Open Journal of Biophysics
Vol.05 No.02(2015), Article ID:56029,17 pages
10.4236/ojbiphy.2015.52005
Spectroscopic Investigations of β-Amyloid Interactions with Propofol and L-Arginine
Saqer M. Darwish1*, Shurook Y. Aiaidah1, Imtiaz M. Khalid2, Musa M. Abuteir1, Lena Qawasmi3
1Physics Department, Al-Quds University, al-Bireh, Palestine
2University College for Educational Sciences
3Nano - Technology Center
Email: *sdarwish@science.alquds.edu
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 8 February 2015; accepted 28 April 2015; published 29 April 2015
ABSTRACT
Beta amyloid (Aβ) aggregation has been characterized to be responsible for several amyloid diseases. Fourier transform infrared (FTIR) spectroscopy, fluorescence, and atomic force microscopy (AFM) are used to investigate induced changes in the secondary structure of Aβ upon thermal denaturation and interaction with propofol and L-arginine. Spectral analysis has revealed an effective static quenching for the intrinsic fluorescence of Aβ by propofol and l-arginine with binding constants of 2.81 × 102 M−1 for Aβ-propofol and 0.37 × 102 M−1 for Aβ-L-arginine. Fourier self-de- convolution (FSD) technique has been used to evaluate the relative intensity changes in the spectra of the component bands in the amide I and amide II regions at different ligand’s concentration in the protein complex. The analysis showed a decrease in the intensities of the parallel beta bands of propofol and L-arginine interactions with Aβ, accompanied with an increase in the anti- parallel bands for the Aβ-propofol interaction and a decrease for the Aβ-l-arginine interaction. The relative increase in peaks’ intensities at 1694 cm−1 and 1531 cm−1 for the propofol interaction is linked to the formation of oligomers in the protein.
Keywords:
FTIR Spectroscopy, Oligomeric Aβ, Alzheimer’s Disease, Amyloid β-Peptide, Antiparallel β-Sheet

1. Introduction
AD (Alzheimer’s disease) is a wide spread brain disease among older people. The disease is characterized by the formation of Amyloid β-peptides (Aβ) in the form of oligomers and fibrillar amyloid deposition resulted after proteolytic cleavage of the amyloid precursor protein (APP) by β- and g-secretases leading to the actual causes of the disease [1] -[4] .
Several studies have suggested that the accumulation of pathogenic amyloid-β (Aβ) assemblies in the brain can result in the dismantling of synapses, neuronal circuits and networks, which are associated with memory loss and cognitive decline [5] - [7] . In addition it has been reported that an imbalance between the formation rate and the clearance rate of Aβ deposits in the brain increases the progression of AD [8] .
Despite the general interest concerning oligomers toxicity compared to fibrils, there is a real need to understand the structure of these insoluble forms of Aβ. Some studies have shown that fibrillar Aβ was organized in a parallel β-sheet conformation while oligomeric Aβ were attributed to an antiparallel β-sheet structure [9] .
Identification of structural changes in the oligomerization process and explanations of the thermodynamics associated with the conformational changes in the assembly of Aβ will facilitate identification of therapeutic agents. Furthermore, the linkage between oligomerization and cellular dysfunction or toxicity is yet not understood.
Recent reports suggested that neurodegenerative disorders, such as Alzheimer’s and Parkinson’s diseases, might be accelerated by anesthesia, proposing that inhaled anesthetics promote protein oligomerization through occupancy of intermolecular cavities, and, in the case of amyloid-β, that these cavities are most prevalent in the diffusible oligomeric precursor of fibrils, hereafter termed intermediate oligomers [10] - [12] .
This study was undertaken to understand the oligomerization process in terms of its causes, structural characteristic, and to suggest mechanisms that may be used by the protein to prevent aggregation. Fourier transform infrared (FT-IR) and atomic force microscopy (AFM) were used to investigate the changes in the secondary structure of beta Amyloid (1-40) due to its interaction with an anesthetic propofol on one side and L-arginine on the other side.
2. Materials and Methods
Aβ (1-40) in powder form, propofol in solution form (5.390 M) and L-arginine in powder form were all purchased from Sigma Aldrich chemical company and used without further purification.
2.1. Preparation of Stock Solutions
An amount of 0.5 mg of Aβ (1-40) was dissolved in 250 μL of triple distilled water to give a concentration of 0.462 mM as a stock solution. A 1.338 mg of L-arginine was dissolved in 2 ml of PBS (PH 7.4) to prepare 3.84 mM L-arginine stock solution. Then different concentrations of L-arginine (3.84, 2.88, 1.92, 1.44, 0.96, 0.72, and 0.48 mM) were prepared. Propofol solutions with concentrations of (3.82, 2.88, 1.92, 1.44, 0.96, and 0.72 mM) were prepared using phosphate buffer saline (PBS) and the original propofol stock solution (5.390 M).
The final solutions of Aβ-propofol were prepared by mixing equal volumes of Aβ and propofol stock solutions. Aβ concentration in all samples was 0.231 mM, and the propofol concentrations in the final protein-drug solution were (1.92, 1.44, 0.96, 0.72, 0.48, 0.36, and.0.24 mM). For the final Aβ-L-arginine solution, equal volumes of Aβ and L-arginine stock solutions were mixed to give L-arginine concentrations of (1.92, 1.44, 0.96, 0.72, 0.48, 0.36, and 0.24 mM).
