Open Journal of Medicinal Chemistry
Vol.2 No.3(2012), Article ID:22910,11 pages DOI:10.4236/ojmc.2012.23007
Homology Modeling of Human Alpha-Glucosidase Catalytic Domains and SAR Study of Salacinol Derivatives
Department of Pharmaceutical Sciences, Kinki University, Kinki, Japan
Email: *nakas@phar.kindai.ac.jp
Received June 25, 2012; revised July 26, 2012; accepted August 5, 2012
Keywords: Homology Modeling; Docking Study; MM/PBSA; Alpha-Glucosidase; Salacinol; Kotalanol
ABSTRACT
Maltase-glucoamylase (MGAM) and sucrase-isomaltase (SI) belong to human intestinal alpha-glucosidase and their N-terminal side catalytic domains are called NtMGAM and NtSI, and their C-terminal side catalytic domains are called CtMGAM and CtSI. As an antidiabetic, alpha-glucosidase inhibitor is required to bind to all of these domains to inhibit disaccharides hydrolysis. Salacinol and kotalanol isolated from Salacia reticulata are novel seed compounds for alpha-glucosidase inhibitor. Even though the complex structures of NtMGAM or NtSI have been determined experimentally, those of CtMGAM and CtSI have not been revealed. Thus, homology modeling for CtMGAM and CtSI has been performed to predict the binding mode of salacinol and its derivatives for each domain. The binding affinities for these compounds were also calculated to explain the experimental structure-activity relationships (SARs). After a docking study of the derivatives to each catalytic domain, the MM/PBSA method has been applied to predict the binding affinities. The predicted binding affinities were almost consistent with the experimental SARs. The comparison of the complex structures and binding affinities provided insights for designing novel compounds, which inhibit all catalytic domains.
1. Introduction
Starch is one of the most important malnutrition sources. Saccharides after ingestion per oral are digested into oligosaccharides by amylase in saliva and pancreas, and reach the small intestine. Disaccharides such as maltose with alpha-1,4-glycosidic bond, isomaltose with alpha-1,6- glycosidic bond, and sucrose with alpha-1,2-glycosidic bond are digested into monosaccharides by alpha-glucosidase located on the small intestinal brush border membrane.
Two kinds of human intestinal alpha-glucosidase, maltase-glucoamylase (MGAM) and sucrase-isomaltase (SI), have two catalytic domains on the N-terminal side
(NtMGAM, NtSI) and the C-terminal side (CtMGAM, CtSI) [1], as shown in Figure 1. These four catalytic domains are homologous, and every domain catalyzes the maltose hydrolysis at the alpha-1,4-glycosidic bond. In addition, NtSI and CtSI catalyze isomaltose and sucrose hydrolysis, respectively [2].
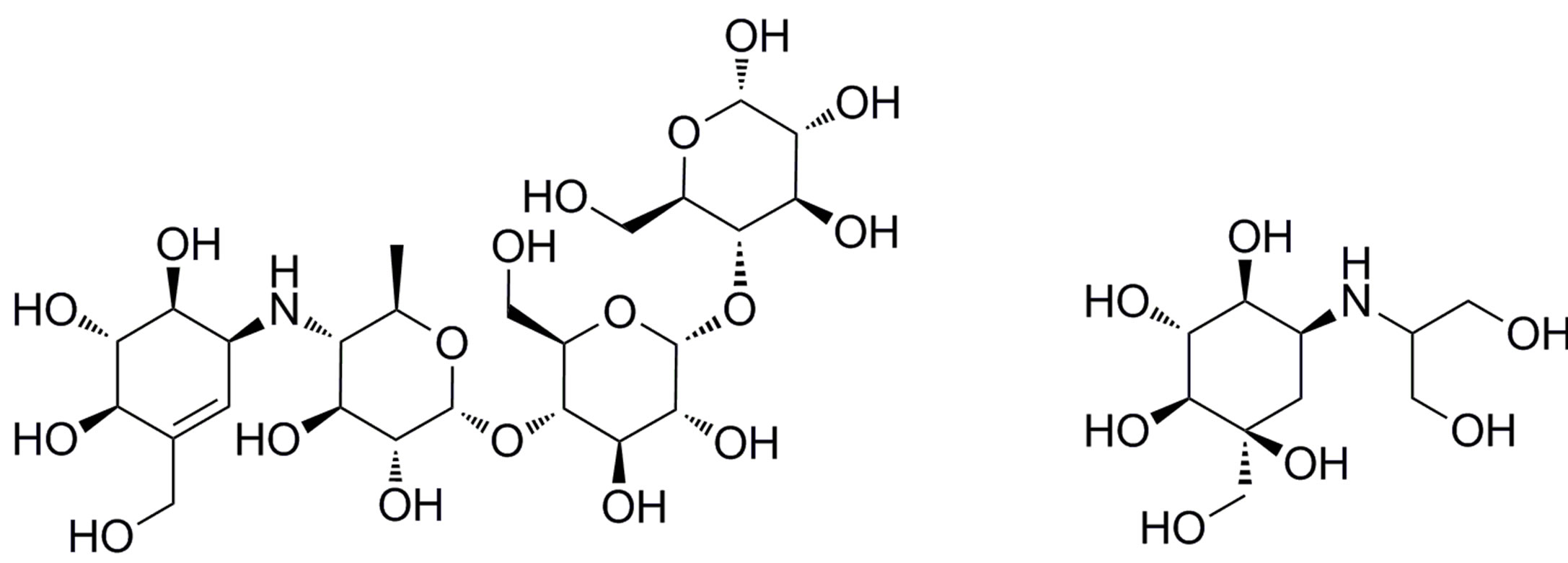
Acarbose (Figure 2(a)) [3] and voglibose (Figure 2(b)) [4] are well-known alpha-glucosidase inhibitors that are used in diabetes therapy. However, abdominal discomfort and hepatotoxicity [5] have been reported for these drugs, and therefore, novel alpha-glucosidase inhibitors without these side effects, especially hepatotoxicity, are required to improve the patients’ quality of life.
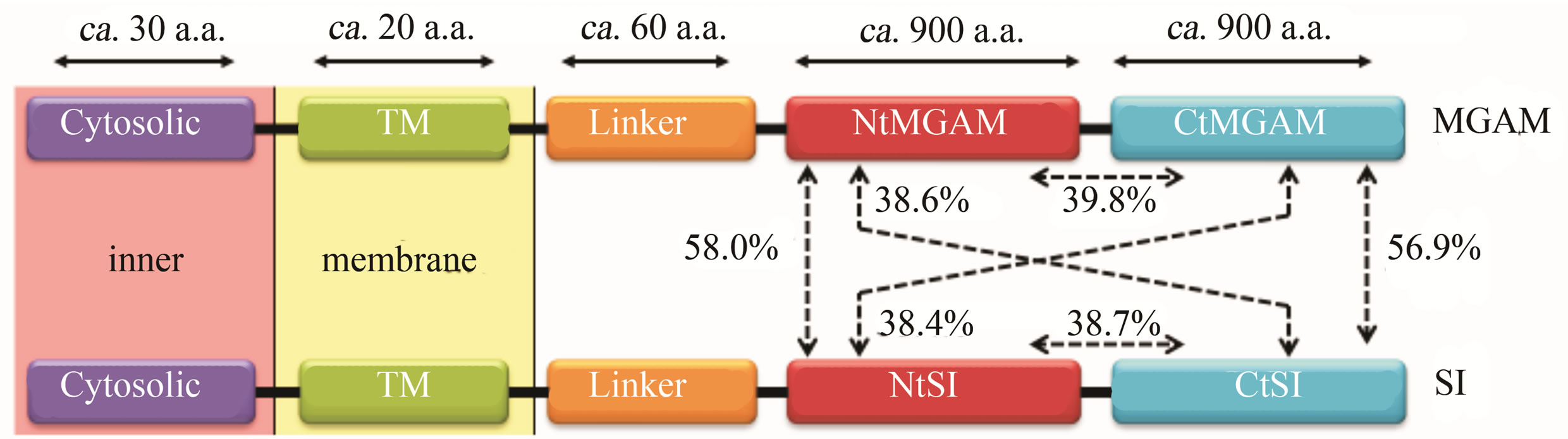
Figure 1. Two kinds of human alpha-glucosidase, Maltase-Glucoamylase (MGAM) and Sucrase-Isomaltase (SI), located on intestinal brush border membrane. Percentage values show the amino acid sequence homology between the two domains.
On the other hand, salacia, a genus of plants, is used to treat diabetes in Ayurveda, the traditional medicine of India and Sri Lanka. Furthermore, kotalanol (Figure 2(c)) [6] and salacinol (1 of Figure 2(d)) [7], isolated from Salacia reticulata have almost the same inhibition acti ity to alpha-glucosidase as acarbose and voglibose [6]. Therefore, several drug discovery studies based on these natural compounds have been carried out [8-13].
Recently, the X-ray crystal structure analysis of NtMGAM or NtSI complexes of these compounds has been reported [8,9]. The structure-activity relationships (SARs) for salacinol derivatives are also vigorously studied [10, 11]. In addition, docking simulations between salacinol derivatives and NtMGAM have been performed [12] and provided the guidelines for optimization studies, such as the replacement of the sulfate group of salacinol and kotalanol with the hydrophobic group. Indeed, several compounds that enhanced the inhibitory activities have been discovered [13].
Though current studies have focused on NtMGAM, they need to take into account other catalytic domains such as NtSI, CtMGAM, and CtSI. The ideal alphaglucosidase inhibitors are expected to simultaneously bind these four domains. The rational design of ideal inhibitors requires clarifying the structural differences among the four catalytic domains.
The structures of the two C-terminal catalytic domains have not been experimentally determined; however, it is possible to construct model structures of both domains by the homology modeling method if the structure of a homologous protein is known. As shown in Figure 1, the sequence homologies of amino acid residues for the four catalytic domains is almost 40%, which is over the threshold value of 30% that the homology modeling method is applicable [14]. Therefore, the model structures of CtMGAM and CtSI are constructed using the crystal structures of NtMGAM and NtSI.
