Journal of Materials Science and Chemical Engineering
Vol.05 No.05(2017), Article ID:76472,13 pages
10.4236/msce.2017.55002
Reaction Rate Constant Evaluation of Thermal Isomerization of α-Pinene
Wei Zhang1, Ming Li2, Tao Zhang2, Lei Wang1,2*
1The School of Environmental and Chemical Engineering, Shanghai University, Shanghai, China
2Nanjing Institute of Technology, Chinese Academy of Sciences, Nanjing, China

Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: April 27, 2017; Accepted: May 23, 2017; Published: May 26, 2017
ABSTRACT
Bayesian inference is applied in this study to evaluate the posterior distribution of rate consents for a thermal isomerization of α-pinene by considering the uncertainty associated with rate constant parameters and kinetic model structural error. The kinetic model of the thermal isomerization of α-pinene is shown to have a mathematically ill-conditioned system that makes it difficult to apply gradient-based optimization methods for rate constant evaluation. The Bayesian inference relates the posterior probability distribution of the rate constants to the likelihood probability of modeled concentration of reaction products meeting the experimentally measured concentration and the prior probability distribution of the parameters. A Markov chain Monte Carlo (MCMC) is used to draw samples from posterior distribution while the Bayesian inference relationship is considered. Multinomial random walk Metropolis-Hastings is applied in this study to construct the histograms of rate constants as well as the confidence intervals and the correlation coefficient matrix. Results showed that the Bayesian approach can successfully apply to estimate the confidence interval of rate constants of reaction model by taking into consideration the uncertainty.
Keywords:
Bayesian Inference, Markov Chain Monte Carlo, Kinetic Model, Rate Constant, Uncertainty, α-Pinene