The solutions were incubated for one hour at 20˚C before spectroscopic measurements were taken.
2.2. UV-Absorption Spectra
The absorption spectra were obtained by using a NanoDrop ND-100 spectrophotometer. The spectra were recorded for 1 - 2 μL liquid samples of free Aβ and for its complexes with propofol solutions with the concentrations of (0.36 mM, 0.48 mM, 0.72 mM, 0.96 mM, 1.44 mM, and 1.92 mM). The same measurements were taken for Aβ-L-arginine complexes with similar concentrations. Repeated measurements were done for all the samples and no significant differences were observed.
2.3. Fluorescence Spectroscopy
The fluorescence measurements were performed by a NanoDrop® ND-3300 Fluorospectrometer at 25˚C. The excitation source comes from one of three solid-state light emitting diodes (LED’s). The excitation source options include: UV LED with maximum excitation 365 nm, Blue LED with excitation 470 nm, and white LED from 500 - 650 nm excitation. A 2048-element CCD array detector covering the range of 400 - 750 nm is connected by an optical fiber to the optical measurement surface. The excitation is done on the wavelength of 350 nm and the maximum emission wavelength is at 431 nm.
2.4. FTIR Spectroscopic Measurements
The FT-IR measurements were obtained on a Bruker IFS 66/S spectrophotometer equipped with a liquid nitrogen-cooled MCT detector and a KBr beam splitter. The spectrometer was continuously purged with dry air during the measurements. Samples were prepared after two hours of incubation for the Aβ-propofol and Aβ- L-arginine solutions at room temperature, four drops of each sample were placed on a certain area on a silicon window plate and left to dry at room temperature. The hydrated films on one side of a silicon window plate of the samples containing different concentrations of propofol or L-arginine with the same protein content.
The absorption spectra were obtained in the wave number range of 400 - 4000 cm−1. A spectrum was taken as an average of 60 scans to increase the signal to noise ratio, and the spectral resolution was at 4 cm−1. The aperture used in this study was 8 mm, which was found that to gives the best signal to noise ratio. Baseline correction, normalization and peak areas calculations were performed for all the spectra by OPUS software. The peak positions were determined using the second derivative by OPUS software.
The infrared spectra of Aβ-propofol complex were obtained in the featured region of 1800 - 1000 cm−1. The FT-IR spectrum of free Aβ was acquired by subtracting the absorption of the buffer solution from the spectrum of the protein solution. For the net interaction effect, the difference spectra {(protein solution + propofol solution) ? (protein solution)} were generated using the featureless region of the protein solution 1800 - 2200 cm−1 as an internal standard [13] . The accuracy of this subtraction method is tested using several control samples with the same protein or drug concentrations, which resulted into a flat base line formation. The spectral differences were used to investigate the nature of the Aβ-ligand interaction.
For the temperature dependence studies, ligand-protein complexes on silicon windows were placed into an infrared cell window. The temperature in the cell was controlled by an external water path which was increased gradually from 20˚C to 80˚C at 3˚C per 10 minute scan rate.
2.5. AFM Measurements
The prepared stock solutions of Aβ-propofol complex were incubated for 2 hrs at room temperature. A 10 µl of the solution was placed on certain area of a freshly cleaved mica substrate, and left to dry at room temperature. Free Aβ, Aβ-propofol and Aβ-L-arginine solutions were characterized by AFM equipped with 4 × 4 µm2 piezoelectric scanner. Analyses were carried out using tapping mode at room temperature using AFM cantilevers with a spring constant 2 N/m (Micro cantilever, OLYMPUS). All scanned images were analyzed using WSxM 5.0 Develop 3.2 software.
3. Results and Discussion
3.1. UV-Absorption Spectroscopy
The Aβ-propofol and Aβ L-arginine binding constants were determined using UV absorption spectroscopy as reported for several ligand-protein complexes [14] - [16] . The absorption spectra for different concentrations of propofol and L-arginine in Aβ are shown in Figure 1. The excitation has been done at 210 nm, while the absorption is recorded at 280 nm. The UV absorbance intensity of Aβ has increased with increasing propofol concentration, while it has decreased with increasing L-arginine concentration.
The absorption data was treated using linear reciprocal plots based on the following equation.
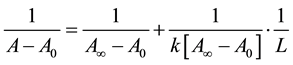 (1)
(1)
where A0 corresponds to the initial absorption of protein at 280 nm in the absence of ligands, A∞ is the final absorption of the protein ligand complex, and A is the recorded absorption at different propofol concentrations (L). The double reciprocal plot of 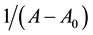 vs.
vs. 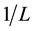 is linear (Figure 2) and the binding constant (K) for Aβ-
is linear (Figure 2) and the binding constant (K) for Aβ-


Figure 1. UV absorbance spectra of Aβ-propofol (A) and of Aβ-arginine (B) at the following concentrations for both cases (a: 0.0 mM, b: 0.36 mM, c: 0.48 mM, d: 0.72 mM, e: 0.96 mM, f: 1.44 mM, and g: 1.92 mM).