Binding mode analysis is an essential process for structure-based drug design, and typically, the binding modes of the ligands are experimentally determined by X-ray crystallography. However, recent progress in the computational docking method enabled us to predict the ligand-binding mode with a positional error less than 2 Å. In this study, the binding modes and binding affinities of the salacinol derivatives (compounds 2-4, shown in Figure 2(d)) to the four catalytic domains have been computationally predicted and the structure-activity relationships (SARs) were analyzed in detail.
The enzyme inhibition activity of the ligand can be computationally predicted by estimating the binding affinity as the binding free energy between the enzyme and the ligand. The binding affinity can be calculated by various computational methods such as the linear interaction energy method [15], the molecular mechanics/ Poisson-Boltzmann surface area (MM/PBSA) method [16]. In this study, the MM/PBSA method was applied to predict the binding affinities of the salacinol derivatives for each catalytic domain. Then, we examined if the predicted binding affinity values could explain the experimental SARs for salacinol derivatives against alphaglucosidase inhibition activity.
2. Materials and Methods
2.1. Homology Modeling
Homology modeling is a method to construct a threedimensional (3D) structure of a target protein from the experimental 3D structure of a homologous protein as
 (a)
(a)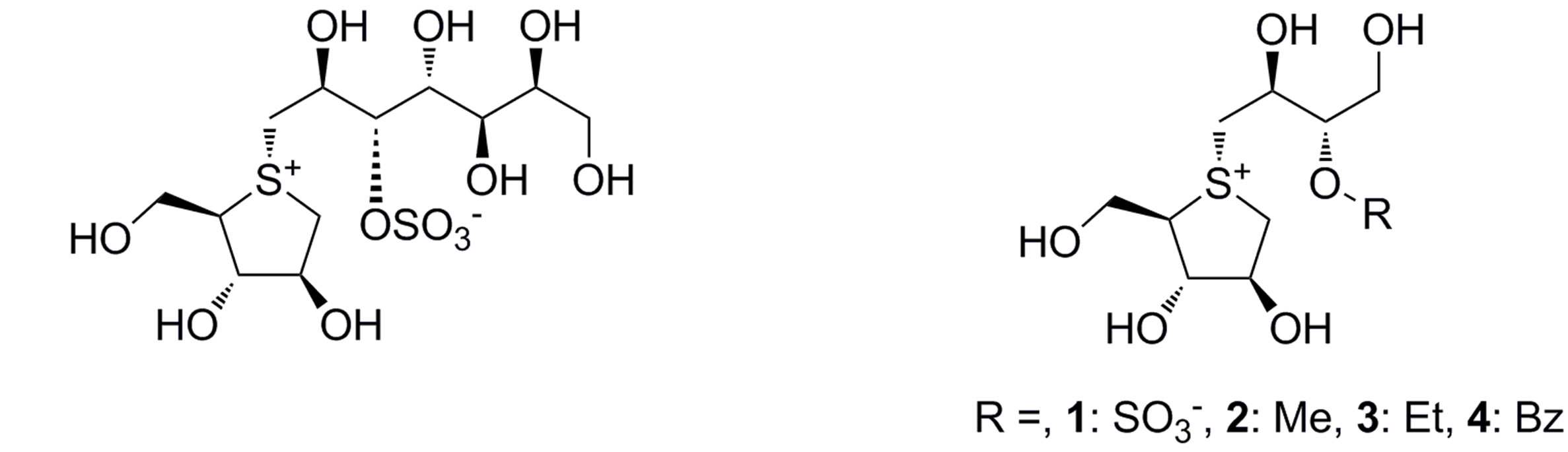 (b)
(b)
Figure 2. Chemical structures of the known alpha-glucosidase inhibitors: (a) Acarbose; (b) Voglibose; (c) Kotalanol; and (d) Salacinol (1) and its derivatives (2-4).
a template. To obtain a reliable model structure, a homology greater than the 30% of the amino acid sequences between a target protein and a template protein is required [14]. As the discrepancy of one amino acid residue pair on the sequence alignment causes a 4 Å displacement of the position [14], the alignment should be performed with care to minimize this error, and a multiple sequence alignment using more than three amino acid sequences is desirable for this purpose.
The amino acid sequences of human CtMGAM and CtSI were retrieved from NCBI-GeneID: 8972 and 6476 [17], respectively. First, the multiple sequence alignment for the four catalytic domains has been performed using the BLOSUM62 score [18]. Then, homology modeling of the two C-terminal catalytic domains has been performed using MODELLER [19].
Prior to the 3D structure construction of the two domains, the calculation conditions of MODELLER were examined by the prediction of the structure of NtSI from the experimental structure of NtSI itself (PDB-ID: 3LPP). Ramachandran plot [20] and the Profile-3D [21] score validated the reliabilities of the built structure models. The calculation conditions that reproduced the experimental structure of NtSI were most appropriately adopted for modeling the unknown structures.
Model structures of CtMGAM and CtSI were built using the experimental structure of the NtMGAM complex with kotalanol (PDB-ID: 3L4V) as a template by MODELLER according to the alignment data and the verified calculation conditions. The positions of the three disulfide bonds were taken from the NtMGAM domain. To consider the hydrogen bond network between kotalanol and the target protein during model building, kotalanol and several water molecules within 10 Å from kotalanol on the binding pocket were also included as a template. Model structures were built two times and loop structures were created three times in each simulation. Therefore, six model structures were proposed. The CHARMM22 force field parameter [22] was used in the model construction.
After the model construction by MODELLER, the hydrogen atoms were assigned to all models using MOE [23]. The geometries of the models were gradually optimized using CHARMM [24]. First, the positions of the hydrogen atoms were optimized, and then, the side chain atoms were relaxed, and finally, all atoms were optimized. At each step, energy minimizations up to 20,000 cycles using the steepest descent and the conjugate gradient method was performed until the RMS gradient was below 0.01 kcal/mol/Å. The model with the most stable potential energy was selected as the representative model structure from the six candidates.
2.2. Binding Mode Prediction
Experimental binding modes for salacinol and/or kotalanol to the N-terminal domains have been already reported [8,9]. In the narrow binding pockets, salacinol and kotalanol formed many hydrogen bonds and a salt bridge with the protein. As the binding modes of the salacinol derivatives may vary according to the shape of each domain mentioned above, docking studies for salacinol and its derivatives (Figure 1(d)) to four catalytic domains have been performed.
For the protein structure to dock, the experimental structures from PDB-ID, 3L4Z and 3LPP, were used for Nt-MGAM and NtSI, respectively, and the predicted model structures were used for CtMGAM and CtSI. The ligand structures were also created based on salacinol from the 3L4Z coordinates. The positions of hydrogen atoms were appropriately assigned using MOE.
Docking studies for each domain have been performed using the MOE-Dock module. The ligand binding site definition was done using the MOE-Alpha SiteFinder module. Before the pose prediction, the conformation generation for the ligands was performed under fixed bond lengths and bond angles. The top 50 poses by the London dG [23] score were chosen from all generated poses for each ligand. These poses were further optimized by the MMFF94x parameter [25] with the generalized born/volume integral solvation model [26] (MM/ GBVI). The pose with the best MM/GBMV score was adopted as its binding mode. The most appropriate docking scheme was explored to reproduce the binding mode observed in the experimental structure, using the NtMGAM complex with salacinol (PDB-ID: 3L4Z).
Recently, the experimental structure of CtMGAM in complex with acarbose has been determined (PDB-ID: 3TOP) [27]. Therefore, the experimental structure of CtMGAM was also employed in the binding mode prediction for comparison purposes. On the other hand, for the modeling of the CtSI structure, using the experimenttal structure of CtMGAM as a template may be more appropriate than using that of NtMGAM, because the sequence homology of CtMGAM to CtSI is much higher than that of NtMGAM (Figure 1). In this study, however, the experimental structure of NtMGAM in complex with kotalanol was adopted to consider an induced-fit of the enzyme to the thiosugar moiety in kotalanol and salacinol derivatives.
2.3. Binding Affinity Prediction
To analyze the ligand affinities considering the thermodynamic behavior of molecules, molecular dynamics (MD) simulations have been performed for each complex. For the MD simulations in a solvent, a spherical cluster of water molecules was generated around 30 Å from the ligand coordinate center using InsightII [28]. Before the MD simulation, the structure optimization was carried out until the RMS gradient was below 0.01 kcal/mol/Å. To relax the molecule system, heating from 0 K to 300 K was performed in 50 ps. After 2 ns in the equilibration stage, 100 snapshots were sampled from a 500 ps simulation. The following MD conditions were used: a time step of 1 fs; a dielectric constant of 1; and a cutoff distance for a nonbonded interaction of 15 Å. SHAKE [29] was applied to fix the bond length in all hydrogen atoms. The surface of the spherical water cluster was constrained by 100 kcal/mol/Å to prevent the water molecules from evaporating. The amino acid residues outside the water cluster were also harmonically constrained to the original position by 100 kcal/mol/Å2. All dynamics simulations were performed using CHARMM [24] with the CHARMm.cfrc parameters [28].
Binding affinities for salacinol and its derivatives were estimated using 100 snapshots for each MD simulation by the MM/PBSA method. The van der Waals and electrostatic interaction energies between the ligand and protein for every snapshot were calculated by CHARMM and their averages were used as the MM interaction term. Desolvation energies for the solvent effect were calculated by DelPhi [30] as the PB term for the polar solvent effect, and by MSMS [31] as the SA term for the nonpolar solvent effect. The dielectric constants of 4 and 80 were used for the solute and solvent, respectively. The ion strength was set to 0.145 M. The PARSE parameters [32] for the atomic radius in the PB and SA calculations were used. To calculate the nonpolar SA term, the following parameters were adopted for the PARSE radius surface: the surface-scale factor was 0.00542 and the constant was 0.92 [32].
