Open Journal of Blood Diseases
Vol.3 No.3(2013), Article ID:36933,9 pages DOI:10.4236/ojbd.2013.33018
SNPs and TFBS Associated with High Altitude Sickness*
![]()
1Department of Pediatrics, University of Washington, Seattle, USA; 2Division of Cardiology, Seattle Children’s Hospital, Institute, Foundation, Seattle, USA; 3Laboratory of Hypoxia Physiology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China; 4People’s Hospital of the Tibet Autonomous Region, Lhasa, China; 5Center for Cardiovascular Biology and Regenerative Medicine, University of Washington, Seattle, USA.
Email: #nburoker@u.washington.edu
Copyright © 2013 Norman E. Buroker et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received May 7th, 2013; revised June 9th, 2013; accepted June 17th, 2013
Keywords: High Altitude Sickness; rSNPs; TFBS
ABSTRACT
The rSNPs for the genes AKT3 (rs4590656), EGLN1 (rs480902), eNOS3 (rs1007311), and VEGFA (rs699947, rs13207311, rs1570360, rs2010963) have been significantly associated with the physiological parameters in high altitude sickness Han or Tibetan Chinese patients at the Qinghai-Tibetan plateau. The alleles of each rSNP have been found to create unique transcriptional factor binding sites for transcription factors that affect the process of hypoxia gene expression in this high altitude hypoxia environment.
1. Introduction
GWAS over the last decade have identified nearly 6500 diseases or trait-predisposing simple nucleotide polymerphisms (SNPs) where only 7% of these are located in protein-coding regions of the genome [1,2] and the remaining 93% are located within non-coding areas [3,4] such as regulatory or intergenic regions. Expression quantitative mapping (eQLT) [5] has been used to identify SNPs that influence gene expression in regulatory regions [6] and to study large intergenic non-coding RNA expression [4]. eQLT SNPs in regulatory regions have been predicted to alter potential transcriptional factor binding sites (TFBS) implicated in the pathogenesis of chronic lymphocytic leukemia [7]. SNPs in the regulatory region of the prion protein (PRNP) gene may play an important role in the pathogenesis of sporadic Creutzfeldt-Jakob disease [8]. SNPs which occur in the putative regulatory region of a gene where a single base change in the DNA sequence of a potential TFBS may affect the process of gene expression are drawing more attention [9-11]. In this report, we coalesce our previous work to draw attention to seven SNPs in four genes (AKT3, ENGL1, eNOS3 and VEGFA) which alter punitive TFBS and are significantly (p < 0.05) associated with physiological parameters of individuals with high altitude sickness (HAS) [12-14].
HAS arises from two different diseases which are acute and chronic mountain sickness. Acute mountain sickness (AMS) is very common in lowlanders who ascend from sea level to altitudes greater than 2600 meters and is characterized by headache, lightheadedness, breathlessness, fatigue, insomnia, anorexia, and nausea [15, 16]. Symptoms begin two to three hours after ascent. The condition is generally self limiting; most symptoms disappear after two to three days, although insomnia may persist [17]. AMS must be treated as an emergency; the illness will resolve if no further altitude is gained however in some cases descent to a lower altitude may be necessary in order to reverse the condition. Chronic mountain sickness (CMS) is characterized by polycythemia, excessive erythrocytosis and severe hypoxemia, which is reversible upon descent from high altitudes [18,19]. Hematologic, neurologic, cardiac and respiratory symptoms are manifestations of the disease. The most common symptoms are bone and muscle pain, headaches, dizziness, dyspnea, insomnia, tinnitus, mental fatigue, and a loss of appetite. The severity of the condition increases with advancing age. CMS is a syndrome resulting from the loss of human adaptation to high altitude and can occur in permanent residents residing in this environment [20,21]. The precise pathogenesis of AMS and CMS is not well understood, but hypoxia is likely to be a major factor [22-26]. This raises the question of why some individuals are susceptible to AMS and CMS while others are not, under the same hypoxic conditions. Tibetans may be one of the oldest high-altitude adapted ethnic groups in the world with origins from the Neolithic period based on current genetic data [27-30]. Although AMS and CMS are different diseases and are treated differently, they both arise in humans at high altitudes.
SNPs that affect gene expression by impacting gene regulatory sequences such as promoters, enhances, and silencers are known as regulatory SNPs (rSNPs) [9,10,31, 32]. A rSNP in a TFBS can have multiple consequences. Most often the rSNP does not change the TFBS interaction nor does it alter gene expression since a transcriptional factor (TF)will usually recognize a number of different binding sites in the gene. In some cases the rSNP may increase or decrease the TF binding which results in allele-specific gene expression. In rare cases, a rSNP may eliminate the natural binding site or generate a new binding site. In which cases the gene is no longer regulated by the original TF. Therefore, functional rSNPs in TFBS may result in differences in gene expression, phenotypes and susceptibility to environmental exposure [11]. Examples of rSNPs associated with diseases susceptibility are numerous and several reviews have been published [11,33-35]. The purpose of this report is to hi-lite the most recent rSNPs associated with HAS in Han and Tibetan Chinese at the Qinghai-Tibetan plateau.