Figure 2. (A) the plot of 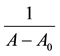 vs
vs  for Aβ with different concentrations of propofol, and (B) the plot of
for Aβ with different concentrations of propofol, and (B) the plot of 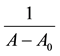 vs
vs  for Aβ with different concentrations of L-arginine.
for Aβ with different concentrations of L-arginine.
propofol is calculated to be 2.13 × 102 M−1, and for Aβ-L-arginine complexes is calculated to be 0.35 × 102 M−1. The binding constant for Aβ-L-arginine is lower than that of Aβ-propofol. Drugs with low binding constants are known to be more effective in their interactions with protein [17] .
The UV absorbance intensity of Aβ has increased with increasing propofol concentration, while it has decreased with increasing L-arginine concentration. The increase of band intensity due to the increase of propofol concentration can be related to an increase in the number of absorption sites as a result of structural changes of the Aβ-propofol complex. The reason for the decrease of intensity in Aβ-L-arginine can be attributed to the weak stability due to the presence of mainly hydrogen bonding between protein donor atoms and the ligands polar groups or an indirect ligand-protein interaction through water molecules [18] .
3.2. Fluorescence Spectroscopy
The intrinsic fluorescence In Aβ (1-40) is mainly due to tyrosine residue. This is because no tryptophan is found in the structure of Aβ (1-40) and because phenylalanine has very low quantum yield [19] [20] . The fluorescence spectra of Aβ free and at various concentrations of propofol (0.24, 0.36, 0.48, 0.96, 1.44, and 1.92 mM) are shown in (Figure 3(A)). The emission fluorescence spectra of Aβ with various L-arginine concentrations (0.24, 0.36, 0.48, 0.96, 1.44, and 1.92 mM) are shown in (Figure 3(B)). The excitation has been performed at 350 nm while the emission occurred at 431 nm.
The fluorescence intensity of Aβ decreases regularly with the increase of ligand’s concentration. Other factors can affect fluorescence intensity of a compound such as molecular rearrangements, exited state reactions, energy transfer, ground state complex formation, and collisional quenching [21] - [23] . The Dynamic quenching process can be described by the Stern-Volmer equation [24] - [26]
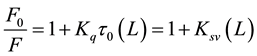 (2)
(2)
where F and F0 are the fluorescence intensity with and without quencher, Kq is the quenching rate constant of the biomolecule, Ksv is the Stern-Volmer quenching constant, t0 is the average lifetime of the biomolecule without quencher, and (L) is the concentration of propofol or L-arginine. As can be seen from Figure 4, the Stern- Volmer plots are linear and the slopes are equal to the Stern-Volmer quenching constants for Aβ-propofol and Aβ-L-arginine complexes with these values: 1.21 × 103 L∙mol−1 and 1.49 × 102 L∙mol−1 respectively. Fluores-

Figure 3. The fluorescence emission spectra of Aβ with various concentrations of propofol in (A) and with various concentrations of L-arginine in (B), the concentration are: (a = 0.0, b = 0.24, c = 0.36, d = 0.48, e = 0.96, f = 1.44, and g = 1.92 mM).

Figure 4. The plot of 
cence quenching can be induced by different mechanisms, which are usually classified into dynamic and static quenching. Dynamic quenching arises from collisional encounters between the fluorophore and quencher, and static quenching results from the formation of a ground state complex between the fluorophore and the quencher [27] .
The quenching rate constants for Aβ-propofol and Aβ-arginine were then calculated and found to be 1.1 × 1012 L∙Mol−1∙s−1 and 1.35 × 1012 L∙Mol−1∙s−1 respectively. The obtained values of the quenching rate constants for propofol and L-arginine are larger than the maximum dynamic quenching constants for various quenchers with biopolymers (2 × 1010 L∙Mol−1∙s−1) which confirms that static quenching is dominant in these complexes [28] [29] . When the static quenching equation is used [30] .

where K is the binding constant of the ligand with Aβ. The value of K can be determined from the slope and the y-intercept of the straight line in Figure 5. The two binding constants are found to be 2.81 × 102 M−1 for Aβ- propofol and 0.37 × 102 M−1 for Aβ-L-arginine which agrees well with the values obtained earlier by UV spectroscopy and supports the effective role of static quenching.
The highly effective quenching rate constants in these cases have led to a lower value of binding constants between the ligands and Aβ due to an effective hydrogen bonding between the ligands and Aβ.
Fluorescence quenching is attributed to changes in the environment of the protein fluorophores caused by the presence of the ligand [31] and is likely to be associated with amyloid fibrils [32] .
3.3. FTIR Spectroscopy
The FTIR absorption spectra in the amide I and amide II regions are taken for different concentrations of Aβ- propofol and Aβ-L-arginine samples. The analysis of the secondary structure of Aβ in the amide I which occurs between 1600 cm−1 and 1700 cm−1 yields several bands. The peaks of these bands correspond to the C=O stretching vibrations of the amide group, coupled to the C-N stretching and C-C-N deformation mode [28] . The peaks in the amide II (1600 - 1480) cm−1 region are due to the coupling of the N-H in-plane bending and C-N stretching modes [33] .
The difference spectra of [(Aβ + propofol or L-arginine) − Aβ] were obtained to show the intensity variation, and the results are shown in (Figure 6 and Figure 7). For the [(Aβ + propofol) − Aβ] case, Figure 6 shows a strong positive feature at 1630 cm−1 in the amide I region in addition to another weak positive feature at 1670 cm−1. For amide II region there is a strong positive feature at 1550 cm−1. These features were obtained at propofol concentrations of (0.48, 0.72, and 1.44 mM).