3. Results and Discussion
3.1. Homology Modeling
The sequence alignment of the amino acids in the four catalytic domains is shown in Figure 3. Homology modeling has been performed using these alignment pairs. The remodeling structure of NtSI and its crystal structure (PDB-ID: 3LPP) were compared and shown in Figure 4. The RMSD value of the main chain atoms between the model and the crystal structure was 1.8 Å and that of all heavy atoms within 5 Å from kotalanol was 0.7 Å. These values are small enough to consider that the model structure

Figure 3. Multiple sequence alignment of the amino acid residues in the four human alpha-glucosidase catalytic domains. Homologous pairs in the alignment are shown in blue background. Blue color depth stands for the characteristic similarity of the amino acid residues.
reproduces the experimental structure. However, the directions of the side chain atoms of several amino acid residues were not reproduced (Figure 4(b)). Most of these amino acid residues are hydrophobic and do not affect the hydrogen bond network in ligand binding. Such positional differences in these residues are also observed between the experimental structures of NtMGAM and NtSI, the most homologous pairs among the four domains (Figure 1).
Also, the predicted CtMGAM structure was compared to the experimental CtMGAM structure (PDB-ID: 3TOP) [27]. The RMSD value of the main chain atoms between the two structures was 3.6 Å, and this large displacement was originated in the long insertion loop near the substrate binding site (Figure 3). Predicting the structure of such a long insertion loop by MODELLER is extremely difficult without reference coordinates in the template structure. However, the RMSD value of 1.2 Å for all heavy atoms within 5 Å from kotalanol indicates that the model structure around the ligand binding site reproduces the experimental structure well.
The structural validity of the model was also examined by using the Ramachandran plot and the Profile-3D method. The φ-ψ angle distribution was normal except for several amino acid residues located at the loop region, which is distant from the active site. The qualitative validation of the model structure was confirmed by Profile-3D [21]. Although the threshold score for NtSI was 17 - 8.22, the calculated score of 369.80 for the NtSI model indicated that the model had no significant structural problem. As for the modeling scheme adopted here, it reproduced the experimental structure well and the structures of the two C-terminal domains were predicted with the same scheme.
The predicted CtMGAM and CtSI models were evaluated by the Ramachandran plot as well as the Profile-3D method. No abnormal φ-ψ angle distributions were observed in the Ramachandran plot of both models. The Profile-3D score for the CtMGAM and CtSI models was 366.45 and 371.46, respectively, and these values exceeded the corresponding threshold values of 185.50 and 184.46. Therefore, the model structures are considered valid for use in the comparison study that investigated the shape of the active site.
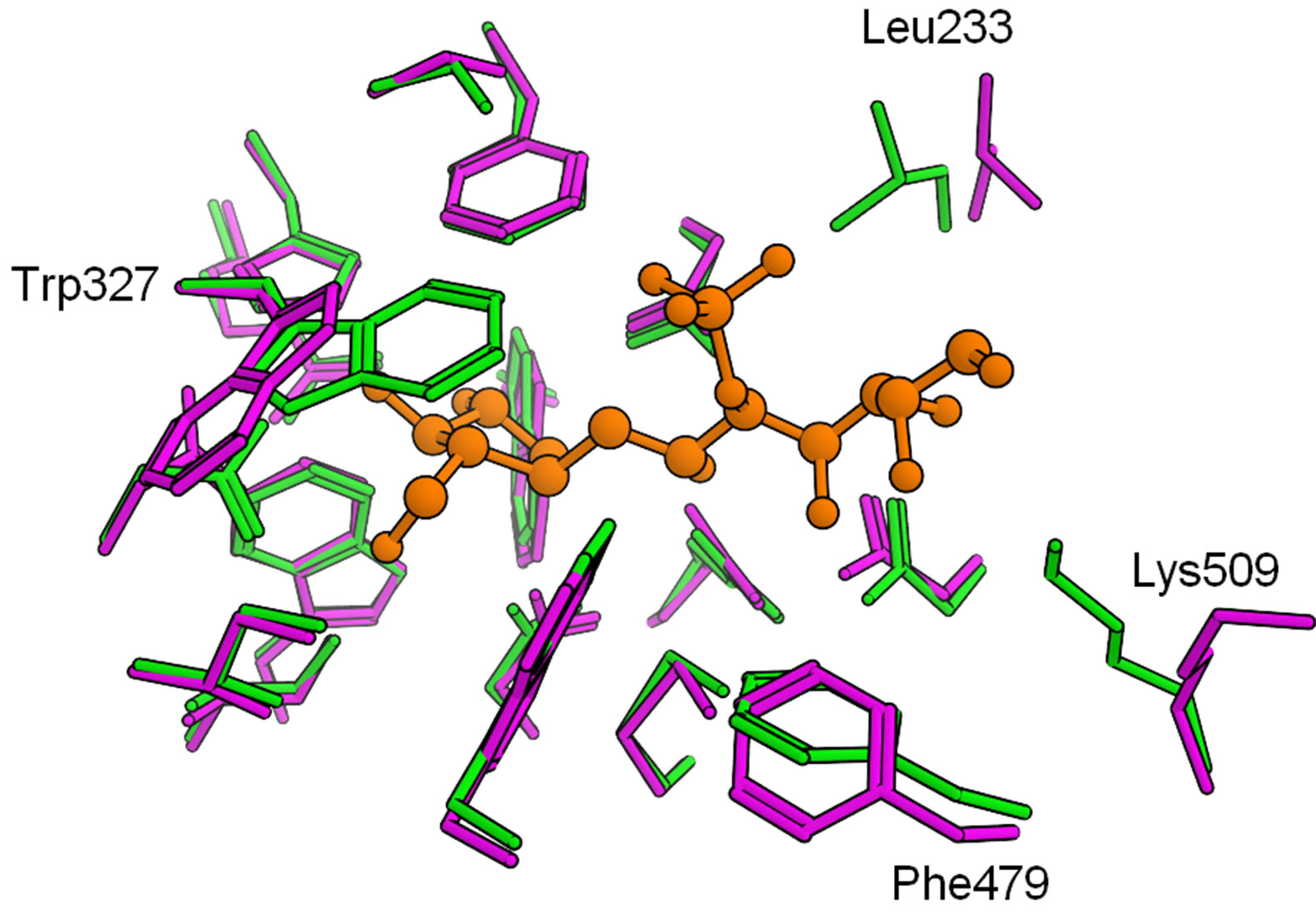
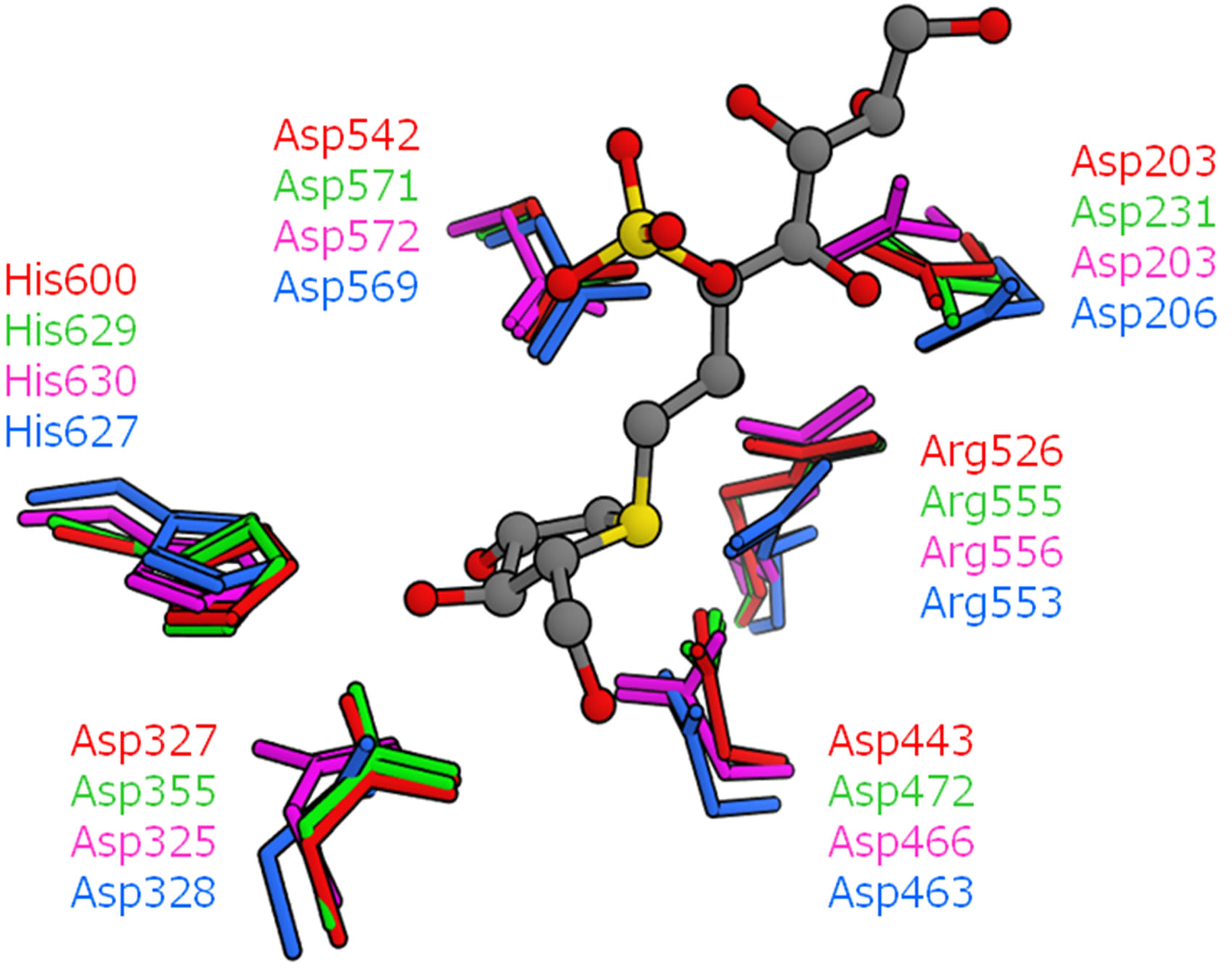