2. Materials and Methods
2.1. Study Groups
The two Chinese ethnic groups studied were the Han who are considered upward migrants from low altitudes, and the Tibetans who are high altitude natives. AMS was studied in association with the Han while CMS was studied in association with the Tibetans resulting in two different HAS groups compared with their respective ethnic controls. All mountain sickness patients in this study had been hospitalized and diagnosed at the Lhasa People Hospital (Tibet, China at 3670 M above sea level) from 2002 to 2008. The CMS patients and Tibetan controls normally live at 3600 - 4400 M. AMS was diagnosed by using the current consensus of mountain sickness in Tibet (Diagnosis and Therapeutics for Mountain Sickness, Xizang Autonomous Region), which is in accord with the Lake Louise scoring system [36]. The Lake Louise consensus on the definition and quantification of altitude illness [36] was the Qinghai diagnostic criteria for measuring CMS. We sampled Han AMS patients from the hospital with symptoms of acute pulmonary edema as diagnosed by a cough accompanied with pink frothy sputum. Moist or bubbling rales in the lungs was suggestive of pulmonary oedema, showing a characteristic shadow on chest X-rays. In addition to the characteristic symptoms of severe acute mountain response, acute cerebraledema was diagnosed by ataxia, disturbance of consciousness or coma, abnormal plantar reflexes and paplilledema. The AMS Han patients were new comers from the low land and acquired the illness within two days after arriving at the higher altitude of Tibet. We also sampled Tibetan CMS patients as diagnosed by erythropoiesis, pulmonary hypertension and/or high arterial blood pressure, right ventricular hypertrophy or right and left ventricular hypertrophy. AMS patients had an average age of 35.3 years, while CMS patients had an average age of 53.6 years. Patients with other diseases having similar clinical manifestations were excluded. Healthy Tibetan and Han people from the Lhasa area were randomly selected to serve as control subjects. All patients and controls sampled in the study signed an informed consent approved by the Human Ethics Committee of the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
2.2. TFBS
The JASPAR CORE database [37,38] and ConSite [39] were used to identify the TFBS in this study. JASPAR is a collection of transcription factor DNA-binding preferences used for scanning genomic sequences where ConSite is a web-based tool for finding cis-regulatory elements in genomic sequences.
3. Results
3.1. Physiological Study
The average physiological parameters for the mountain sickness (AMS and CMS) and normal control study groups are listed (Table 1). The arterial oxygen saturation (SaO2) was significantly lower in both the AMS and CMS compared to the normal study group. The hemoglobin (Hb) concentration, red blood cell (RBC) count and hematocrit (Hct) were significantly higher among CMS patients compared to the AMS patients and the normal controls (Table 1). The heart rate (Hr) was significantly higher for the AMS patients than either the CMS patients or normal study groups. There was no significant difference in heart rate between the sexes. All the measured values of controls are in the normal ranges that are currently being used in the Lhasa region. Most AMS patients consist of new comers or short time residents to the high altitude environment of Tibet where the average age was 35.3 ± 1 years, while most CMS patients were permanent residents of the high altitude environment with a average age of 53.6 ± 1.7 years. It should be noted that the average age is 30.1 for the Tibetan control group.