For [(Aβ + L-arginine) − Aβ] interaction, Figure 7 shows the difference spectra for Aβ with different L-argi- nine concentrations. In the amide I region there is a strong negative feature at 1629 cm−1, and another weak negative feature at 1668 cm−1, while in amide II region there is a strong negative feature at 1538 cm−1 and another weak negative feature at 1493 cm−1.
It is clearly shown in Figure 6 that the strong positive features in amide I and amide II regions become stronger as the propofol concentration increased. On the other hand the strong negative features in amide I and amide II regions become stronger with increasing L-arginine concentration.
The observed positive features are attributed to the increase in the intensity of the amide I bands at 1630, and 1670 cm−1, and amide II band at 1550 cm−1 as a result of propofol interaction with Aβ. This increase in the intensity is due to the increase in the number of β-sheet bonds [26] . The negative featured noticed in the difference spectra of [(Aβ-L-arginine) − Aβ] are due to the decrease in the intensity with increasing L-arginine concentration. This decrease in the intensity is due to the decrease in the number of β-sheet bonds.

Figure 5. The plot of 

Figure 6. (A) FTIR spectra for Aβ free and Aβ-propofol complex (1.44 mM), and (B) the difference spectra of Aβ free and Aβ-propofol at different concentrations.
Figure 7. (A) FTIR spectra for Aβ free and Aβ-L-arginine complex 0.72 mM, and (B) the difference spectra of Aβ free and Aβ-L-arginine at different concentrations.
The component bands of the amide I and amide II regions were determined using Fourier self-deconvolution (FSD) and second derivative resolution with curve fitting procedures. This procedure has provided a basis for the quantitative estimation of protein secondary structure [27] [30] [34] and [35] .
Based on several previous proteins’ studies [9] [19] , the amide I bands are assigned as follows: (1600 - 1612 cm−1) to anti-parallel β-sheet, (1612 - 1630 cm−1) to parallel β-sheet, (1630 - 1650 cm−1) to random coil, (1650 - 1670 cm−1) to α-helix, (1670 - 1685 cm−1) to β-turns, and (1685 - 1700 cm−1) to anti-parallel β-sheet. Similarly for the amide II region, the absorption spectra is composed of six bands assigned in the following order: (1480 - 1500 cm−1) to parallel β-sheet, (1500 - 1520 cm−1) to random coil, (1520 - 1538 cm−1) to anti parallel β-sheet, (1538 - 1560 cm−1) to α-helix, (1560 - 1580 cm−1) to β-turns, and (1580 - 1600 cm−1) to anti-parallel β-sheet.
In this work a quantitative analysis of the protein secondary structure for Aβ free, Aβ-propofol and Aβ-L-ar- ginine complexes in dehydrated films are determined from the shape of amide I and amide II bands. Figure 8 shows the spectra and its second derivative for the amide I and the amide II regions. The curve-fitted graphs for the amide I and amide II regions are shown in Figure 9 for Aβ-propofol complex and in Figure 10 for Aβ-L-
Figure 8. (A) the second derivative spectra of Aβ free, and the spectra of both Aβ-propofol (B) and Aβ-L-arginine (C) at the following concentrations (a = 0.0 mM, b = 0.48 mM, c = 0.72 mM, d = 1.44 mM, and e = 1.92 mM).


Figure 9. Curve fitted graphs for Aβ free in amide I (A), Aβ-propofol complex with 1.44 mM concentration in amide I (B), Aβ free in amide II (C) and Aβ-propofol complex with 1.44 mM concentration in amide II (D).


Figure 10. Curve fitted graphs for Aβ free in amide I (A), Aβ-L-arginine complex with 0.72 mM concentration in amide I (B), Aβ free in amide II (C) and Aβ-L-arginine complex with 0.72 mM concentration in amide II (D).
arginine complex. The curve fitted graphs show the secondary structure determinations of the Aβ free and Aβ- propofol and Aβ-L-arginine complexes.
The exact frequency of the vibrations depend on the nature of the hydrogen bonding involving the amide group, and by the particular secondary structure adopted by the protein.
Table 1 listed the peak positions of the separate components of the amide I and amide II regions using both second derivative and FSD procedures. The quantitative estimation of protein secondary structure is based on the assumption that the total absorption due to the carbonyl (C=O) vibration can be represented by the sum of the individual absorptions due to the vibrations of the separate elements of the band [27] .
The percentages of each secondary structure of the Aβ-propofol and Aβ-L-arginine complexes were calculated from the integrated areas of the component bands in amide I and amide II respectively. The relative intensity of each secondary structure component of Aβ before and after the interaction with propofol and L-arginine at different concentrations is shown in Table 2 and Table 3.
The changes in band intensities as a result of changing propofol and L-arginine concentrations while keeping Aβ concentration constant in the amide I and amide II regions are shown in Figure 11. These changes show a decrease in the parallel β-sheet intensity percentage accompanied with an increase in anti-parallel β-sheet structures. This behavior is believed to be caused by the misfolded protein in the presence of propofol as a result of H-bonding formation between propofol and Aβ [34] . The parallel β-sheet has a weak structural arrangement, because the geometry of the individual amino acid molecules forces the hydrogen bonds to occur at angles. Introducing non-planarity in the inter-strand hydrogen bonding make them longer and thus weaker. Contrarily, in the anti-parallel arrangement the hydrogen bonds are aligned directly opposite to each other allowing the inter-
Table 1. Bands assignment in the absorbance spectra of Aβ with different propofol and L-Arginine concentrations for Amide I - II regions.