The detailed structural comparisons among the four catalytic domains are shown in Figure 5. The amino acid residues, which form hydrogen bonds or a salt bridge with kotalanol, were also compared (Figure 5(a)). These residues were conserved among the four catalytic domains and no significant structural difference was observed. That is, hydrogen bonds and salt-bridge interacttion that are essential for kotalanol binding are common among all the catalytic domains. This means that such interactions would be conserved in the binding of salacinol and its derivatives. Then, the mutated amino acid residues around the kotalanol binding site were compared (Figure 5(b)). Subsequently, we focus our discussion on the five amino acid residues: The205, Tyr299, Trp406, Ala576, and Gly602 of NtMGAM. The residues corresponding to Thr205 in the NtMGAM and located near the sulfate group were deemed distant enough not to affect the ligand interaction; however, the large hydrophobic residue Leu233 in NtSI would make the pocket narrow and may show a different ligand recognition and specificity. Tyr-299 in NtMGAM mutates to Trp327 in NtSI and is conserved in the other domains. As the hydroxy groups of the conserved tyrosine residues interact with the other residues, there are no polar interactions with the ligand. The larger tryptophan residue in NtSI would form a smaller binding pocket. The position of Trp406 at the sulfate group-binding pocket in NtMGAM was occupied with the proline residue in CtSI. Even though the tryptophan residue interacted through the CH-O interaction with the sulfate group of kotalanol, the proline residue cannot interact with the same manner and it is supposed to interact weakly. Ala576 in NtMGAM corresponded to
 (a)
(a)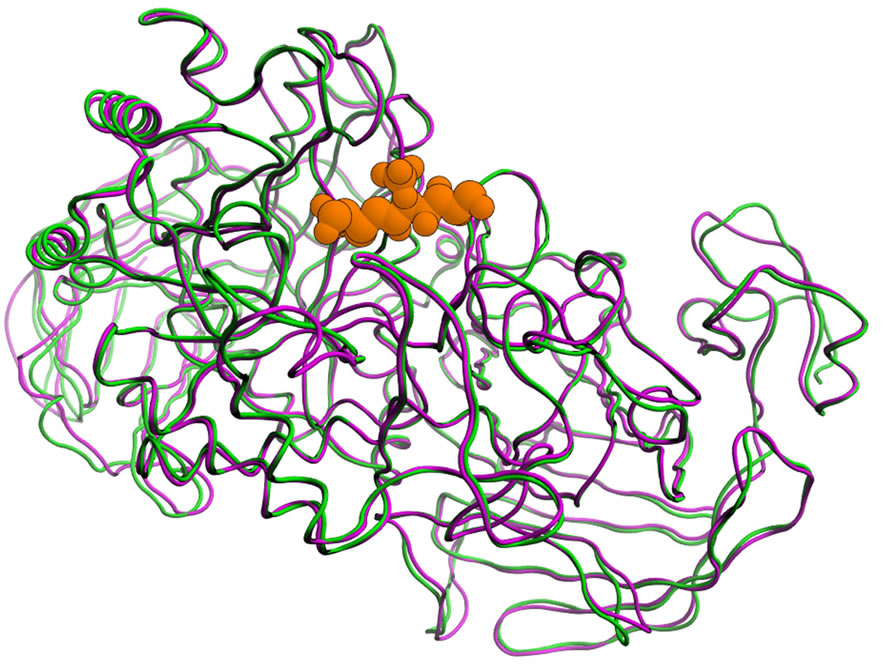 (b)
(b)
Figure 4. Comparison of the NtSI structure (magenta: homology model, green: experiment), and kotalanol in 3LPP is shown in orange. (a) Overall folding of the main chain and (b) Active site, amino acid residues with relatively large displacement are labeled.
 (a)
(a)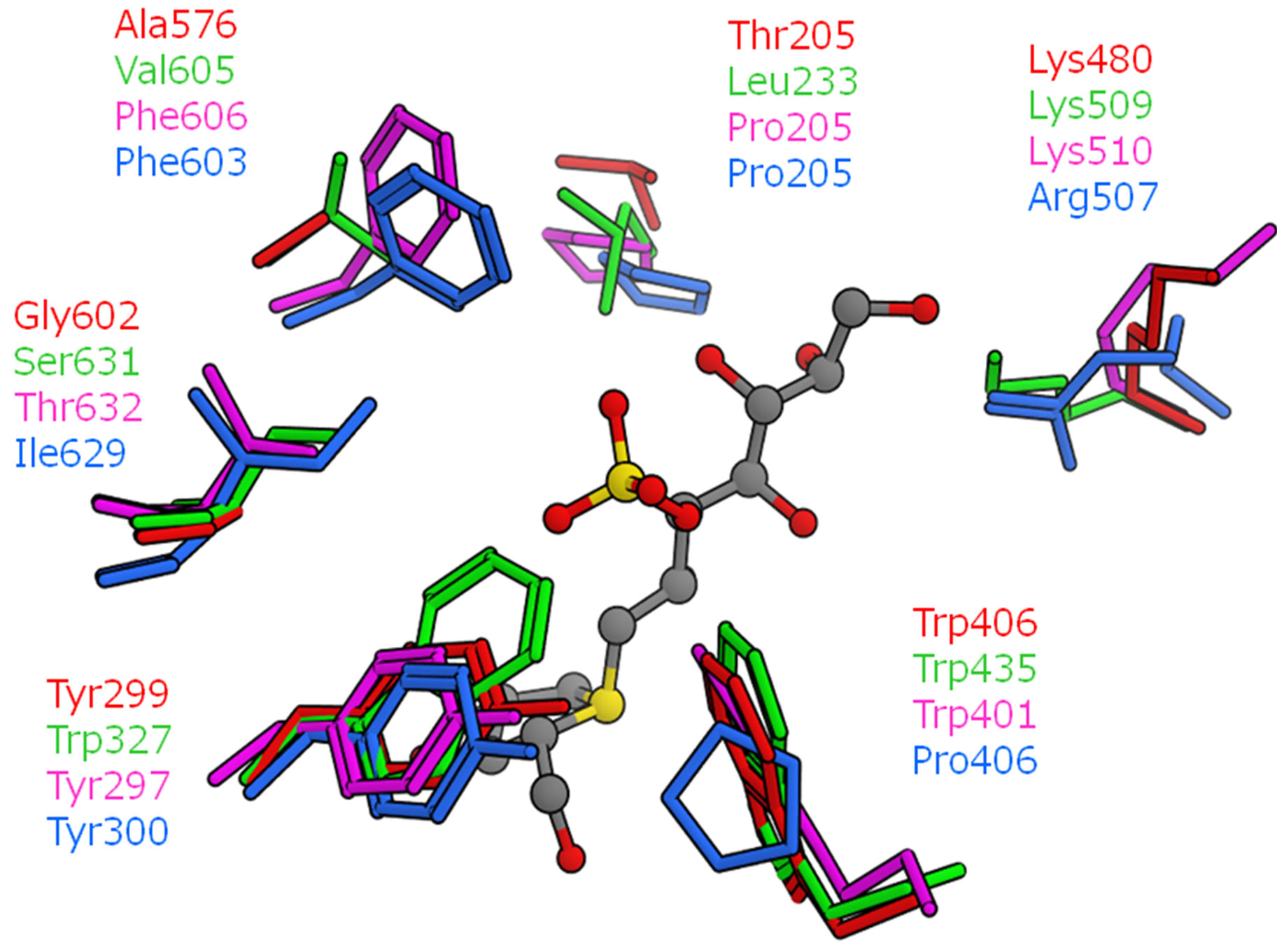 (b)
(b)
Figure 5. Superposition of the four catalytic domains (red: NtMGAM, green: NtSI, magenta: CtMGAM, cyan: CtSI). The position of kotalanol in the NtMGAM complex is shown as a ball-stick model. (a) Amino acid residues related to hydrogen bonds and a salt bridge with kotalanol; (b) Neighboring amino acid residues with mutation.
valine in NtSI, phenylalanine in CtMGAM and CtSI. Although this residue is slightly apart from kotalanol, the binding pockets of both the C-terminal domains are narrower than the N-terminal domains. Gly602 in NtMGAM varied among the domains. For NtSI and CtMGAM, this residue is mutated by the serine and threonine residues, respectively. As for CtSI, the isoleucine residue is located in this position. These residues do not significantly contribute to the ligand-binding interaction because this residue is located behind Tyr299 and Phe575, and is not exposed to the active site surface.
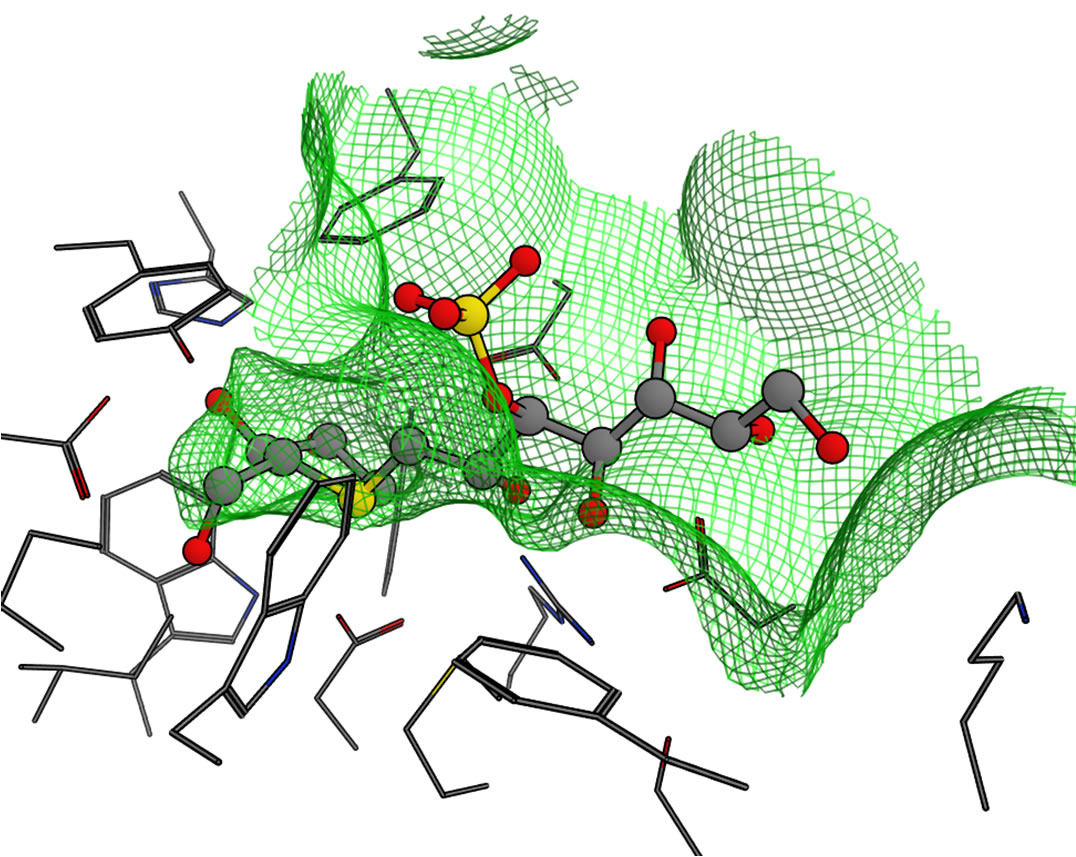
To compare the shape of the binding pockets, the molecular surfaces of the kotalanol binding site (around 5 Å from kotalanol in NtMGAM) are shown in Figure 6. It was predicted that the pockets were deeper in the order of NtMGAM, NtSI, CtMGAM, and CtSI. In particular, in CtSI, the overlaid sulfate group of kotalanol seemed to be entirely covered by the pocket and might contact with the wall (Figure 6(d)). This means that the van der Waals interaction between the sulfate group of salacinol and each domain might be stronger in the same order of the pocket depth except for CtSI.