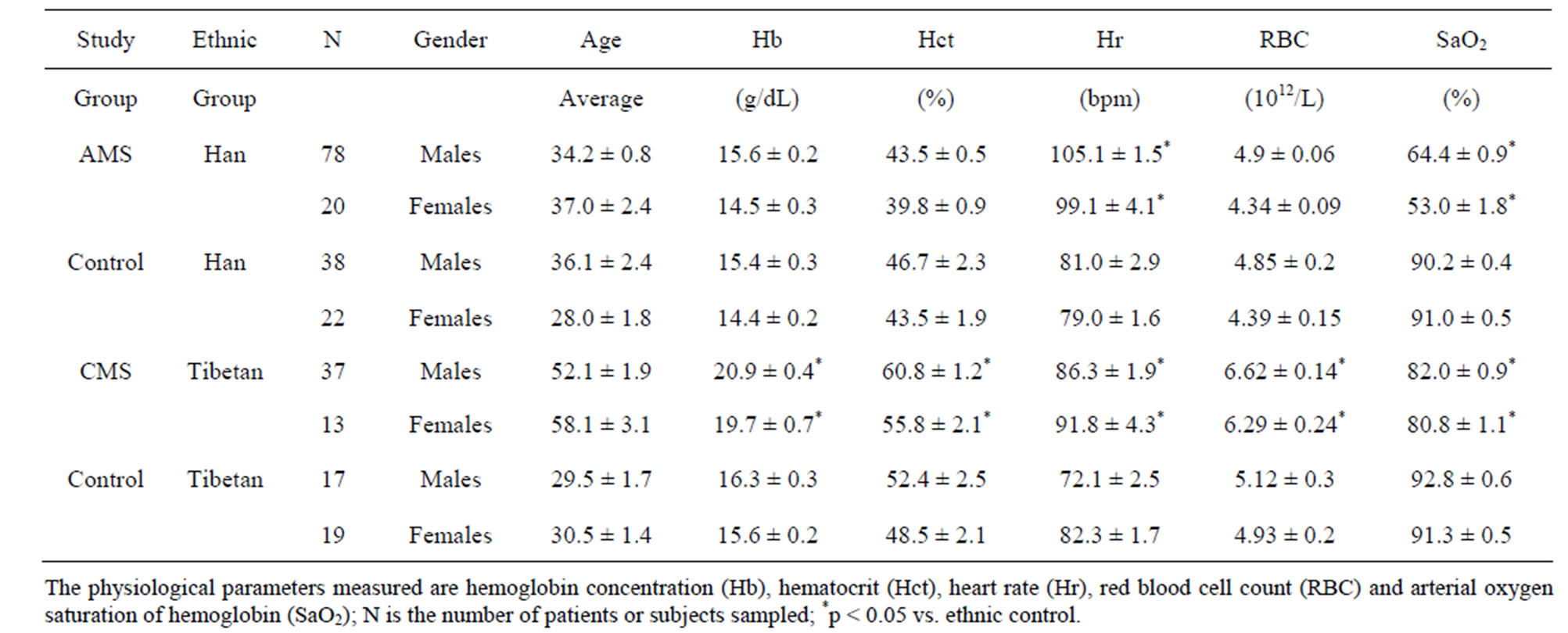
Table 1. Listed are the gender, average ages and physiological parameters with standard errors for the AMS Han patients, CMS Tibetans patients and the corresponding control subjects.
The measured physiological values of the control study groups fall within the normal ranges of residents at the Lhasa region.
3.2. Physiological Parameters, SNPs and TFBS
A list of physiological parameters obtained from HAS patients which were found to be significantly (p < 0.05) associated with seven rSNPs in four genes are presented in Table 2 [12-14]. Also listed are the SNP alleles which are unique to the adjacent TFBS. The SNPs were also present in other TFBS which were not changed by the presence of the SNP and will not be discussed. A description of the TFs that bind the sites in Table 2 can be found in the supplement.
The v-akt murine thymoma viral oncogene homolog 3 (AKT3) gene transcribes is a serine/threonine kinase that plays a key role in regulating cell survival, insulin signaling, angiogenesis and tumor formation. The AKT3 SNP (rs4590656) has been found to be significantly associated with Hb and Hct in Tibetan Chinese with CMS [13]. The AKT3-C allele creates two unique TFBS for the ARNT:AHR and HIF1α:ARNT TFs which are involved xenobiotic metabolism and cellular and systemic responses to hypoxia, respectively. The AKT3-T allele creates two other unique TFBS for the HNF4 and PAX2 TFs which are involved in the expression of several hepatic genes and the conserved DNA-binding paired box, respectively (Table 2 & Supplement).
The egl nine homolog1 (EGLN1) gene is a key oxygen sensor that negatively regulates the activity of the hypoxia-inducible factor-1alpha (HIF1α). Hypoxia leads to the inactivation of EGLN1 thereby increasing HIF activity that induces the expression of genes which mediates the adaptive responses through glycolytic enzymes, hemeoxygenase, vascular endothelial growth factor and erythropoietin (EPO) [40]. The EGLN1 SNP (rs480902) has been found to be significantly associated with Hct, Hr and SaO2 in Han Chinese with AMS [12]. The EGLN1- C allele does not generate any known TFBS while the EGLN1-T allele creates three unique TFBS for the NFE2L1:MAFG, GATA3 and FOXL1 TFs which are involved with the antioxidant response element (ARE) and cell differentiation of erythrocytes, zinc-finger TFs and fork head box L1 TF, respectively (Table 2 & Supplement).
The endothelial cell nitric oxide synthase 3 (eNOS3) gene encodes an enzyme that produces nitric oxide (NO) and is implicated in vascular smooth muscle relaxation. NO mediates vascular endothelial growth factor (VEGF)- induced angiogenesis in coronary vessels and promotes blood clotting through the activation of platelets. The eNOS3 SNP (rs1007311) has been found to be significantly associated with Hb in Tibetan Chinese with CMS [13]. The eNOS3-A allele creates one TFBS for the AP2 TF which is involved in recruiting transcription machinery. The eNOS3-G allele creates two TFBS for the MIZF and RUNX1 TFs which are involved with transcription repression and development, respectively (Table 2 & Supplement).