Table 2. Secondary structure determination for Amide I and Amide II regions in Aβ and its propofol complexes.
Table 3. Secondary structure determination for Amide I and Amide II regions in Aβ and its L-arginine complexes.


Figure 11. Relative intensity variation for different concentrations of Aβ-propofol complexes in amide I (A), Aβ-propofol complexes in amide II (B), Aβ-L-arginine complexes in amide I (C) and Aβ-L-arginine complexes in amide II (D).
strand hydrogen bonding between carbonyl group and the hydrogen atoms of the amines groups to be planar making a planar inter-strand hydrogen bonding and resulting in a shorter (stronger) and more stable bonds [36] - [38] .
The observed increase in the intensity of anti-parallel β-sheet band as a result of Aβ interaction with propofol is an indication of oligomers or fibrils formation [19] [39] . These changes in intensities can also be noted directly from the fitted curves in (Figure 9). Previous studies using circular dichroism (CD), FTIR spectroscopy, and X-ray diffraction studies confirmed that amyloid fibrils have a β-pleated sheet structure [40] .
The analysis of the absorption spectra of Aβ-L-arginine complexes in amide I and amide II, both parallel and anti-parallel β-sheet intensities shows a gradual decrease with increasing L-arginine concentration as shown in (Figure 11(C), and Figure 11(D)). This decrease is an indication of L-arginine effect in decreasing oligomerization by converting the insoluble (β-sheet rich) fibrils or oligomers into more soluble entities.
3.4. Atomic Force Microscopy (AFM)
AFM technique has been used as an additional method to investigate the effect of propofol and L-arginine on Aβ by taking images for Aβ-free solution, Aβ-propofol complex, and Aβ-arginine complex as shown in (Figures 12(A)-(C) respectively). The heights of the clusters in each of these complexes by scanning the surface of the



Figure 12. AFM image of Aβ solution (A), Aβ-Propofol (0.96 mM) complex (B), and Aβ-L-arginine (0.96 mM) complex (C). The graphs on the right side show the height changes in the surface of the sample.
samples of Aβ-free, Aβ-propofol, and Aβ-L-arginine are shown respectively to the right of each image.
It is clear from AFM images that Aβ specimens have clustered in a certain area as a result of its interaction with propofol, while in the case of Aβ-L-arginine interaction showed little or no change in cluster formation. From the graph shown in (Figure 12(A)) the height of the clusters in Aβ-free sample was found to be between (30 - 35 nm), while for Aβ-propofol sample it has been found to be (250 - 300 nm) as shown in (Figure 12(B)) and for Aβ-L-arginine sample, the height has been found to be (25 - 50 nm) as shown in (Figure 12(C)).
These results clearly indicate the effect of propofol in increasing the sizes of the oligomers which are shown as spherical shaped objects or clusters. This confirms the results already found by FTIR spectroscopy that propofol enhances oligomers formation.
A little change in the heights of Aβ complex structures are noticed upon interaction with L-arginine as shown in (Figure 12(A), and Figure 12(C)) meaning, L-arginine interacts with Aβ in a way different from that of propofol. These results support the results already found by FTIR spectroscopy.
4. Concluding Remarks
The formation of oligomers in the precursor protein induces structural changes leading to a disturbance in the primary function of the protein. Although the molecular detail underlying formation and growth of amyloid oligomers has eluded structural approaches at this time, exact causes are unrevealed. One possible scenario is to assume that information and images are saved in memory at a particular structural state associated with a certain frequency. When a certain electrical signal is provided by neurons an electromagnetic field is produced in the protein which can be reproduced at any later time if the same electrical signal is provided. It seems that propofol interacts with Aβ in the brain to produce oligomers which behave similar to polarized dielectric material that is sandwiched between different beta-sheets or within the folded β-sheet to create a capacitor-like structure. Inserting this material between different beta-sheets or within the folded β-sheet will increase the internal energy of the folded arrangement leading to a more stable complex. Anesthetic molecules are small, hydrophobic molecules that bind principally in internal protein cavities. The interactions between anesthetic molecules and the cavities within the protein restore Van der Waals contacts and thus provide additional free energy for oligomerization [41] . Similarly the interactions between Aβ and propofol within the β-sheet of the protein can lead to more rigid stable oligomers formations. The resulting solid structure can hinder the reproduction of the original frequencies related to the saved information in the memory of the brain, and prevent electrical and chemical signals’ transmittance which is vital for carrying information among different regions of the brain. Consequently, retrieving related existing images and information becomes very challenging and demanding process. In addition, processing new signals and information may slow down.
The direct cause leading to synaptic and neuronal loss in the brain is not completely clear yet; however, some studies suggest that nerve damage might result from the conversion of nontoxic soluble monomers to non-so- luble toxic oligomers and protofibrils [42] .