3.2. Binding Mode Prediction of Salacinols
The structure of the calculated NtMGAM complex with salacinol was compared with that of the experiments to validate the docking condition. The RMSD value for non- hydrogen atoms was 0.4 Å, which encouraged us to predict the binding modes of other ligand complexes by this condition.
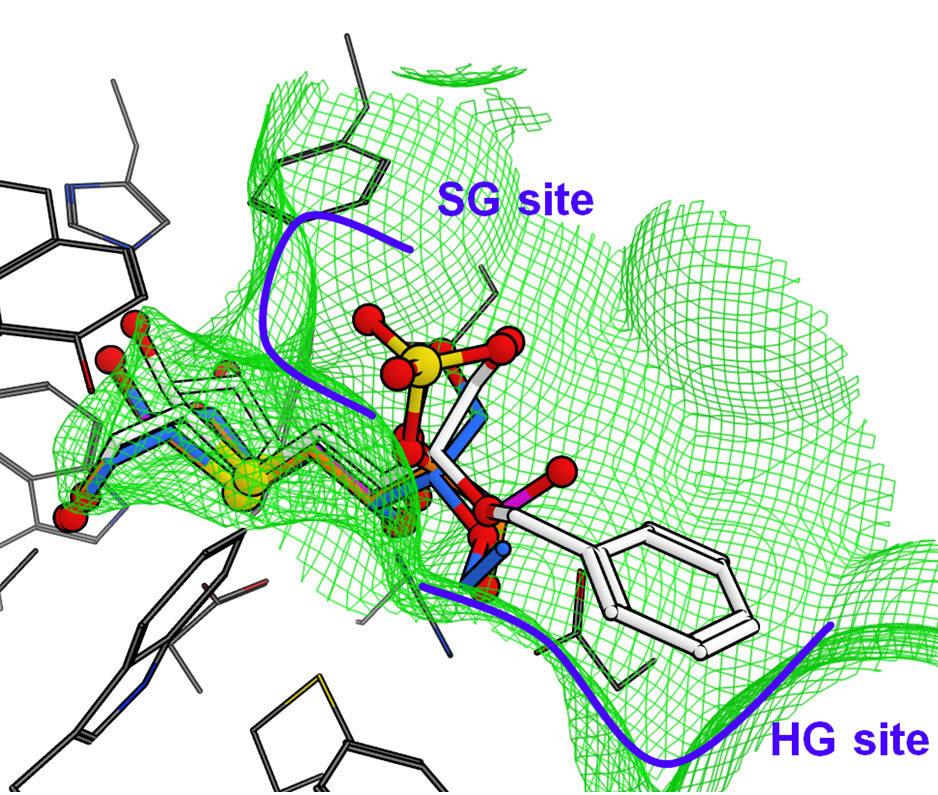
The docked structure for salacinol and its derivatives to NtMGAM is shown in Figure 7(a). Interestingly, the methyl group of 2 is located on a similar position of the sulfate group of salacinol (1), and the ethyl group of 3 and the benzyl group of 4 are located where the C4’ hydroxy group exists. In the following discussion, the sulfate group-binding site and the C4’ hydroxy groupbinding site are referred to as the SG site and HG site, respectively.
In NtMGAM, the SG site consists of many aromatic residues such as Phe575, Tyr299, and Trp406; the HG site consists of various kinds of residues such as Phe450, Asp203, and Lys480. This indicates that the hydrophobic group is likely to bind at the SG site, but it conflicts with the docking study that the hydrophobic ethyl and phenyl group of compound 2 and 3 bind at the HG site. However, the HG site is formed by the aromatic ring and the hydrophobic methylene chains of the hydrophilic amino acid residues, and the environment may be considerably hydrophobic. It is assumed that shape complementarity is important for ligand recognition. For example, the size of the phenyl ring of 4 is sufficient to bind either to the SG site or the HG site. However, the whole phenyl ring interacts with the HG site, whereas part of the ring can interact with the SG site.
The predicted binding mode of the salacinol derivatives to NtSI is shown in Figure 7(b). The NtSI complex structure with salacinol (1) has not been reported but that with kotalanol was known [9]. The hydrogen bond network and the salt-bridge formation were nearly common between both ligands on the NtMGAM complex [8]. Thus, the binding mode of salacinol on NtSI was supposed to be the same as that of kotalanol. Similar to the NtMGAM case, the methyl group of 2 was bound to the SG site. The SG site of NtSI consists of many aromatic amino acid residues similar to NtMGAM, but the