighbor, bilinear, bicubic, and Lanczos interpolations to compare the ScSR and interpolation methods (Figure 3). The Lanczos interpolation method is reported to be superior to other linear interpolation methods because it outperforms them in detecting edge and linear features [14] . Lanczos kernel 2 is used in the Lanczos interpolation method. Subsequently, we measured the peak signal-to-noise ratio (PSNR) [15] and structural similarity (SSIM) [16] , two metrics reflecting image quality. The PSNR measures the image quality based on the pixel difference between two images. The SSIM is used to evaluate the similarity between two images as a means to assess the perceptual image quality. As SSIM is generally calculated in a local region of the target image to assess structural similarity locally, we measured 5 regions of interest in each test image and evaluated the mean SSIM of these 5 regions of interest.
2.2.4. Statistical Analysis
The statistical differences in the PSNR and SSIM between the interpolation and ScSR methods were analyzed by one-way analysis of variance and Tukey’s post-hoc test. For all analyses, p-values < 0.05 were considered significant. All statistical analyses were performed using R version 3.2.2 (The R Foundation for Statistical Computing, Vienna, Austria).
3. Results
3.1. Comparison of Image Quality
3.1.1. T1
Figure 4 and Figure 5 show the PSNRs and SSIMs, respectively, of the five
Figure 3. Overview of the experiments.
Figure 4. Comparisons of the peak signal-to-noise ratio (PSNR) of each magnification method for the 4 magnetic resonance imaging sequences: (a) T1; (b) T2; (c) fluid attenuated inversion recovery (FLAIR); (d) diffusion-weighted imaging (DWI). ScSR, sparse-coding super-resolution.
Figure 5. Comparisons of the structural similarity (SSIM) of each magnification method for the 4 magnetic resonance imaging sequences: (a) T1; (b) T2; (c) fluid attenuated inversion recovery (FLAIR); (d) diffusion-weighted imaging (DWI). Abbreviation: ScSR, sparse-coding super-resolution.
magnification schemes for the 4 different MRI sequences. For T1-weighted imaging, the mean ± SD PSNRs of the nearest neighbor, bilinear, bicubic, Lanczos, and ScSR methods were 66.57 ± 2.26, 67.90 ± 2.46, 69.73 ± 2.59, 69.82 ± 2.60, and 72.66 ± 2.49 dB, respectively (Figure 4(a)); the corresponding mean ± SD SSIMs were 0.99958 ± 0.00020, 0.99970 ± 0.00016, 0.99979 ± 0.00012, 0.99979 ± 0.00012, and 0.99989 ± 0.00006, respectively (Figure 5(a)).
Table 1 and Table 2 show the statistical results of the PSNR and SSIM, respectively. In brief, the PSNR was significantly higher for ScSR than for all interpolation methods (p < 0.001 for all) (Table 1). Similarly, ScSR showed significantly better results than the linear interpolation methods in terms of the SSIM (p < 0.01 for all) (Table 2).
3.1.2. T2
The mean ± SD PSNRs for the nearest neighbor, bilinear, bicubic, Lanczos, and ScSR methods were 66.26 ± 2.22, 67.23 ± 2.35, 68.83 ± 2.40, 68.90 ± 2.40, and 71.47 ± 2.37 dB, respectively, for T2-weighted imaging (Figure 4(b)). The corresponding SSIMs were 0.99946 ± 0.00027, 0.99957 ± 0.00022, 0.99969 ± 0.00017, 0.99970 ± 0.00016, and 0.99983 ± 0.00010, respectively (Figure 5(b)).
Table 1. Comparisons of the peak signal-to noise ratio (PSNR) of each magnification method for the 4 MRI sequences; T1-weighted imaging (T1), T2-weighted imaging (T2), fluid attenuated inversion recovery imaging (FLAIR), and diffusion-weighted imaging (DWI).
Abbreviations: ScSR, Sparse-coding Super-Resolution; CI, Confidence Interval.
Table 2. Comparisons of the structural similarity (SSIM) of each magnification method for the 4 MRI sequences: T1-weighted imaging (T1), T2-weighted imaging (T2), fluid attenuated inversion recovery imaging (FLAIR) imaging, and diffusion-weighted imaging (DWI).
Abbreviations: ScSR, Sparse-coding Super-Resolution; CI, Confidence Interval.
The results for T2-weighted imaging were similar to those for T1: for both the PSNR and SSIM, the ScSR method was significantly better than the interpolation methods (Table 1 and Table 2, respectively).