Evidence are provided within this manuscript for a novel biophysical interaction leading to structural changes in Aβ-propofol interaction. These changes are evident by the observed variations in the intensities of the absorption spectra of the β-sheet bands in the amide I and amide II regions. Recent NMR investigations of Aβ-propofol interaction reaffirm that smaller molecular sized anesthetics do play a leading role in Aβ oligomerization [43] , and a previous study of pentobarbital-transthyretin interactions have yielded similar results and showed protein oligomerization [28] .
Many studies have proven that amyloid oligomers or fibrils which are found in the brains of patients with Alzheimer disease are rich in β-sheet especially the more stable anti-parallel β-sheet [44] - [46] . Since increasing propofol concentration was shown to increase the intensity of the absorption band of anti-parallel β-sheet, one can deduce that propofol increases the rate of oligomerization or fibrils formation. On the other hand arginine shows an opposite effect as increasing its concentration in Aβ-L-arginine complex causes a decrease in the intensity of anti-parallel β-sheet. It can be concluded that L-arginine not only does not induce oligomerization, but it also may decrease the rates of oligomerization.
In addition, the AFM images did not show any changes with regard to cluster formation for L-arginine interaction with Aβ, while a remarkable increase in heights of the clusters is observed for propofol interaction with Aβ.
While the mechanism of L-arginine’s interaction with Aβ is not yet well understood, it seems to be related to its binding to aromatic groups in partially unfolded states of proteins acting as an effective inhibitory agent for a wide variety of amyloid forming proteins. The strong binding with Aβ peptide monomers results in stable complexes and prevents the formation of soluble oligomers produced by hydrophobic interaction.
Acknowledgements
This work is supported by the German Research Foundation DFG grant No. DR228/24-2.
References
- Roher, A.E., Chaney, M.O., Kuo, Y.M., Webster, S.D., Stine, W.B., Haverkamp, L.J., Woods, A.S., Cotter, R,J., Tuohy, J.M., Krafft, G.A., Bonnell, B.S. and Emmerling, M.R. (1996) Morphology and Toxicity of Abeta-(1-42) Dimer Derived from Neuritic and Vascular Amyloid Deposits of Alzheimer’s Disease. The Journal of Biological Chemistry, 271, 20631-20635. http://dx.doi.org/10.1074/jbc.271.34.20631
- Klein, W.I., Krafft, G.A. and Finch, C.E. (2001) Targeting Small Aβ Oligomers: The Solution to an Alzheimer’s Conundrum. Trends in Neurosciences, 24, 219-224. http://dx.doi.org/10.1016/S0166-2236(00)01749-5
- Dahlgren, K.W., Manelli, A.M., Stine, W.B., Baker, L.K., Kraft, G.K. and LaDu, M.J. (2002) Oligomeric and Fibrillar Species of Amyloid-Beta Peptide Differentially Affect Neuronal Viability. The Journal of Biological Chemistry, 277, 32046-32053. http://dx.doi.org/10.1074/jbc.M201750200
- Kayed, R., Head, E., Thompson, J.L., Mcintire, T.M., Milton, S.C., Cotman, C.W. and Glabe, C.G. (2003) Common Structure of Soluble Amyloid Oligomers Implies Common Mechanism of Pathogenesis. Science, 300, 486-489. http://dx.doi.org/10.1126/science.1079469
- Hsia, A.Y., et al. (1999) Plaque-Independent Disruption of Neural Circuits in Alzheimer’s Disease Mouse Models. Proceedings of the National Academy of Sciences of the United States of America, 96, 3228-3233. http://dx.doi.org/10.1073/pnas.96.6.3228
- Chapman, P.F., et al. (1999) Impaired Synaptic Plasticity and Learning in Aged Amyloid Precursor Protein Transgenic Mice. Nature Neuroscience, 2, 271-276. http://dx.doi.org/10.1038/6374
- Walsh, D.M., Klyubin, I., Fadeeva, J.V., Cullen, W.K., Anwyl, R., Wolfe, M.S., Rowan, M.J. and Selkoe, D.J. (2002) Naturally Secreted Oligomers of Amyloid β Protein Potently Inhibit Hippocampal Long-Term Potentiation in Vivo. Nature, 41, 535-539. http://dx.doi.org/10.1038/416535a
- Hardy, J. and Selkoe, D.J., (2002) The Amyloid Hypothesis of Alzheimer’s Disease: Progress and Problems on the Road to Therapeutics. Science, 297, 353-356. http://dx.doi.org/10.1126/science.1072994
- Cerf, E., et al. (2007) Antiparallel β-Sheet: A Signature Structure of the Oligomeric Amyloid β-Peptide. Biochemical Journal, 421, 415-423. http://dx.doi.org/10.1042/BJ20090379
- Carnini, A., Lear, J.D. and Eckenhoff, R.G. (2007) Inhaled Anesthetic Modulation of Amyloid Aβ1-40 Assembly and Growth. Current Alzheimer Research, 4, 233-241. http://dx.doi.org/10.2174/156720507781077278
- Bohnen, N.I., Warner, M.A., Kokmen, E., Beard, C.M. and Kurland, L.T. (1994) Alzheimer-Disease and Cumulative Exposure to Anesthesia―A Case-Control Study. Journal of the American Geriatrics Society, 42, 198-201.