difference from Tyr299 in NtMGAM to Trp327 in NtSI made the SG site a little smaller as previously described. Furthermore, Ser448 around the HG site in NtMGAM changed into the basic and more hydrophilic Lys509 in NtSI, and the HG site became narrow as shown in Figure 7(b). Therefore, the hydrophobic substituent of 3 and 4 bind to the SG site and gain more van der Waals interacttions with the hydrophobic SG site than those with the HG site, whose hydrophobicity decreased. These results suggest that the introduction of the acidic group at the C4’ position to interact with the basic Lys509 residue would enhance the binding affinity to NtSI.
The docking of salacinol derivatives to the CtMGAM model is shown in Figure 7(c). The binding mode of 1, 2, and 3 in CtMGAM were similar to those in NtMGAM and NtSI. On the other hand, the interaction between the phenyl ring of 4 and the HG site wall seen in NtMGAM was lost. The phenyl ring of 4 was bound to the SG site benefiting from the many van der Waals interactions to
 (a)
(a)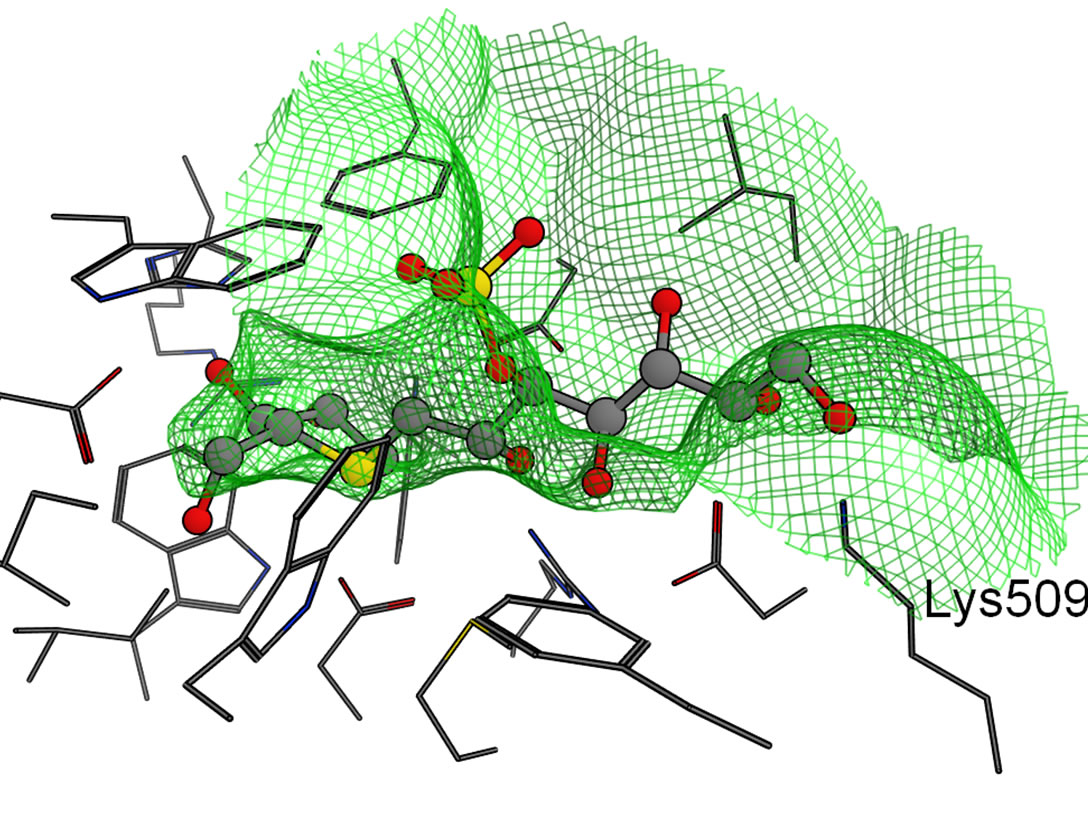 (b)
(b)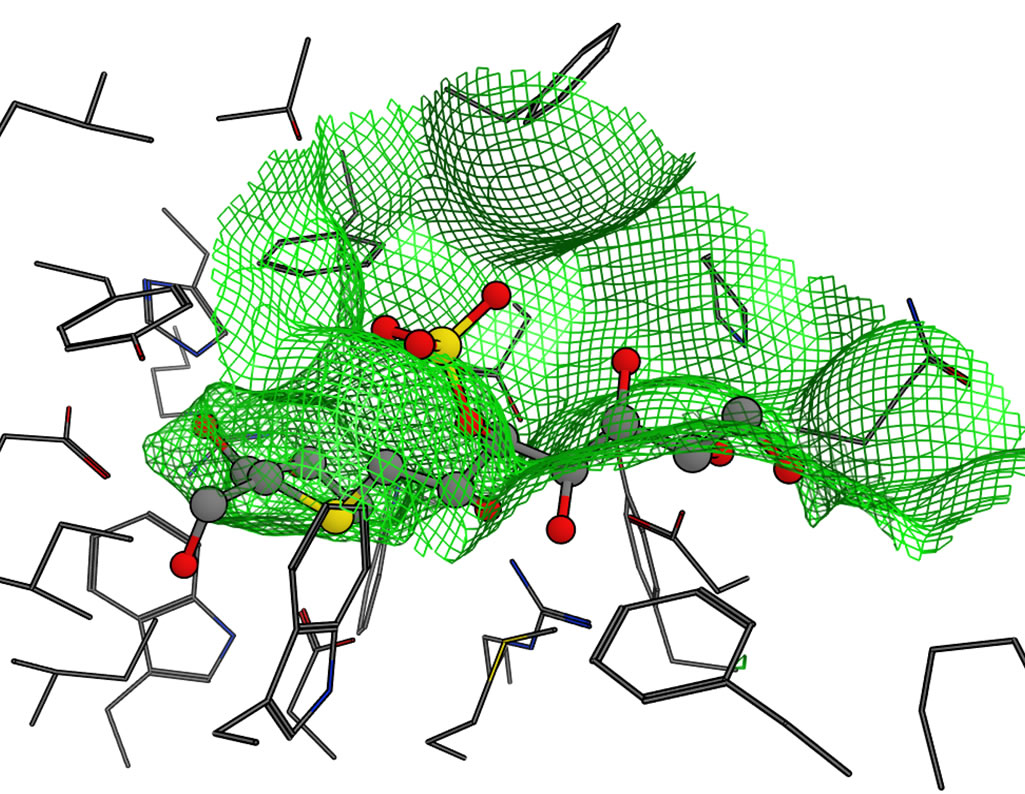 (c)
(c)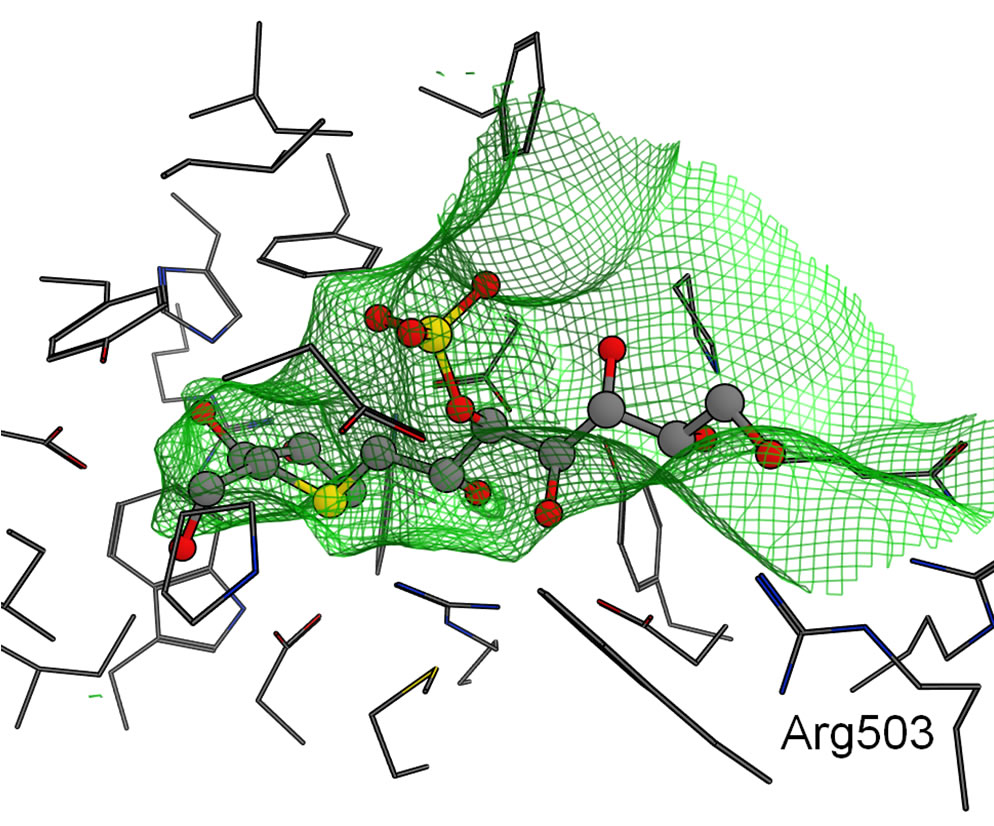 (d)
(d)
Figure 6. Molecular surfaces of the active sites in the four domains. The green mesh shows the van der Waals contact surface. The position of kotalanol in the NtMGAM complex is overlaid as a ball-stick model: (a) NtMGAM; (b) NtSI; (c) CtMGAM; and (d) CtSI.
 (a)
(a)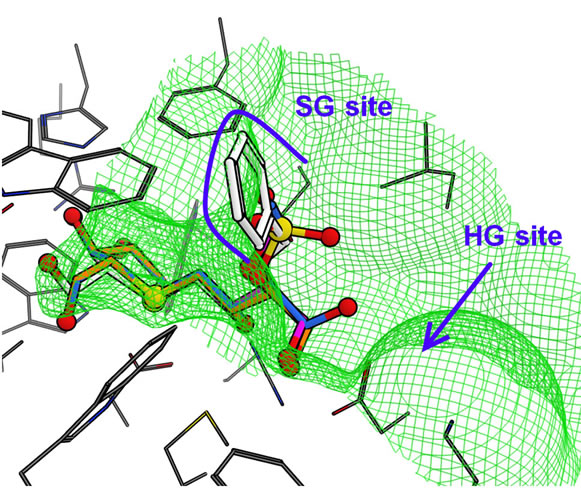 (b)
(b)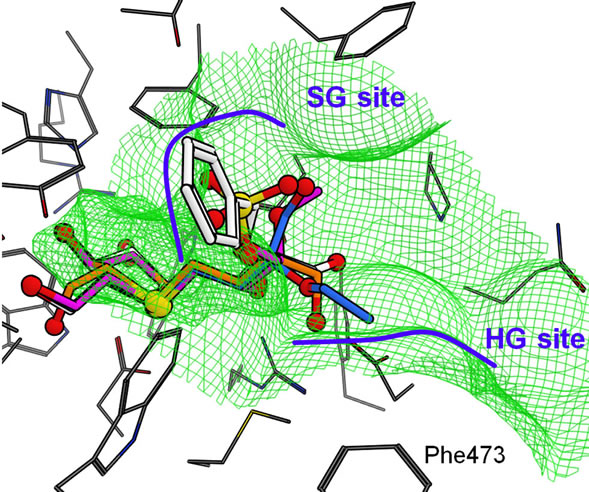 (c)
(c)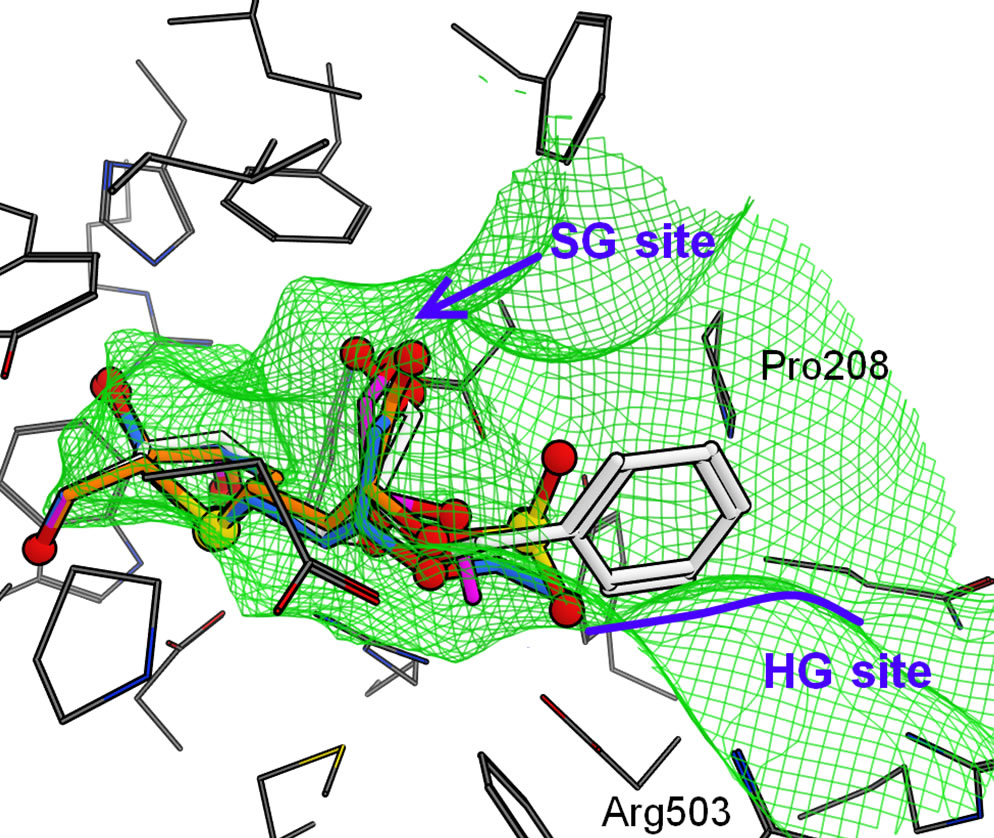 (d)
(d)
Figure 7. Predicted binding modes of salacinol and its derivatives for each catalytic domain: (a) NtMGAM, (b) NtSI, (c) CtMGAM, and (d) CtSI. Compounds 1, 2, 3, and 4 are shown in orange, magenta, blue, and white, respectively, by the stick model with a partially ball model for noncarbon atoms. The green mesh shows van der Waals contact surface (same as Figure 6). Blue curves or arrows depict the sulfate group-binding site (SG site) and the C4’ hydroxy group-binding site (HG site).
the hydrophobic amino acid residues (Tyr297, Trp401, and Ile633) because the HG site became shallower by Phe473, which moved to the HG site that was being affected by the long insertion loop in CtMGAM.