The vascular endothelial growth factor (VEGFA) gene is part of a signaling pathway where the VEGFA protein is a growth factor activator for angiogenesis, vasculogenesis and endothelial cell growth. The VEGFA gene has four SNPs in the promoter region that are significantly associated with SaO2 in Han Chinese with AMS and one SNP (rs1570360) which is significantly associated with RBC in Tibetan Chinese with CMS [13,14]. The VEGFA-A allele for the rs13207351 SNP creates two TFBS for the MIZF and NKX3-2 TFs which are involved

Table 2. Physiological parameters and rSNPs significantly (p < 0.05) associated with either AMS or CMS. Genes whose rSNPs alter the transcriptional factor binding sites (TFBS) in non-coding regulatory regions of the gene. The rSNP alleles are found only within the adjacent TFBS. Allele in bold indicates common allele for HAS.
with DNA methylation and transcription repression. The VEGFA-G allele creates two TFBS for the NFIC and TFCP2L1 TFs which are involved with DNA methylation and transcription repression (Table 2 & Supplement). The VEGFA-A allele for the rs1570360 SNP creates two TFBS for the SP1 and ZNG354C TFs which are involved with activation and repression of transcription in response to physiological and pathological stimuli (Table 2 & Supplement). The VEGFA-C allele for the rs2010963 SNP creates TFBS for the GABPα and IRF1, 2 TFs which are involved with cytochrome oxidase expression and interferon regulation, respectively. The VEGFA-G allele creates the TFBS for the SP1 TF which is involved with the activation and repression of transcription in response to physiological and pathological stimuli (Table 2 & Supplement). The VEGFA-C allele for the rs699947 SNP creates three TFBS for the GATA3, HIF1α:ARNT and TAL1:TCF3 TFs which are involved with regulating proliferation of hematopoietic and endocrine cell lineages, systemic responses to hypoxia and regulator of erythroid differentiation. The VEGFA-A allele creates the TFBS for the NFIC TF which is involved with activating transcription and replication (Table 2 & Supplement).
4. Discussion
HAS arises in humans that transcending from sea level to high altitude environments [41,42]. Ascent to high altitudes requires adaptation to a hypobaric hypoxic environment, while failure to adapt results in AMS [43]. Although the mechanisms of AMS are still a matter of active investigation, rapid ascent to altitude is the firmly established cause [41,42,44,45]. In our reports [12-14] Han Chinese that acquired AMS after arriving at the Qinghai-Tibetan plateau had a significantly higher Hr and lower SaO2 (Table 1). These physiological parameters have been significantly linked to rSNPs in the EGLN1 and VEGF A genes (Table 2). The EGLN1 gene is a key oxygen sensor that negatively regulates the activity of HIF1α. Hypoxia leads to the inactivation of EGLN1 there by increasing HIF activity that induces the expression of VEGF and EPO [40]. The EGLN1-T allele of the SNP (rs480902) creates the TFBS for the NFE2L1:MAFG TF which is involved with the antioxidant response element (ARE) and cell differentiation of erythrocytes (Table 2 & Supplement). The EGLN1-C allele which generates no known TFBS has a high incidence in AMS patients [Table 2, [12]]. The VEGFA gene, a growth factor activator for angiogenesis, vasculogenesis and endothelial cell growth, has four rSNPs significantly associated with SaO2 in AMS patients [Table 2, [13,14]]. The VEGFA alleles of the four rSNPs create unique TFBS in the promoter (Table 2). The common SNP alleles in bold create TFBS for GATA3, HIF1:ARNT, KLF4, MIZF, NFIC, SP1, TAL1:TCF3 and TFCP2L1 TFs that are prevalent in AMS patients. The stimulating protein-1 (SP1) participates in the transcription regulation of Kruppel-like factor-4 (KLF4), a zinc finger-containing TF and together they are part of a (SP1/KLF) family of TFs which play a role in diverse cellular processes, including vascular smooth muscle cell (VSMC) proliferation, cell differentiation, apoptosis and, on cogenic processes [46,47], induction of pluripotent stem cells [48] and control of gene transcription [49-52]. There have been at least 20 KLFs identified in mammals with each individually participating in one of the above biological functions [51]. KLF4 has been shown to play a key role in pathological vascular processes and is considered a molecular switch in regulating VSMC function [51]. The histone H4 (MIZF), nuclear factor I/C type (NFIC) and CP2-like1 (TFCP2L1) TFs are involved with transcription activation and suppression.