- Muravchick, S. and Smith, D.S. (1995) Parkinsonian Symptoms during Emergence from General-Anesthesia. Anesthesiology, 82, 305-307. http://dx.doi.org/10.1097/00000542-199501000-00039
- Surewicz, W.K., Mantsch, H.H. and Chapman, D. (1993) Determination of Protein Secondary Structure by Fourier Transform Infrared Spectroscopy: A Critical Assessment. Biochemistry, 32, 389-394. http://dx.doi.org/10.1021/bi00053a001
- Stephanos, J.J. and Inorg, J. (1996) Drug-Protein Interactions. Two-Site Binding of Heterocylclic Ligands to a Monomeric Hemoglobin. Journal of Inorganic Biochemistry, 62, 155-169. http://dx.doi.org/10.1016/0162-0134(95)00144-1
- Klotz, M.I. and Hunston, L.D. (1971) Properties of Graphical Representations of Multiple Classes of Binding Sites. Biochemistry, 10, 3065-3069. http://dx.doi.org/10.1021/bi00792a013
- Klotz, M.I. (1982) Numbers of Receptor Sites from Scatchard Graphs: Facts and Fantasies. Science, 217, 1247-1249. http://dx.doi.org/10.1126/science.6287580
- Bhattacharya, A.A., Gruene, T. and Curry, S. (2000) Crystallographic Analysis Reveals Common Modes of Binding of Medium and Long-Chain Fatty Acids to Human Serum Albumin. Journal of Molecular Biology, 303, 721-732. http://dx.doi.org/10.1006/jmbi.2000.4158
- Purcell, M., Neault, J.F. and Tajmir-Riahi, H.A. (2000) Interaction of Taxol with Human Serum Albumin. Biochimica et Biophysica Acta, 1478, 61-68. http://dx.doi.org/10.1016/S0167-4838(99)00251-4
- Sarroukh, R., Cerf, E., Derclaye, S., Dufrêne, Y.F., Goormaghtigh, E., Ruysschaert, J.-M. and Raussens, V. (2011) Transformation of Amyloid β(1-40) Oligomers into Fibrils Is Characterized by a Major Change in Secondary Structure. Cell. Cellular and Molecular Life Sciences, 68, 1429-1438. http://dx.doi.org/10.1007/s00018-010-0529-x
- Amaro, M., Kubiak-Ossowska, K., Birch, D.J.S. and Rolinski, O.J. (2013) Initial Stages of Beta-Amyloid Aβ1-40 and Aβ1-42 Oligomerization Observed Using Fluorescence Decay and Molecular Dynamics Analyses of Tyrosine. Methods and Applications in Fluorescence, 1, Article ID: 015006. http://dx.doi.org/10.1088/2050-6120/1/1/015006
- Turro, N.J. (1991) Modern Molecular Photochemistry. University Science Books, Sausalito.
- Jianghong, T., Ning, L., Xianghong, H. and Guohua, Z. (2008) Investigation of the Interaction between Sophoricoside and Human Serum Albumin by Optical Spectroscopy and Molecular Modeling Methods. Journal of Molecular Structure, 889, 408-414. http://dx.doi.org/10.1016/j.molstruc.2008.02.031
- Li, J., Ren, C., Zhang, Y., Liu, X., Yao, X. and Hu, Z. (2008) Human Serum Albumin Interaction with Honokiol Studied Using Optical Spectroscopy and Molecular Modeling Methods. Journal of Molecular Structure, 881, 90-96. http://dx.doi.org/10.1016/j.molstruc.2007.08.039
- Sheehan, D. (2009) Physical Biochemistry: Principles and Applications. 2nd Edition, John Wiley & Sons, Hoboken.
- Tian, J.N., Liu, J.Q., Zhang, J.Y., Hu, Z.D. and Chen, X.G. (2003) Fluorescence Studies on the Interactions of Barbaloin with Bovine Serum Albumin. Chemical & Pharmaceutical Bulletin, 51, 579-582. http://dx.doi.org/10.1248/cpb.51.579
- Lakowicz, J.R. (2006) Principles of Fluorescence Spectroscopy. 3rd Edition, Springer Science, New York.
- Kong, J. and Yu, S. (2007) Fourier Transform Infrared Spectroscopic Analysis of Protein Secondary Structures. Acta Biochimica et Biophysica Sinica, 39, 549-559. http://dx.doi.org/10.1111/j.1745-7270.2007.00320.x
- Darwish, S.M., Ghithan, J., Abuteir, M.M., Faroun, M. and Abu-Hadid, M.M. (2013) Spectroscopic Investigation of Pentobarbital Interaction with Transthyretin. Journal of Spectroscopy, 2013, Article ID: 927962.