The docking simulation to the experimental CtMGAM structure was also performed. The structural difference of the long insertion loop near the ligand binding region between the experimental and the model structures did not affected the binding mode of salacinol derivatives. That is, the binding mode of each derivative to the both structures was quite similar. However, attention would be required in docking much larger compounds such as acarbose.
Finally, the binding modes for CtSI are shown in Figure 7(d). Interestingly, the sulfate group of 1 was predicted to bind to the HG site like the phenyl ring of 4 in NtMGAM as mentioned above because (a) the SG site of CtSI surrounded by Phe603, Ile629, and Leu407 were much narrower than the other domains (Figures 5 and 7) and (b) positively charged Arg503 was located at the HG site. Therefore, the negatively charged sulfate group was attracted by Arg503 in the HG site. As the size of the SG site was fitted to the hydroxy group at the C4’ position, the methyl group of 2 and the ethyl group of 3 were located toward the HG site. The HG site also became narrow because of the existence of Arg503, and then, the phenyl ring of 4 was caught in a narrow groove formed by Arg503 and Pro208.
Thus, the binding mode of salacinol and its derivatives for the four domains has been deduced by the docking study. The substituent groups were not uniformly bound and their location varied according to the shape and properties of the active site in each catalytic domain.
3.3. Binding Affinity Prediction
The experimental inhibition activities of the three substrates [7,13] and calculated binding affinities of salacinol derivatives for the four domains are shown in Table 1. As for the calculated binding affinities for NtMGAM, three compounds (1, 2, and 3) have almost the same strengths and 4 was much stronger than others in all four domains. For CtMGAM(model) and NtSI, 4 was the strongest but 1 was weaker than 2 and 3. In CtSI, however, 2 was much weaker than 1.
The binding affinities of the compounds in CtMGAM (X-ray) were relatively weaker than those in CtMGAM (model). The induced-fit of the enzyme to the thiosugar moiety would have arisen this difference and affected the affinity order. However, it is noted that 4 was still strongest.
It is known that maltose hydrolysis, which is most important for diabetes, is related to all domains, whereas isomaltose and sucrose hydrolysis are related to NtSI and CtSI, respectively [9,33]. The calculated affinities for NtSI agreed quite well with the isomaltase inhibition activity. For maltase, the inhibitions by 3 and 4 are three times and ten times as strong as those by 1 and 2, respectively. It is difficult to evaluate the maltase inhibition simply due to the effect of the four domains. That is, the maltase inhibitory activity should be considered for all catalytic domains. For example, the weak affinity of 2 for NtMGAM and CtSI could be compensated by the strong affinities for NtSI and CtMGAM. Thus, maltose hydrolysis can be explained by the calculated binding affinities.
As described above, the inhibition activities of maltose and isomaltose hydrolysis were consistent with the predicted binding affinities. However, for sucrose, the calculated values for 3 were inconsistent with the experiments. This means that the current MM/PBSA scheme, including the model structure construction of the C-terminal domains, leaves room for improvement. For example, the recent alpha-glucosidase study showed the existence of splicing variants on CtMGAM [34]. As some
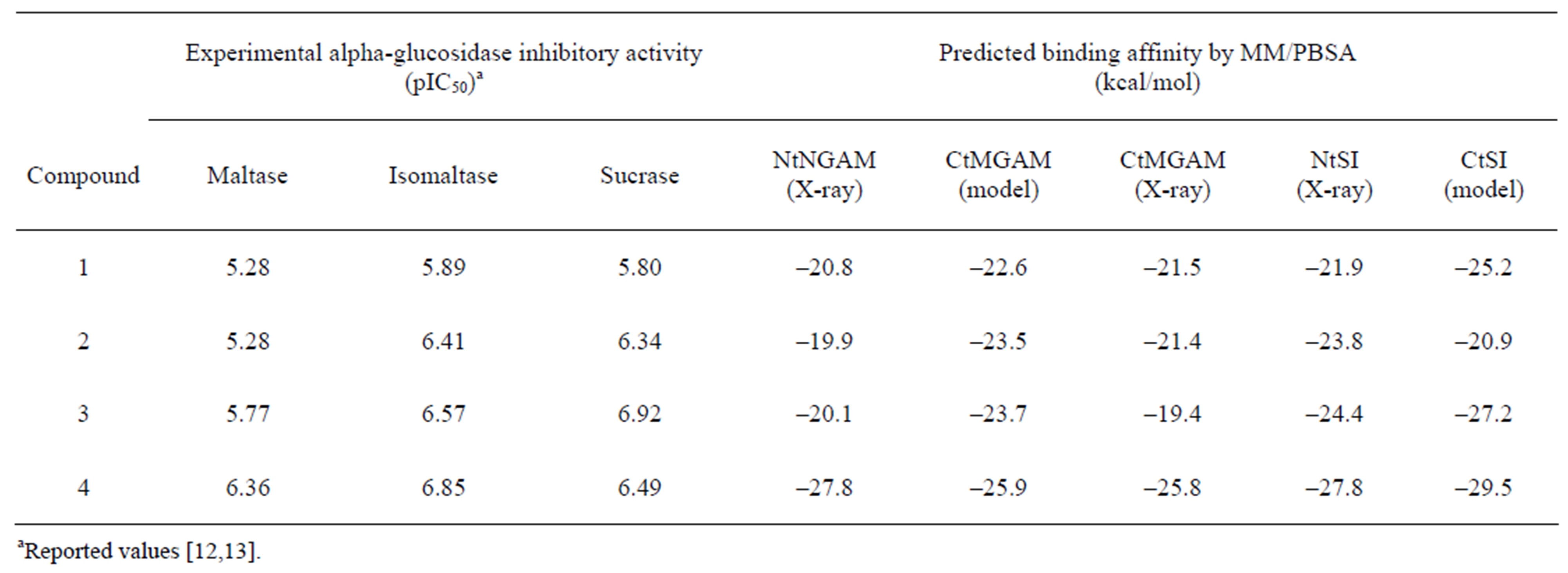
Table 1. Experimental inhibitory activity and estimated binding affinities of salacinol and its derivatives.
amino acid residues of the active site might mutate, we have to consider such effects.
4. Conclusions
In this study, the inhibition of alpha-glucosidase, a target protein in diabetic therapy, has been computationally investigated. As the four catalytic domains of human alpha-glucosidase have been known, the shapes and the characters of their binding pockets were analyzed. Furthermore, the binding modes and the affinities of the salacinol derivatives have been compared.
Because the structures of CtMGAM and CtSI were unknown at the time we started this study, they have been predicted using the homology modeling method. The comparison of the ligand-binding pockets among the four catalytic domains indicated few differences on the salt bridge and hydrogen bond network for the ligand binding observed in the NtMGAM complex with salacinol. However, several amino acid residues mutated, resulting in differences in the shape and physicochemical properties of the SG site and HG site of each catalytic domain.
The binding modes of salacinol and its derivatives to the four domains have been predicted by the docking study. The locations of the substituent groups in the active site varied according to the shape features of the active site in each catalytic domain.
To predict the binding affinity quantitatively, the binding affinities of salacinol and its derivatives for each domain were estimated by the MM/PBSA method. The inhibition activities of the maltose and isomaltose hydrolysis were consistent with the experimentally determined binding affinities, whereas that of sucrose hydrolysis was partly inconsistent.
Though more accuracy is required for the model construction and the affinity prediction, several designed compounds based on the model structures and SARs have improved the activities of all alpha-glucosidase [35].
5. Acknowledgements
This work was supported by the “High-Tech Research Center Project”: Matching Fund Subsidy for Private Universities from MEXT (Ministry of Education, Culture, Sports, Science and Technology), 2007-2011.
REFERENCES
- H. A. Ernst, L. L. Leggio, M. Willemoës, G. Leonard, P. Blum and S. Larsen, “Structure of the Sulfolobus Solfataricus Alpha-Glucosidase: Implications for Domain Conservation and Substrate Recognition in GH31,” Journal of Molecular Biology, Vol. 358, No. 4, 2006, pp. 1106-1124. doi:10.1016/j.jmb.2006.02.056
- G. M. Gray, B. C. Lally and K. A. Conklin, “Action of Intestinal Sucrase-Isomaltase and Its Free Monomers on an α-Limit Dextrin,” The Journal of Biological Chemistry, Vol. 254, No. 13, 1979, pp. 6038-6043.