1. Introduction
Isomerization of α-pinene is to produce other monoterpene hydrocarbons that have wide application in the cosmetic, perfume, aromatic polymer, and other domestic chemicals industries [1] . Thermal isomerization of α-pinene in liquid-phase [2] , gas-phase [3] , and supercritical solvents-fluids [1] is studied to obtain higher production yields and selectivity. Reaction kinetic modeling is a vital tool for the simulation of the thermal isomerization of α-pinene and for industrial reactor design. Reaction modeling consists of two main steps: first is to model the reaction pathways known as a kinetic model that needs an insight understanding of mechanisms in the reaction background and second is to calibrating the kinetic model by using experimental data [4] .
Kinetic models often involve a number of rate constants to be estimated. The value of these parameters is heavily dependent on the reaction conditions such as temperature, pressure, reaction media, the assumed reaction pathway, and the quantity and quality of the measured experimental data. These are called uncertainty sources and can significantly affect the estimated value of the reaction rates.
Least-square regressions are extensively used to find the best-fitted curve to the experimental data with a given kinetic model. Classical gradient-based optimization methods such as gradient descent and evolutionary algorithms (EA) and Genetic algorithm are applied to minimize the square error between the measured and modeled data points. In an ill-conditioned system of equations, the gradient-based algorithms can trap in local minimums and are usually failed to find the global minimum error [5] . However, the EA methods result in a global optimum. Kinetic model of the thermal isomerization of α-pinene is studied by Tjoa et al. [6] who applied local estimator methods for the rate constant estimation and found that these local methods need good initial guesses to be able to find the optimum rate constants. Yermakova et al. [1] applied gradient- based Gauss-Marquardt iteration method for the kinetic model identification of the thermal isomerization of α-pinene and in order to overcome to the ill-con- ditioning conditions, they applied regularization techniques that can result in a biased estimation of reaction constants. Rodriguez-Fernandez et al. [5] studied the same reaction system and successfully applied a global optimization based on the Scatter Search method and obtained the point estimate of rate constants and the global minimum of the least-square objective function. However, these approaches result in a point estimate of reaction rates without taking into account the uncertainty. Therefore, other statistical techniques such as analysis of variance (ANOVA), t-distribution tests, Fisher Information Matrix, jackknifing, and bootstrapping methods need to be used as complementary methods to obtain the parameter covariance matrix.
Bayesian inference is a logical statistical framework that applies experimental measured data as evidence points to increase the degree of confidence for the targeted parameters with a given prior knowledge. In the kinetic model calibration, the Bayesian inference can be used to quantify the uncertainty associated with the rate constant values as well as model structural error. Markov chain Monte Carlo (MCMC) methods such as Metropolis-Hastings algorithm are usually used to construct the posterior distribution of the Bayesian inference. This method results in an evidence base confidence interval of the rate constants while the correlation between parameters is taken into consideration. MCMC methods need to take large enough samples from the posterior distribution to result in a converged histogram of parameters. Therefore, applying MCMC methods are computationally expensive. Galagali et al. [7] proposed an adaptive MCMC to alleviate a portion of this computational burdensome. High-perfor- mance computing can largely enhance the performance of MCMC sampling methods to deal with uncertainties. For example, Alikhani et al. [8] [9] showed an application of adaptive ODE solution algorithm in parallel-based GPU to accelerate the solution of mass balance rate equations in the kinetic models. They showed that applying these improvements can reduce the overall computation time by up to 50%. Altogether, the nowadays high-speed computers plus high- performance computing algorithms made it more feasible to use MCMC methods in the uncertainty evaluation.
Moles et al. [4] studied a non-linear biochemical model and applied different optimization method to estimate 36 model parameters and concluded that only stochastic algorithms were able to successfully solve the problem. Sathyamoorthy et al. [10] applied the Generalized Uncertainty Estimation (GLUE) technique for the uncertainty evaluation of a biological nitrification model. Sun et al. [11] applied the Bayesian inference with MCMC to obtain rate constants of Phytate (an organic phosphorus compound in agricultural soils) degradation in three different pathways. The Bayesian inference is applied in other studies in different fields for uncertainty analysis and parameter estimation [12] [13] [14] [15] [16] .
This study is aiming at applying the Bayesian inference to numerically evaluate the uncertainty associated with the reaction rate constants and their correlation matrix for the thermal isomerization of α-pinene over the experimental data obtained from Fuguitt et al. [2] . The obtained results are compared with the results in other studies obtained by using other methods to demonstrate the strength of proposed method.
2. Materials and Methods
Kinetic Model
Experimental data of liquid-phase thermal isomerization of α-pinene in 189.5°C obtained by Fuguitt et al. [2] is presented in Table 1. This reaction produces four products that are presented in weight fraction in eight intervals each with duplicated results. Hunter et al. [17] proposed a kinetic model for this reaction as it is shown in Figure 1 followed by Equations (1) to (5). In this scheme α-pinene (C1) converts to dipentene (C2) and allo-ocimen (C3) in two parallel paths; dipentene is the end-point component but allo-ocimen yields α- and β-pyronene (C4) and involves in an equilibrium reaction with other dimer compounds (C5).
In this kinetic model, each reaction pathway follows by a first order reaction.
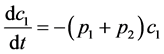 (1)
(1)
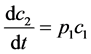 (2)
(2)
Figure 1. Kinetic model of the thermal isomerization of α-pinene where C1 is α-pinene, C2 is dipenetene, C3 is allo-ocimene, C4 is pyrone, and C5 denotes for other dimer compounds [5] .
Table 1. Experimental data of thermal isomerization of α-pinene at 189.5 °C [2] .
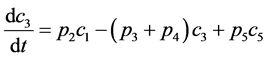 (3)
(3)
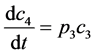 (4)
(4)
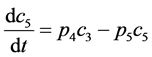 (5)
(5)
The kinetic model can be simplified to show the system of ordinary differential equations (ODEs):
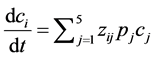 (6)
(6)
where j denotes each reaction pathway and 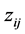 is the stoichiometery of component i in path j. By introducing
is the stoichiometery of component i in path j. By introducing 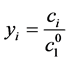 as normalized concentration where
as normalized concentration where
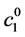 is the initial concentration of C1 (α-pinene), the Equation (6) can be rewritten as [1] :
is the initial concentration of C1 (α-pinene), the Equation (6) can be rewritten as [1] :
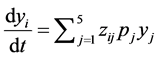 (7)
(7)
With the following initial conditions: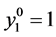 , and other
, and other 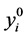 equal to zero.
equal to zero.
Model calibration is determining the numerical value of rate constants (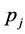 ) considering the experimental data.
) considering the experimental data.
3. Model Calibration
The idea of applying the Bayesian inference in the rate constant evaluation of a kinetic model is based on recent work by Alikhani et al. [18] , which they proposed a systematic way to apply the Bayesian inference for parameter estimation and uncertainty quantification of a large-sized kinetic model.
Alikhani et al. [18] introduced a general form of the kinetic model in a vector form:
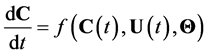 (8)
(8)
where f represents the interactions between species by applying mass balance equation while 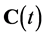 is the concentrations of all the species,
is the concentrations of all the species, 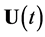 is the external forcing input, and
is the external forcing input, and 