CMS results from the loss of human adaptation to high altitude and occurs in permanent residence residing in this environment for along period of time [20,21]. In our reports [12-14] Tibetan Chinese which acquired CMS after living at the Qinghai-Tibetan plateau had a significantly higher Hb, Hct, Hr, RBC and lower SaO2 (Table 1). The Hb, Hct and RBC physiological parameters have been significantly linked to the rSNP in the AKT3, eNOS3 and VEGFA genes. The AKT3 gene plays a key role in regulating cell survival, insulin signaling, angiogenesis. The AKT3 rSNP (rs4590656) has been significantly linked to Hb and Hct in CMS patients [Table 2, [13]] and the common AKT3-C allele associated with CMS creates a TFBS for the ARNT:AHR and HIF1α: ARNT TFs which are involved in xenobiotic metabolism and responds to hypoxia, respectively. The eNOS3 gene is implicated in vascular smooth muscle relaxation. The eNOS3 rSNP (rs1007311) has been significantly linked to Hb in CMS patients [Table 2, [13]] and the common eNOS3-A allele associated with CMS creates the TFBS for the AP2 TF which is involved with recruiting transcription machinery. The VEGFA rSNP (rs1570360) has been significantly linked to RBC in CMS patients [Table 2, [13]] and the common VEGFA-G allele associated with CMS creates the TFBS for the KLF4 and MIZF TFs which are discussed above.
In conclusion, we show that seven rSNPs from four genes are significantly associated with HAS in Han and Tibetan Chinese at the Qinghai-Tibetan plateau. These rSNPs alter the TFBS for TFs and create unique binding sites for TFs that affect the process of gene expression in this high altitude hypoxia environment.
5. Acknowledgements
This study was supported in part by the grants from Children’s Hospital and Regional Medical Center (HR5836), National Natural Science Foundation (No. 38970307 and No. 30393130) and National Basic Research Program of China “973” (No. 2006CB504100).
REFERENCES
- E. Pennisi, “The Biology of Genomes, Disease Risk Links to Gene Regulation,” Science, Vol. 332, No. 6033, 2011, p. 1031. doi:10.1126/science.332.6033.1031
- V. Kumar, C. Wijmenga and S. Withoff, “From GenomeWide Association Studies to Disease Mechanisms: Celiac Disease as a Model for Autoimmune Diseases,” Seminars in Immunopathology, Vol. 34, No. 4, 2012, pp. 567-580. doi:10.1007/s00281-012-0312-1
- L. A. Hindorff, P. Sethupathy, H. A. Junkins, E. M. Ramos, J. P. Mehta, F. S. Collins and T. A. Manolio, “Potential Etiologic and Functional Implications of GenomeWide Association Loci for Human Diseases and Traits,” Proceedings of the National Academy of Sciences, Vol. 106, No. 23, 2009, pp. 9362-9367. doi:10.1073/pnas.0903103106
- V. Kumar, H. J. Westra, J. Karjalainen, D. V. Zhernakova, T. Esko, B. Hrdlickova, R. Almeida, A. Zhernakova, E. Reinmaa, U. Vosa, M. H. Hofker, R. S. Fehrmann, J. Fu, S. Withoff, A. Metspalu, L. Franke and C. Wijmenga, “Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression,” PLoS Genetics, Vol. 9, No. 1, 2013, Article ID: e1003201. doi:10.1371/journal.pgen.1003201
- W. Cookson, L. Liang, G. Abecasis, M. Moffatt and M. Lathrop, “Mapping Complex Disease Traits with Global gene expression,” Nature Reviews Genetics, Vol. 10, No. 3, 2009, pp. 184-194. doi:10.1038/nrg2537
- T. Pastinen, “Genome-Wide Allele-Specific Analysis: Insights into Regulatory Variation,” Nature Reviews Genetics, Vol. 11, No. 8, 2010, pp. 533-538. doi:10.1038/nrg2815
- F. C. Sille, R. Thomas, M. T. Smith, L. Conde and C. F. Skibola, “Post-GWAS Functional Characterization of Susceptibility Variants for Chronic Lymphocytic Leukemia,” PLoS One, Vol. 7, No. 1, 2012, Article ID: e29632. doi:10.1371/journal.pone.0029632
- J. Bratosiewicz-Wasik, J. Smolen-Dzirba, C. Watala, A. J. Rozemuller, C. Jansen, W. Spliet, G. H. Jansen, T. J. Wasik and P. P. Liberski, “Association of the PRNP Regulatory Region Polymorphisms with the Occurrence of Sporadic Creutzfeldt-Jakob Disease,” Folia Neuropathologica, Vol. 50, No. 1, 2012, pp. 68-73.