- Wang, H., Duennwald, M.L., Roberts, B.E., Rozeboom, L.M., Zhang, Y.L., Steele, A.D., et al. (2008) Direct and Selective Elimination of Specific Prions and Amyloids by 4,5-Dianilinophthalimide and Analogs. Proceedings of the National Academy of Sciences, 105, 7159-7164. http://dx.doi.org/10.1073/pnas.0801934105
- Cordeiro, Y., Kraineva, J., Suarez, M.C., Tempesta, A.G., Kelly, J.W., Silva, J.L., Winter, R. and Foguel, D. (2006) Fourier Transform Infrared Spectroscopy Provides a Fingerprint for the Tetramer and for the Aggregates of Trans- thyretin. Biophysical Journal, 91, 957-967. http://dx.doi.org/10.1529/biophysj.106.085928
- Sulkowaska, A. (2002) Interaction of Drugs with Bovine and Human Serum Albumin. Journal of Molecular Structure, 614, 227-232. http://dx.doi.org/10.1016/S0022-2860(02)00256-9
- Garzon-Rodriguez, W., Vega, A., Sepulveda-Becerra, M., Milton, S., Johnson, D.A., Yatsimirsky, A.K. and Glabe, C.G. (2000) A Conformation Change in the Carboxyl Terminus of Alzheimer’s Aβ(1-40) Accompanies the Transition from Dimer to Fibril as Revealed by Fluorescence Quenching Analysis. Journal of Biological Chemistry, 275, 22645- 22649. http://dx.doi.org/10.1074/jbc.M000756200
- Sirotkin, V.A., Zinatullin, A.N., Solomonov, B.N., Faizullin, D.A. and Fedotov, V.D. (2001) Calorimetric and Fourier transform Infrared Spectroscopic Study of Solid Proteins Immersed in Low Water Organic Solvents. Biochimica et Biophysica Acta, 1547, 359-369. http://dx.doi.org/10.1016/S0167-4838(01)00201-1
- Zandomeneghi, G., Krebs, M.R.H., Mccammon, M.G. and Fandrichi, M. (2004) FTIR Reveals Structural Differences between Native b-Sheet Proteins and Amyloid Fibrils. Protein Science, 13, 3314-3321. http://dx.doi.org/10.1110/ps.041024904
- Shan-Yang, L. and Horng-Lun, C. (2003) Fourier Transform Infrared Spectroscopy Used to Evidence the Prevention of β-Sheet Formation of Amyloid β(1-40) Peptide by a Short Amyloid Fragment. International Journal of Biological Macromolecules, 32, 173-177. http://dx.doi.org/10.1016/S0141-8130(03)00051-5
- Perczel, A., Gáspári, Z. and Csizmadia, I.G. (2005) Structure and Stability of β-Pleated Sheets. Journal of Computational Chemistry, 26, 1155-1168. http://dx.doi.org/10.1002/jcc.20255
- Irie, K., Murakami, K., Masuda, Y., Morimoto, A., Ohigashi, H., Ohashi, R., et al. (2005) Structure of Beta-Amyloid Fibrils and Its Relevance to Their Neurotoxicity: Implications for the Pathogenesis of Alzheimer’s Disease. Journal of Bioscience and Bioengineering, 99, 437-447.
- Perczel, A., Gáspári, Z. and Csizmadia, I.G. (2005) Structure and Stability of β-Pleated Sheets. Journal of Computational Chemistry, 26, 1155-1168. http://dx.doi.org/10.1002/jcc.20255
- Juszczyk, P., Kołodziejczyk, A.S. and Grzonka, Z. (2009) FTIR Spectroscopic Studies on Aggregation Process of the β-Amyloid 11-28 Fragment and Its Variants. Journal of Peptide Science, 15, 23-29. http://dx.doi.org/10.1002/psc.1085
- Lomaki, A., Chung, D.S., Benedek, G.B., Kirschner, D.A. and Teplow, D.B. (1996) On the Nucleation and Growth of Amyloid β-Protein Fibrils: Detection of Nuclei and Quantitation of Rate Constants. Proceedings of the National Academy of Sciences of the United States of America, 93, 1125-1129. http://dx.doi.org/10.1073/pnas.93.3.1125
- Eckenhoff, R.G., Johansson, J.S., Wei, H.F., Carnini, A., Kang, B.B., Wei, W.L., et al. (2004) Inhaled Anesthetic Enhancement of Amyloid-Oligomerization and Cytotoxicity. Anesthesiology, 101, 703-709. http://dx.doi.org/10.1097/00000542-200409000-00019
- Hashimoto, M., Rockenstein, E., Crews, L. and Masliah, E. (2003) Role of Protein Aggregation in Mitochondrial Dysfunction and Neurodegeneration in Alzheimer’s and Parkinson’s Diseases. NeuroMolecular Medicine, 4, 21-36. http://dx.doi.org/10.1385/NMM:4:1-2:21
- Mandal, P.K., Bhavesh, N.S., Chauhan, V.S. and Fodale, V. (2010) NMR Investigations of Amyloid-β Peptide Interactions with Propofol at Clinically Relevant Concentrations with and without Aqueous Halothane Solution. Journal of Alzheimer’s Disease, 21, 1303-1309.
- Lindgren, M. and Hammarstrom, P. (2010) Amyloid Oligomers: Spectroscopic Characterization of Amyloidogenic Protein States. FEBS Journal, 277, 1380-1388. http://dx.doi.org/10.1111/j.1742-4658.2010.07571.x
- Murphy, M.R. (2002) Peptide Aggregation in Neurodegenerative Disease. Annual Review of Biomedical Engineering, 4, 155-174. http://dx.doi.org/10.1146/annurev.bioeng.4.092801.094202
- Kirkitadze, M.D., Condron, M.M. and Teplow, D.B. (2001) Identification and Characterization of Key Kinetic Intermediates in Amyloid Beta-Protein Fibrillogenesis. Journal of Molecular Biology, 312, 1103-1119. http://dx.doi.org/10.1006/jmbi.2001.4970
NOTES
*Corresponding author.


