3.1.3. FLAIR
The mean ± SD PSNRs for the nearest neighbor, bilinear, bicubic, Lanczos, and ScSR methods were 70.52 ± 2.38, 71.17 ± 2.39, 72.53 ± 2.37, 72.59 ± 2.37, and 74.72 ± 2.21 dB, respectively for FLAIR imaging (Figure 4(c)); the corresponding SSIMs were 0.99982 ± 0.00010, 0.99985 ± 0.00009, 0.99988 ± 0.00006, 0.99989 ± 0.00006, and 0.99993 ± 0.00003, respectively (Figure 5(c)). For both the PSNR and SSIM measures, the SR methods were significantly better than the interpolation methods (PSNR, p < 0.001; SSIM, p < 0.01) (Table 1 and Table 2, respectively).
3.1.4. DWI
For DWI, the mean ± SD PSNRs for the nearest neighbor, bilinear, bicubic, Lanczos, and ScSR methods were 63.81 ± 0.14, 63.73 ± 0.10, 63.94 ± 0.10, 63.95 ± 0.10, and 64.25 ± 0.10 dB, respectively (Figure 4(d)); the mean ± SD SSIMs were 0.999965 ± 0.000016, 0.999966 ± 0.000014, 0.999981 ± 0.000006, 0.999982 ± 0.000006, and 0.999990 ± 0.000003, respectively (Figure 5(d)). For both the PSNR and SSIM, ScSR was significantly better than the interpolation methods (PSNR, p < 0.001; SSIM, p < 0.05) (Table 1 and Table 2, respectively).
3.2. Visual Examples
Figures 6-9 show examples of the resulting high-resolution images by all five magnification methods for 2× magnifications of the 4 different MRI sequences. ScSR generated clearly higher quality images (i.e., more similar to the original image) compared with the interpolation methods for all 4 sequences. The ScSR method also produced clearer edges compared to the other methods, without any obvious artifacts.
Figure 6. Representative reconstructed high-resolution T1-weighted images. (a) nearest neighbor, (b) bilinear, (c) bicubic, (d) Lanczos, (e) sparse-coding super-resolution, and (f) original region (ground-truth) image.
Figure 7. Representative reconstructed high-resolution T2-weighted images. (a) nearest neighbor; (b) bilinear; (c) bicubic; (d) Lanczos; (e) sparse-coding super-resolution; and (f) original region (ground-truth) image.
Figure 8. Representative reconstructed high-resolution fluid attenuated inversion recovery images. (a) nearest neighbor; (b) bilinear; (c) bicubic; (d) Lanczos; (e) sparse-coding super-resolution; and (f) original region (ground-truth) image.
Figure 9. Representative reconstructed high-resolution diffusion-weighted images. (a) nearest neighbor, (b) bilinear, (c) bicubic, (d) Lanczos, (e) sparse-coding super-resolution, and (f) original region (ground-truth) image.
4. Discussion
In the present study, we used ScSR to improve the quality of 16-bit DICOM MR images when magnifying the images, and compared the resulting image quality to that of other common interpolation methods. As a result, the ScSR method yielded a significantly higher image quality than the interpolation methods using 16-bit DICOM, for two different image quality metrics.
Previous studies using 8-bit medical images have indicated the effectiveness of ScSR [3] [4] [7] ; however, the performance of ScSR with 16-bit DICOM has not been evaluated to date, even though most medical images are digitized with a resolution of 16-bit. As not only ScSR but also other example-based SR techniques often use 8-bit image format, loss of information is a concern when performing SR.
Our experimental results showed that the ScSR scheme also yields a significantly higher image quality than the interpolation methods using 16-bit DICOM in 4 kinds of MRI datasets (PSNR: p < 0.001, SSIM: p < 0.05, respectively), suggesting that the ScSR does not need to convert images in 16-bit DICOM format to 8-bit image format. Whether the improved performance of ScSR with 16-bit DICOM images is due to using other medical images, or a different factor altogether, will require further study.
There are several limitations in this study that need to be acknowledged. In ScSR, to minimize the l1-norm to effectively seek optimal image bases, single or double precision floating point numbers need to be converted from the original image format. This conversion will cause a loss of information of 16-bit DICOM. Consequently, further improvement will be needed to obtain higher image quality as a means to sustain the information of the original image. Additionally, we used 8-bit non-medical images as the training images. Although this study revealed the ability to use a 16-bit DICOM input in the testing phase of the ScSR, as compared to conventional interpolation methods, in the training phase, dictionaries were generated after conversion of the training images to a single or double precision floating point number from 8-bit image format. Further, loss of information when converting the training image format is another concern, despite using 16-bit DICOM images in the training phase as well. Further studies will be needed to identify whether 16-bit DICOM images would be effective for training the ScSR to enhance the resolution of 16-bit DICOM images.
5. Conclusion
Herein, we applied and evaluated the ScSR method for improvement of image quality of magnified MR images (T1-weighted, T2-weighted, FLAIR, and DWI images) in16-bit DICOM format. Our results suggest that the ScSR scheme using 16-bit DICOM outperformed the current interpolation methods used to enhance the image resolution of MRI.
Conflicts of Interest
The authors report no conflicts of interest related to this study.