- J. C. Knight, “Functional Implications of Genetic Variation in Non-Coding DNA for Disease Susceptibility and Gene Regulation,” Clinical Science, Vol. 104, No. 5, 2003, pp. 493-501. doi:10.1042/CS20020304
- X. Wang, D. J. Tomso, X. Liu and D. A. Bell, “Single Nucleotide Polymorphism in Transcriptional Regulatory Regions and Expression of Environmentally Responsive Genes,” Toxicology and Applied Pharmacology, Vol. 207, No. 2, 2005, pp. 84-90. doi:10.1016/j.taap.2004.09.024
- B. N. Chorley, X. Wang, M. R. Campbell, G. S. Pittman, M. A. Noureddine and D. A. Bell, “Discovery and Verification of Functional Single Nucleotide Polymorphisms in Regulatory Genomic Regions: Current and Developing Technologies,” Mutation Research/Reviews in Mutation Research, Vol. 659, No. 1-2, 2008, pp. 147-157. doi:10.1016/j.mrrev.2008.05.001
- N. E. Buroker, X. H. Ning, Z. N. Zhou, K. Li, W. J. Cen, X. F. Wu, W. Z. Zhu, C. R. Scott and S. H. Chen, “EPAS1 and EGLN1 Associations with High Altitude Sickness in Han and Tibetan Chinese at the Qinghai-Tibetan Plateau, Blood Cells,” Molecules & Diseases, Vol. 49, No. 2, 2012, pp. 67-73.
- N. E. Buroker, X. H. Ning, Z. N. Zhou, K. Li, W. J. Cen, X. F. Wu, W. Z. Zhu, C. R. Scott and S. H. Chen, “AKT3, ANGPTL4, eNOS3, and VEGFA Associations with High Altitude Sickness in Han and Tibetan Chinese at the Qinghai-Tibetan Plateau,” International Journal of Hematology, Vol. 96, No. 2, 2012, pp. 200-213.
- N. E. Buroker, X. H. Ning, Z. N. Zhou, K. Li, W. J. Cen, X. F. Wu, W. Z. Zhu, C. R. Scott and S. H. Chen, “VEGFA SNPs and Transcriptional Factor Binding Sites Associated with High Altitude Sickness in Han and Tibetan Chinese at the Qinghai-Tibetan Plateau,” The Journal of Physiological Sciences, Vol. 63, No. 3, 2013, pp. 183-193. doi:10.1007/s12576-013-0257-8
- P. H. Hackett and R. C. Roach, “High-Altitude Illness,” The New England Journal of Medicine, Vol. 345, No. 2, 2001, pp. 107-114. doi:10.1056/NEJM200107123450206
- P. Bartsch, D. M. Bailey, M. M. Berger, M. Knauth and R. W. Baumgartner, “Acute Mountain Sickness: Controversies and Advances,” High Altitude Medicine & Biology, Vol. 5, No. 2, 2004, pp. 110-124. doi:10.1089/1527029041352108
- X. H. Ning and S. P. Li, “Health Care at High Altitude—Self-Care Universal Health Book,” Shanghai Science and Technology Publishing House, Shanghai, 2006, pp. 66-68.
- C. Monge, “Chronic Mountain Sickness,” Physiological Reviews, Vol. 23, No. 1, 1943, pp. 166-184.
- R. M. Winslow and C. C. E. Monge, “Hypoxia, Polycythemia, and Chronic Mountain Sickness,” Johns Hopkins University Press, 1987.
- T. Y. Wu, W. S. Li, L. Y. Wei, et al., “A Preliminary Studies on the Diagonosis of Chronic Mountain Sickness in Tibetan Populations,” Press Commmittee of the 3rd World congress on Mountain Medicine and High Altitude Physiology, Matsumoto, 1998.