- J. L. Chiasson, R. G. Josse, R. Gomis, M. Hanefeld, A. Karasik and M. Laakso, “Acarbose for Prevention of Type 2 Diabetes Mellitus: The STOP-NIDDM Randomised Trial,” Lancet, Vol. 359, No. 9323, 2002, pp. 2072-2077. doi:10.1016/S0140-6736(02)08905-5
- B. Göke, H. Fuder, G. Wieckhorst, U. Theiss, E. Stridde, T. Littke, P. Kleist, R. Arnold and P. W. Lücker, “Voglibose (AO-128) Is an Efficient α-Glucosidase Inhibitor and Mobilizes the Endogenous GLP-1 Reserve,” Digestion, Vol. 56, No. 6, 1995, pp. 493-501. doi:10.1159/000201282
- R. J. Andrade, M. Lucena, J. L. Vega, M. Torres, F. J. Salmeron, V. Bellot, M. D. Garcia-Escano and P. Moreno, “Acarbose-Associated Hepatotoxicity,” Diabetes Care, Vol. 21, No. 11, 1998, pp. 2029-2030. doi:10.2337/diacare.21.11.2029
- M. Yoshikawa, T. Murakami, K. Yashiro and H. Matsuda, “Kotalanol, a Potent α-Glucosidase Inhibitor with Thiosugar Sulfonium Sulfate Structure, from Antidiabetic Ayurvedic Medicine Salacia reticulata,” Chemical and Pharmaceutical Bulletin, Vol. 46, No. 8, 1998, pp. 1339- 1340. doi:10.1248/cpb.46.1339
- M. Yoshikawa, T. Murakami, H. Shimada, H. Matsuda, J. Yamahara, G. Tanabe and O. Muraoka, “Salacinol, Potent Antidiabetic Principle with Unique Thiosugar Sulfonium Sulfate Structure from the Ayurvedic Traditional Medicine Salacia reticulata in Sri Lanka and India,” Tetrahedron Letters, Vol. 38, No. 48, 1997, pp. 8367-8370. doi:10.1016/S0040-4039(97)10270-2
- L. Sim, K. Jayakanthan, S. Mohan, R. Nasi, B. D. Johnston, B. M. Pinto and D. R. Rose, “New Glucosidase Inhibitors from an Ayurvedic Herbal Treatment for Type 2 Diabetes: Structures and Inhibition of Human Intestinal Maltase-Glucoamylase with Compounds from Salacia reticulata,” Biochemistry, Vol. 49, No. 3, 2010, pp. 443- 451. doi:10.1021/bi9016457
- L. Sim, C. Willesma, S. Mohan, Y. H. Naim, B. M. Pinto and D. R. Rose, “Structural Basis for Substrate Selectivity in Human Maltase-Glucoamylase and Sucrase-Isomaltase N-Terminal Domains,” The Journal of Biological Chemistry, Vol. 285, No. 23, 2010, pp. 17763-17770. doi:10.1074/jbc.M109.078980
- R. Eskandari, K. Jones, D. R. Rose and B. M. Pinto, “Probing the Active-Site Requirements of Human Intestinal N-Terminal Maltase Glucoamylase: The Effect of Replacing the Sulfate Moiety by a Methyl Ether in Ponkoranol, a Naturally Occurring α-Glucosidase Inhibitor,” Bioorganic & Medicinal Chemistry Letters, Vol. 20, No. 19, 2010, pp. 5686-5689. doi:10.1016/j.bmcl.2010.08.020
- M. Yoshikawa, F. Xu, S. Nakamura, T. Wang, H. Matsuda, G. Tanabe and O. Muraoka, “Salaprionol and Ponkoranol with Thiosugar Sulfonium Sulfate Structure from Salacia prinoides and α-Glucosidase Inhibitory Activity of Ponkoranol and Kotalanol Desulfate,” Heterocycles, Vol. 75, No. 6, 2008, pp. 1397-1405. doi:10.3987/COM-07-11315
- S. Nakamura, K. Takahira, G. Tanabe, T. Morikawa, M. Sakano, K. Ninomiya, M. Yoshikawa, O. Muraoka and I. Nakanishi, “Docking and SAR Studies of Salacinol Derivatives as α-Glucosidase Inhibitors,” Bioorganic & Medicinal Chemistry Letters, Vol. 20, No. 15, 2010, pp. 4420- 4423. doi:10.1016/j.bmcl.2010.06.059
- G. Tanabe, T. Otani, W. Cong, T. Minematsu, K. Ninomiya, M. Yoshikawa and O. Muraoka, “Biological Evaluation of 3’-O-Alkylated Analogs of Salacinol, the Role of Hydrophobic Alkyl Group at 3’ Position in the Side Chain on the α-Glucosidase Inhibitory Activity,” Bioorganic & Medicinal Chemistry Letters, Vol. 21, No. 10, 2011, pp. 3159-3162. doi:10.1016/j.bmcl.2011.02.109
- N. Eswar, B. Webb, M. A. Marti-Renom, M. S. Madhusudhan, D. Eramian, M. Shen, U. Pieper and A. Šali “Comparative Protein Structure Modeling Using MODELLER,” Current Protocols in Protein Science, 2007, pp. 2.9.1-2.9.31. doi:10.1002/0471140864.ps0209s50
- J. Aqvist, C. Medina and J. E. Samuelsson, “New Method for Predicting Binding-Affinity in Computer-Aided Drug Design,” Protein Engineering Design and Selection, Vol. 7, No. 3, 1994, pp. 385-391. doi:10.1093/protein/7.3.385
- P. A. Kollman, I. Massova, C. Reyes, B. Kuhn, S. Huo, L. Chong, M. Lee, T. Lee, Y. Duan, W. Wang, O. Donini, P. Cieplak, J. Srinivasan, D. A. Case and T. E. Cheatham III, “Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models,” Accounts of Chemical Research, Vol. 33, No. 12, 2000, pp. 889-897. doi:10.1021/ar000033j
- B. L. Nichols, S. Avery, P. Sen, D. M. Swallow, D. Hahn and E. Sterchi, “The Maltase-Glucoamylase Gene: Common Ancestry to Sucrase-Isomaltase with Complementary Starch Digestion Activities,” Proceedings of the National Academy of Sciences, Vol. 100, No. 3, 2003, pp. 1432-1437. doi:10.1073/pnas.0237170100
- S. R. Eddy, “Where Did the BLOSUM62 Alignment Score Matrix Come From?” Nature Biotechnology, Vol. 22, No. 8, 2004, pp. 1035-1036. doi:10.1038/nbt0804-1035
- A. Šali and T. L. Blundell, “Comparative Protein Modelling by Satisfaction of Spatial Restraints,” Journal of Molecular Biology, Vol. 234, No. 3, 1993, pp. 779-815. doi:10.1016/S1357-4310(95)91170-7
- S. S. Sheik, P. Sundararajan, A. S. Z. Hussain and K. Sekar, “Ramachandran Plot on the Web,” Bioinformatics, Vol. 18, No. 11, 2002, pp. 1548-1549. doi:10.1093/bioinformatics/18.11.1548
- R Lüthy, J. U. Bowie and D. Eisenberg, “Assessment of Protein Models with Three-Dimensional Profiles,” Nature, Vol. 356, No. 6364, 1992, pp. 83-85. doi:10.1038/356083a0
- A. D. MacKerell Jr., D. Bashford, M. Bellott, R. L. Dunbrack Jr., J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher III, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiórkiewicz-Kuczera, D. Yin and M. Karplus, “All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins,” The Journal of Physical Chemistry B, Vol. 102, No. 18, 1998, pp. 3586-3616. doi:10.1021/jp973084f
- MOE ver.2010, Chemical Computing Group Inc., Montreal. http://www.chemcomp.com/
- B. R. Brooks, R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan and M. Karplus, “CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations,” Journal of Computational Chemistry, Vol. 4, No. 2, 1983, pp. 187-217. doi:10.1002/jcc.540040211
- T. A. Halgren and R. B. Nachbar, “Merck Molecular Force Field. IV. Conformational Energies and Geometries for MMFF94,” Journal of Computational Chemistry, Vol. 17, No. 5-6, 1996, pp. 587-615. doi:10.1002/(SICI)1096-987X(199604)17:5/6<587::AID-JCC4>3.0.CO;2-Q
- P. Labute, “The Generalized Born/Volume Integral Implicit Solvent Model: Estimation of the Free Energy of Hydration Using London Dispersion Instead of Atomic Surface Area,” Journal of Computational Chemistry, Vol. 29, No. 10, 2008, pp. 1693-1698. doi:10.1002/jcc.20933
- L. Ren, X. Qin, X. Cao, L. Wang, F. Bai, G. Bai and Y. Shen, “Structural Insight into Substrate Specificity of Human Intestinal Maltase-Glucoamylase,” Protein Cell, Vol. 2, No. 10, 2011, pp. 827-836. doi:10.1007/s13238-011-1105-3
- InsightII ver.2000, Accelrys Inc. San Diego. http://accelrys.com/
- J. P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, “Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes,” Journal of Computational Physics, Vol. 23, No. 3, 1977, pp. 327-341. doi:10.1016/0021-9991(77)90098-5
- M. K. Gilson, K. Sharp and B. Honig, “Calculating the Electrostatic Potential of Molecules in Solution: Method and Error Assessment,” Journal of Computational Chemistry, Vol. 9, No. 4, 1988, pp. 327-335. doi:10.1002/jcc.540090407
- M. Sanner, A. J. Olson and J. C. Spehner, “Reduced Surface: An Efficient Way to Compute Molecular Surfaces,” Biopolymers, Vol. 38, No. 3, 1996, pp. 305-320. doi:10.1002/(SICI)1097-0282(199603)38:3<305::AID-BIP4>3.0.CO;2-Y
- D. Sitkoff, K. A. Sharp and B. Honig, “Accurate Calculation of Hydration Free Energies Using Macroscopic Solvent Models,” The Journal of Physical Chemistry, Vol. 98, No. 7, 1994, pp. 1978-1988. doi:10.1021/j100058a043
- L. Ren, X. Cao, P. Geng, F. Bai and G. Bai, “Study of the Inhibition of Two Human Maltase-Glucoamylases Catalytic Domains by Different α-Glucosidase Inhibitors,” Carbohydrate Research, Vol. 346, No. 17, 2011, pp. 2688- 2692. doi:10.1016/j.carres.2011.09.012
- K. Jones, L. Sim, S. Mohan, J. Kumarasamy, H. Liu, S. Avery, H. Y. Naim, R. Quezada-Calvillo, B. L. Nichols, B. M. Pinto and D. R. Rose, “Mapping the Intestinal Alpha-Glucogenic Enzyme Specificities of Starch Digesting Maltase-Glucoamylase and Sucrase-Isomaltase,” Bioorganic & Medicinal Chemistry, Vol. 19, No. 13, 2011, pp. 3929-3934. doi:10.1016/j.bmc.2011.05.033
- G. Tanabe, S. Nakamura, N. Tsutsui,G. Balakishan, W, Xie, S. Tsuchiya, J. Akaki, T. Morikawa, K. Ninomiya, I. Nakanishi, M. Yoshikawa and O. Muraoka, “In Silico Design, Synthesis and Evaluation of 3’-O-Benzylated Analogs of Salacinol, a Potent-Glucosidase Inhibitor from Ayurvedic Traditional Medicine ‘Salacia’,” Chemical Communications, 2012, in Press. doi:10.1039/c2cc34144a
NOTES

*Corresponding author.