In the normalized form followed by Equation (7), the model output concentrations become only a function of model parameters:

By using this notation, model calibration defines as estimating the values of 




where 






By this assumption (normal distribution for errors and a constant independent standard error), the square error minimization becomes a specific case of maximum likelihood method as minimizing the numerator of the exponential in Equation (11) maximizes the



Stochastic model calibration is to find all set of rate constants that meet the experimental data. In order to find the

where 


The prior distribution can take a uniform distribution within the lower and upper range of parameters, a normal distribution with its prior mean and standard deviation, or another type of distributions. For example, Alikhani et al. [18] assumed that log-normal distribution is a better representation of the prior distribution of the parameters in their kinetic model. In this study, to be consistent with the likelihood distribution, all the prior distributions are assumed to be a normal distribution.
MCMC is a class of techniques to sample from multidimensional joint probability distributions [7] [19] [20] [21] . Random walk Metropolis-Hastings (MH) algorithm [22] [23] is a subset of MCMC algorithms that is used in this study to estimate the probability histogram of parameters, defines as:
if 

where 




where 



where 


where by given 


where 


4. Results and Discussion
To obtain the posterior distribution of the parameters, Equation (13) was implemented in MATLAB and the ODE equation set of Equation (7) was solved by ode45 built-in function. Alikhani et al. [18] suggested throwing away the initial portion of the random walk samples due to burn-in period. Therefore, after 5000 samples as burn-in samples, the MCMC convergence was examined every 10,000 samples, and it is found that after 180,000 samples the change in the standard deviation of posterior distributions is negligible. Histograms of the five rate constants and the ESDn are shown in Figure 2. The 95% confidence interval of the rate constants is extracted from the posterior distribution and is shown in Table 2.
The deterministic point estimate of the parameters is also obtained by running Genetic algorithm (GA). Comparing the point estimate results shows a very good agreement between the point estimates and the median of the 95% posterior interval of the 5 rate constants. However, the value obtained for the ESD in the deterministic approach with the value of 0.77 is significantly different than its confidence interval of 1.73 - 2.35 obtained by using Bayesian inference application. This is the main difference between the deterministic and the stochastic approaches which the ESD―the representative of the uncertainty―is missing in the deterministic calculations. Therefore, the ESD of the model is not true in the point estimate method.
The ESD of the model is representing both the uncertainty associated with the measurements error and the uncertainty associated with the model structure. It should be noted that for each species in the kinetic model a separate ESD could be considered resulting in a more insight uncertainty assessment. However, adding more parameters to the random walk space needs more experimental data and larger sample size (resulting in higher computational cost) to be converged.
Figure 2. Histograms representing the posterior probability density of the thermal isomerization of α-pinene rate constants (105 min−1) and the error standard deviation.
Rodriguez-Fernandez et al. [5] studied the same kinetic model for the isomerization of α-pinene and applied a global optimization based on the Scatter
Table 2. The confidence interval of the thermal isomerization of α-pinene rate constants (105 min−1) and the error standard deviation of the kinetic model.
Search method for model calibration. They obtained the point estimates of p1 = 5.9259, p2 = 2.9634, p3 = 2.0473, p4 = 27.449, and p5 = 3.9980 (values are times to 105 to be consistent with the units in Table 2). The results of their study are in a close agreement with the results obtained in this study. However, this was not a surprising point because both studies are applied an evolutionary optimization technique on the same set of experimental data. But what distinguishes two studies is the significance evaluation of the parameter values. Rodriguez-Fernandez et al. [5] applied an approximation method based on a local linearization of the model output to evaluate the parameter covariance matrix based on the Fisher information matrix. They obtained the value of 0.07195, 0.06519, 0.33328, 2.7657, and 0.9757 for the standard deviation of p1 to p5, respectively; where comparing with the standard deviation of parameters in Table 2, it shows roughly 86%, 76%, 33%, 38%, and 28% higher values. This point is also mentioned by Rodriguez-Fernandez et al. [5] that the confidence intervals obtained from the Fisher information matrix show a wider confidence interval. It can be concluded that the confidence interval of rate constants obtained by applying Bayesian inference concept is a better representation of uncertainty associated with the rate constants.