- F. Leon-Velarde, R. G. McCullough, R. E. McCullough and J. T. Reeves, “Proposal for Scoring Severity in Chronic Mountain Sickness (CMS). Background and Conclusions of the CMS Working Group,” Advances in Experimental Medicine and Biology, Vol. 543, 2003, pp. 339- 354. doi:10.1007/978-1-4419-8997-0_24
- J. B. West, “The Physiologic Basis of High-Altitude Diseases,” Annals of Internal Medicine, Vol. 141, No. 10, 2004, pp. 789-800. doi:10.7326/0003-4819-141-10-200411160-00010
- R. B. Schoene, “Illnesses at High Altitude,” Chest, Vol. 134, No. 2, 2008, pp. 402-416. doi:10.1378/chest.07-0561
- K. P. Strohl, “Lessons in Hypoxic Adaptation from High-Altitude Populations,” Sleep and Breathing, Vol. 12, No. 2, 2008, pp. 115-121. doi:10.1007/s11325-007-0135-9
- M. H. Wilson, S. Newman and C. H. Imray, “The Cerebral Effects of Ascent to High Altitudes,” The Lancet Neurology, Vol. 8, No. 2, 2009, pp. 175-191. doi:10.1016/S1474-4422(09)70014-6
- D. Martin and J. Windsor, “From Mountain to Bedside: Understanding the Clinical Relevance of Human Acclimatisation to High-Altitude Hypoxia,” Postgraduate Medical Journal, Vol. 84, No. 998, 2008, pp. 622-627. doi:10.1136/pgmj.2008.068296
- B. Su, J. Xiao, P. Underhill, R. Deka, W. Zhang, J. Akey, W. Huang, D. Shen, D. Lu, J. Luo, J. Chu, J. Tan, P. Shen, R. Davis, L. Cavalli-Sforza, R. Chakraborty, M. Xiong, R. Du, P. Oefner, Z. Chen and L. Jin, “Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age,” The American Journal of Human Genetics, Vol. 65, No. 6, 1999, pp. 1718-1724. doi:10.1086/302680
- B. Su, C. Xiao, R. Deka, M. T. Seielstad, D. Kangwanpong, J. Xiao, D. Lu, P. Underhill, L. Cavalli-Sforza, R. Chakraborty and L. Jin, “Y Chromosome Haplotypes Reveal Prehistorical Migrations to the Himalayas,” Human Genetics, Vol. 107, No. , 2000, pp. 582-590. doi:10.1007/s004390000406
- A. Torroni, J. A. Miller, L. G. Moore, S. Zamudio, J. Zhuang, T. Droma and D. C. Wallace, “Mitochondrial DNA Analysis in Tibet: Implications for the Origin of the Tibetan Population and Its Adaptation to High Altitude,” American Journal of Physical Anthropology, Vol. 93, No. 2, 1994, pp. 189-199. doi:10.1002/ajpa.1330930204
- R. Du, C. Xiao and L. L. Cavalli-Sforza, “Genetic Distances between Chinese Populations Calculated on Gene Frequencies of 38 Loci,” Science in China Series C: Life Sciences, Vol. 40, No. 6, 1997, pp. 613-621. doi:10.1007/BF02882691
- J. C. Knight, “Regulatory Polymorphisms Underlying Complex Disease Traits,” Journal of Molecular Medicine, Vol. 83, No. 2, 2005, pp. 97-109. doi:10.1007/s00109-004-0603-7
- X. Wang, D. J. Tomso, B. N. Chorley, H. Y. Cho, V. G. Cheung, S. R. Kleeberger and D. A. Bell, “Identification of Polymorphic Antioxidant Response Elements in the Human Genome,” Human Molecular Genetics, Vol. 16, No. 10, 2007, pp. 1188-1200. doi:10.1093/hmg/ddm066
- L. Prokunina and M. E. Alarcon-Riquelme, “Regulatory SNPs in Complex Diseases: Their Identification and Functional Validation,” Expert Reviews in Molecular Medicine, Vol. 6, No. 10, 2004, pp. 1-15. doi:10.1017/S1462399404007690
- P. R. Buckland, “The Importance and Identification of Regulatory Polymorphisms and Their Mechanisms of Action,” Biochimica et Biophysica Acta (BBA)—Molecular Basis of Disease, Vol. 1762, No. 1, 2006, pp. 17-28. doi:10.1016/j.bbadis.2005.10.004
- W. Sadee, D. Wang, A. C. Papp, J. K. Pinsonneault, R. M. Smith, R. A. Moyer and A. D. Johnson, “Pharmacogenomics of the RNA World: Structural RNA Polymorphisms in Drug Therapy,” Clinical Pharmacology & Therapeutics, Vol. 89, No. 3, 2011, pp. 355-365.
- O. H. Hackett and O. Oelz, “The Diagnoses Accord with the Lake Louise Scoring System,” In: G. C. J. R. Sutton and C. S. Houston, Eds., Hypoxia and Mountainsickness, Pergamon Press, New York, 1992, pp. 327-330.