Cite this paper
Ota, J., Umehara, K., Ishimaru, N. and Ishida, T. (2017) Application of Sparse-Coding Super-Resolution to 16-Bit DICOM Images for Improving the Image Resolution in MRI. Open Journal of Medical Imaging, 7, 144-155. https://doi.org/10.4236/ojmi.2017.74014
References
- 1. Siu, W.C. and Hung, K.W. (2012) Review of Image Interpolation and Super-Resolution. Proceedings of the 2012 Asia Pacific Signal and Information Processing Association Annual Summit and Conference, California, 3-6 December 2012, 1-10.
- 2. Huang, T. and Tsai, R. (1984) Multi-Frame Image Restoration and Registration. In: Advances in Computer Vision and Image Processing, JAI Press, Inc. Greenwich, CT, USA, 317-339.
- 3. Tang, S., Guo, H., Zhou, N., Huang, L. and Zhan, T. (2016) Coupled Dictionary Learning on Common Feature Space for Medical Image Super Resolution. Proceedings of the IEEE Conference on Image Processing, Phoenix, 25-28 September 2016, 574-578. https://doi.org/10.1109/ICIP.2016.7532422
- 4. Ota, J., Umehara, K., Ishimaru, N., Ohno, S., Okamoto, K., Suzuki, T., Shirai, N. and Ishida, T. (2017) Evaluation of the Sparse Coding Super-Resolution Method for Improving Image Quality of Up-Sampled Images in Computed Tomography. Proceedings of SPIE Medical Imaging 2017: Image Processing, 10133, Orlando, 11-16 February, 101331S-1-101331S-9. https://doi.org/10.1117/12.2253582
- 5. Umehara, K., Ota, J., Ishimaru, N., Ohno, S., Okamoto, K., Suzuki, T., Shirai, N. and Ishida, T. (2017) Super-Resolution Convolutional Neural Network for the Improvement of the Image Quality of Magnified Images in Chest Radiographs. Proceedings of SPIE Medical Imaging 2017: Image Processing, 10133, Orlando, 11-16 February, 101331P-1-101331P-7. https://doi.org/10.1117/12.2249969
- 6. Ota, J., Umehara, K., Ishimaru, N., Ohno, S., Okamoto, K., Suzuki, T., Shirai, N. and Ishida, T. (2016) Comparative Evaluation of Super-Resolution Methods Using Sparse Coding and Deep Convolutional Neural Network for Improving Image Quality of Extended Images in Chest Radiograph. Proceedings of RSNA, Chicago, 27 November - 2 December 2016.
- 7. Trinh, D.H., Luong, M., Dibos, F., Rocchisani, J.M., Pham, C.D. and Nguyen, T.Q. (2014) Novel Example-Based Method for Super-Resolution and Denoising of Medical Images. IEEE Transactions on Image Processing, 23, 1882-1895. https://doi.org/10.1109/TIP.2014.2308422
- 8. NEMA (2017) The DICOM Standard. http://dicom.nema.org/medical/dicom/current/output/html/part03.html
- 9. Yang, J., Wright, J., Huang, T. and Ma, Y. (2008) Image Super-Resolution as Sparse Representation of Raw Image Patches. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Anchorage, 23-28 June 2008, 1-8. https://doi.org/10.1109/CVPR.2008.4587647
- 10. Yang, J., Wright, J., Huang, T.S. and Ma, Y. (2010) Image Super-Resolution via Sparse Representation. IEEE Transactions on Image Processing, 19, 2861-2873. https://doi.org/10.1109/TIP.2010.2050625
- 11. Scarpace, L., Flanders, A.E., Jain, R., Mikkelsen, T. and Andrews, D.W. (2015) Data from REMBRANDT. The Cancer Imaging Archive.
- 12. Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., Moore, S., Phillips, S., Maffitt, D., Pringle, M., Tarbox, L. and Prior, F. (2013) The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Journal of Digital Imaging, 26, 1045-1057.
- 13. Fred Kingdom’s Laboratory at McGill Vision Research (2015) McGill Calibrated Colour Image Database. http://tabby.vision.mcgill.ca/html/browsedownload.html
- 14. Iqbal, M.Z., Ghafoor, A. and Siddiqui, A.M. (2013) Satellite Image Resolution Enhancement using Dual-Tree Complex Wavelet Transform and Nonlocal Means. IEEE Geoscience and Remote Sensing Letters, 10, 451-455. https://doi.org/10.1109/LGRS.2012.2208616
- 15. Huynh-Thu, Q. and Ghanbari, M. (2008) Scope of Validity of PSNR in Image/Video Quality Assessment. Electronics Letters, 44, 800-801. https://doi.org/10.1049/el:20080522
- 16. Wang, Z., Bovik, A.C., Sheikh, H.R. and Simoncelli, E.P. (2004) Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Transactions on Image Processing, 13, 600-612. https://doi.org/10.1109/TIP.2003.819861