The random walk Metropolis-Hastings took many samples from the posterior distribution of the rate constants. It is, therefore, is straightforward to find the real correlation between each pair of parameters as it is shown in Figure 3. Except for the p4-p5 pair, the correlation between other rate constants is relatively low. The high correlation factor shows that the current kinetic model cannot be fully identified, and the values of p4 and p5 in Table 2 do not correctly represent the rate constants [18] . From chemistry point, the high correlation coefficient between p4-p5 makes sense because both parameters are belonging to one reaction pathway with equilibrium conditions. In fact, in the equilibrium conditions, the ratio of the rate constants is important. This is why the rate constants of p4 and p5 cannot be identified. This finding is also observed by Yermakova et al. [1] and they suggest for a reduction in the kinetic model by removing the fifth path.
Following the method presented in [24] , to show the level of uncertainty associated with the kinetic model of the thermal isomerization of α-pinene, a Monte
Figure 3. Correlation matrix of the thermal isomerization of α-pinene rate constants.
Carlo simulations performed on the posterior samples. 1000 parameters set were randomly selected from the pool of accepted 
As it is seen in Figure 4, the duplicate experimental points are used in the parameter estimation without taking their average to take into consideration the real measurement errors. In this way, the experimental data provide better information about the parameter values.
5. Conclusion
In this study, Bayesian inference and random walk MCMC are applied to estimate the confidence interval of the rate constants of a kinetic model. Thermal isomerization of α-pinene is considered as a case study and the performance of the proposed model is evaluated by obtaining the posterior distribution of its five rate constants plus the error standard deviation of the proposed kinetic model. The results showed that:
1) The Bayesian inference solved by the random walk Metropolis-Hastings technique can successfully estimate the posterior distribution of the rate constants for a proposed kinetic model while experimental data are given.
Figure 4. The thermal isomerization of α-pinene model output confidence interval by considering the parameter and model structural uncertainties.
2) The 95% confidence interval and standard deviation for each rate constants can be obtained from the posterior distribution and its range represents the uncertainty associated with that parameter.
3) The error standard deviation in the likelihood function treated as an unknown parameter in the stochastic model identification approach and its numerical value literally represents the quantified uncertainty of the model.
4) Correlation between each pair of parameters is considered in the MCMC samples. The results of correlation matrix can provide useful information about the identifiability of the proposed kinetic model.
It can be concluded that the confidence interval of rate constants obtained by applying the Bayesian inference concept is an acceptable representation of uncertainties associated with the kinetic model.
Cite this paper
Zhang, W., Li, M., Zhang, T. and Wang, L. (2017) Reaction Rate Constant Evaluation of Thermal Isomerization of α-Pinene. Journal of Materials Science and Chemical Engineering, 5, 9-21. https://doi.org/10.4236/msce.2017.55002
References
- 1. Yermakova, A., Chibiryaev, A., Kozhevnikov, I., Mikenin, P. and Anikeev, V. (2007) Thermal Isomerization of α-Pinene in Supercritical Ethanol. Chemical Engineering Science, 62, 2414-2421.
- 2. Fuguitt, R.E. and Hawkins, J.E. (1947) Rate of the Thermal Isomerization of α-Pinene in the Liquid Phase. Journal of the American Chemical Society, 69, 319- 322.
https://doi.org/10.1021/ja01194a047 - 3. Gajewski, J.J. and Hawkins, C.M. (1986) Gas-Phase Pyrolysis of Isotopically Stereochemically Labeled α-Pinene. Evidence for a Non-Randomized Intermediate. Journal of the American Chemical Society, 108, 838-839.