- J. C. Bryne, E. Valen, M. H. Tang, T. Marstrand, O. Winther, I. da Piedade, A. Krogh, B. Lenhard and A. Sandelin, “JASPAR, the Open Access Database of Transcription Factor-Binding Profiles: New Content and Tools in the 2008 Update,” Nucleic Acids Research, Vol. 36, Suppl. 1, 2008, pp. D102-D106. doi:10.1093/nar/gkm955
- A. Sandelin, W. Alkema, P. Engstrom, W. W. Wasserman and B. Lenhard, “JASPAR: An Open-Access Database for Eukaryotic Transcription Factor Binding Profiles,” Nucleic Acids Research, Vol. 32, Suppl. 1, 2004, pp. D91-D94. doi:10.1093/nar/gkh012
- A. Sandelin, W. W. Wasserman and B. Lenhard, “ConSite: Web-Based Prediction of Regulatory Elements Using Cross-Species Comparison,” Nucleic Acids Research, Vol. 32, Suppl. 2, 2004, pp. W249-W252. doi:10.1093/nar/gkh372
- S. Aggarwal, S. Negi, P. Jha, P. K. Singh, T. Stobdan, M. A. Pasha, S. Ghosh, A. Agrawal, B. Prasher and M. Mukerji, “EGLN1 Involvement in High-Altitude Adaptation Revealed through Genetic Analysis of Extreme Constitution Types Defined in Ayurveda,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 107, No. 44, 2010, pp. 18961-18966. doi:10.1073/pnas.1006108107
- S. A. Gallagher and P. H. Hackett, “High-Altitude Illness,” Emergency Medicine Clinics of North America, Vol. 22, No. 2, 2004, pp. 329-355. doi:10.1016/j.emc.2004.02.001
- B. Basnyat and D. R. Murdoch, “High-Altitude Illness,” The Lancet, Vol. 361, No. 9373, 2003, pp. 1967-1974. doi:10.1016/S0140-6736(03)13591-X
- S. J. Jackson, J. Varley, C. Sellers, K. Josephs, L. Codrington, G. Duke, M. A. Njelekela, G. Drummond, A. I. Sutherland, A. A. Thompson and J. K. Baillie, “Incidence and Predictors of Acute Mountain Sickness among Trekkers on Mount Kilimanjaro,” High Altitude Medicine & Biology, Vol. 11, No. 3, 2010, pp. 217-222. doi:10.1089/ham.2010.1003
- I. Singh, P. K. Khanna, M. C. Srivastava, M. Lal, S. B. Roy and C. S. Subramanyam, “Acute Mountain Sickness,” New England Journal of Medicine, Vol. 280, No. 4, 1969, pp. 175-184.
- B. Basnyat, J. Lemaster and J. A. Litch, “Everest or Bust: A Cross Sectional, Epidemiological Study of Acute Mountainsickness at 4243 Meters in the Himalayas,” Aviation, Space, and Environmental Medicine, Vol. 70, No. 9. 1999, pp. 867-873.
- M. Nemer and M. E. Horb, “The KLF Family of Transcriptional Regulators in Cardiomyocyte Proliferation and Differentiation,” Cell Cycle, Vol. 6, No. 2, 2007, pp. 117- 121. doi:10.4161/cc.6.2.3718
- Y. Liu, C. Zhang, J. Fan, L. Xiao, B. Yin, L. Zhou and S. Xia, “Comprehensive Analysis of Clinical Significance of Stem-Cell Related Factors in Renal Cell Cancer,” World Journal of Surgical Oncology, Vol. 9, 2011, p. 121. doi:10.1186/1477-7819-9-121
- K. Takahashi, K. Tanabe, M. Ohnuki, M. Narita, T. Ichisaka, K. Tomoda and S. Yamanaka, “Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors,” Cell, Vol. 131, No. 5, 2007, pp. 861-872. doi:10.1016/j.cell.2007.11.019
- T. Suzuki, K. Aizawa, T. Matsumura and R. Nagai, “Vascular Implications of the Kruppel-Like Family of Transcription Factors,” Arteriosclerosis, Thrombosis, and Vascularbiology, Vol. 25, 2005, pp. 1135-1141. doi:10.1161/01.ATV.0000165656.65359.23
- I. S. Kalra, M. M. Alam, P. K. Choudhary and B. S. Pace, “Kruppel-Like Factor 4 Activates HBG Gene Expression in Primary Erythroid Cells,” British Journal of Haematology, Vol. 154, No. 2, 2011, pp. 248-259. doi:10.1111/j.1365-2141.2011.08710.x
- J. H. Shi, B. Zheng, S. Chen, G. Y. Ma and J. K. Wen, “Retinoic Acid Receptor Alpha Mediates All-trans-Retinoic Acid-Induced Klf4 Gene Expression by Regulating Klf4 Promoter Activity in Vascular Smooth Muscle Cells,” The Journal of Biological Chemistry, Vol. 287, 2012, pp. 10799-10811. doi:10.1074/jbc.M111.321836
- P. M. Evans and C. Liu, “Roles of Krupel-Like Factor 4 in Normal Homeostasis, Cancer and Stem Cells,” Acta Biochimica et Biophysica Sinica, Vol. 40, No. 7, 2008, pp. 554-564.
Appendices
Supplement. Transcriptional factor binding sites, TFs and TF discription.



NOTES
*Conflict of interest: The authors have no conflicts of interest or financial ties to disclose.
#Correponding author.