https://doi.org/10.1021/ja00264a047 - 4. Moles, C.G., Mendes, P. and Banga, J.R. (2003) Parameter Estimation in Biochemical Pathways: A Comparison of Global Optimization Methods. Genome Research, 13, 2467-2474.
https://doi.org/10.1101/gr.1262503 - 5. Rodriguez-Fernandez, M., Egea, J.A. and Banga, J.R. (2006) Novel Metaheuristic for Parameter Estimation in Nonlinear Dynamic Biological Systems. BMC Bioinformatics, 7, 483.
https://doi.org/10.1186/1471-2105-7-483 - 6. Tjoa, I.-B. and Biegler, L.T. (1991) Simultaneous Solution and Optimization Strategies for Parameter Estimation of Differential-Algebraic Equation Systems. Industrial & Engineering Chemistry Research, 30, 376-385.
https://doi.org/10.1021/ie00050a015 - 7. Galagali, N. and Marzouk, Y.M. (2015) Bayesian Inference of Chemical Kinetic Models from Proposed Reactions. Chemical Engineering Science, 123, 170-190.
- 8. Alikhani, J., Massoudieh, A. and Bhowmik, U.K. (2016) GPU-Accelerated Solution of Activated Sludge Model’s System of ODEs with a High Degree of Stiffness. 2016 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, 15-17 December 2016, 555-560.
https://doi.org/10.1109/CSCI.2016.0111 - 9. Alikhani, J., Shoghli, B., Bhowmik, U.K. and Massoudieh, A. (2016) An Adaptive Time-Step Backward Differentiation Algorithm to Solve Stiff Ordinary Differential Equations: Application to Solve Activated Sludge Models. American Journal of Computational Mathematics, 6, 298-312.
https://doi.org/10.4236/ajcm.2016.64031 - 10. Sathyamoorthy, S., Vogel, R.M., Chapra, S.C. and Ramsburg, C.A. (2014) Uncertainty and Sensitivity Analyses Using GLUE When Modeling Inhibition and Pharmaceutical Cometabolism during Nitrification. Environmental Modelling & Software, 60, 219-227.
- 11. Sun, M., Alikhani, J., Massoudieh, A., Greiner, R. and Jaisi, D.P. (2017) Phytate Degradation by Different Phosphohydrolase Enzymes: Contrasting Kinetics, Decay Rates, Pathways, and Isotope Effects. Soil Science Society of America Journal, 81, 61-75.
https://doi.org/10.2136/sssaj2016.07.0219 - 12. Gel, A., Shahnam, M. and Subramaniyan, A.K. (2017) Quantifying Uncertainty of a Reacting Multiphase Flow in a Bench-Scale Fluidized Bed Gasifier: A Bayesian Approach. Powder Technology, 311, 484-495.
- 13. Chaudhary, A. and Hantush, M.M. (2017) Bayesian Monte Carlo and Maximum Likelihood Approach for Uncertainty Estimation and Risk Management: Application to Lake Oxygen Recovery Model. Water Research, 108, 301-311.
- 14. Song, Y., Jin, L., Zhu, G. and Ma, M. (2016) Parameter Estimation for a Simple Two-Source Evapotranspiration Model Using Bayesian Inference and Its Application to Remotely Sensed Estimations of Latent Heat Flux at the Regional Scale. Agricultural and Forest Meteorology, 230, 20-32.
- 15. Zhou, F., Lei, B., Liu, Y. and Jiao, R.J. (2017) Affective Parameter Shaping in User Experience Prospect Evaluation Based on Hierarchical Bayesian Estimation. Expert Systems with Applications, 78, 1-15.
- 16. Alderman, P.D. and Stanfill, B. (2016) Quantifying Model-Structure- and Parameter-Driven Uncertainties in Spring Wheat Phenology Prediction with Bayesian Analysis. European Journal of Agronomy.
- 17. Hunter, W. and McGregor, J. (1967) The Estimation of Common Parameters from Several Responses: Some Actual Examples. Unpublished Report.
- 18. Alikhani, J., Takacs, I., Al Omari, A., Murthy, S. and Massoudieh, A. (2017) Evaluation of the Information Content of Long-Term Wastewater Characteristics Data in Relation to Activated Sludge Model Parameters. Water Science and Technology, 75, 1370-1389.
https://doi.org/10.2166/wst.2017.004 - 19. Bretthorst, G.L. (2013) Bayesian Spectrum Analysis and Parameter Estimation. Vol. 48, Springer Science & Business Media, Berlin, Heidelberg.
- 20. Robert, C.P. (2004) Monte Carlo Methods. Wiley, Hoboken.
- 21. Gilks, W.R., Richardson, S. and Spiegelhalter, D. (1995) Markov Chain Monte Carlo in Practice. CRC Press, Boca Raton.
- 22. Gamerman, D. and Lopes, H.F. (2006) Markov Chain Monte Carlo: Stochastic Simulation for Bayesian Inference. 2nd Edition, Taylor & Francis, UK.
- 23. Hastings, W.K. (1970) Monte Carlo Sampling Methods Using Markov Chains and Their Applications. Biometrika, 57, 97-109.
https://doi.org/10.1093/biomet/57.1.97 - 24. Alikhani, J., Deinhart, A.L., Visser, A., Bibby, R.K., Purtschert, R., Moran, J.E., Massoudieh, A. and Esser, B.K. (2016) Nitrate Vulnerability Projections from Bayesian Inference of Multiple Groundwater Age Tracers. Journal of Hydrology, 543, 167-181.
https://doi.org/10.1016/j.jhydrol.2016.04.028







