Open Journal of Genetics
Vol.4 No.1(2014), Article ID:43456,8 pages DOI:10.4236/ojgen.2014.41002
Construction of Vector for Functional Analysis of the Intrinsically Bent DNA in the ACE3 Replicator from Drosophila melanogaster
Douglas Vinícius Bassalobre de Freitas, Fabrícia Gimenes, Adriana Fiorini, Fabiana dos Santos Rando, Quirino Alves de Lima Neto, Maria Aparecida Fernandez
Departamento de Biotecnologia, Genética e Biologia Celular, Universidade Estadual de Maringá, Maringá, Brasil
Email: mafernandez@uem.br
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 3 December 2013; revised 1 January 2014; accepted 30 January 2014
ABSTRACT
DNA replication is a crucial process for species survival, nevertheless it is not clear which factors define origin selection in multicellular eukaryotes. Developmental gene amplification systems, such as the one described during ovarian follicles development in Drosophila melanogaster, are useful tools for studying of DNA replication process in these organisms. We previously described that the well characterized third chromosome amplified domain of D. melanogater displays three intrinsically bent DNA sites: b1, localized at an amplification control element (ACE3), b2 and b3, both localized at the preferential origin ori-β. This proposal aimed to construct a Drosophila transformation vector, which contains a short deletion at the ACE3, in order to reduce the intrinsically bent DNA site b1, and analyze the functional role of this site in the gene amplification process. Through a series of cloning steps, we obtained a Big Parent vector derivative, containing a deletion at the positions 176 - 180 bp, inside the ACE3. The generation of a Drosophila transformation vector displays a reduced intrinsically bent DNA site in the third chromosome amplified domain, it will allow the analysis of the functional role of this curvature in developmental gene amplification, providing new insights on replication initiation in D. melanogaster and the function of intrinsically bent DNA sites.
Keywords:DNA Replication; Gene Amplification; ACE3; Intrinsically Bent DNA; Drosophila melanogaster

1. Introduction
The efficiency of the DNA replication process is essential for species survival. Thus, the genome should be quickly and faithfully duplicated only once per cell division. In bacteria, usually there is only one single origin of replication in the bacterial chromosome called OriC, which presents multiple consensus sequences called DnaA box, which act as interaction sites for protein DnaA, directing the local opening of the double-stranded DNA and recruitment other proteins [1] [2] . In the yeast Saccharomyces cerevisiae, the sites of initiation of replication (Autonomosly Replicating Sequences, ARS) include a short 10 to 11 bp consensus sequence rich in A/T called ACS (ARS consensus sequence) [3] . From this organism, during late M phase and early G1 phase of cell cycle, was isolated the ORC (Origin Recognition Complex), responsible for recognizing the sites of initiation of replication and nuclear components of pre-replicative complex (pre-RC), that are necessary to indicate permission for the initiation of replication in S phase [4] . In multicellular eukaryotes, although the machinery required for the replication process is well-known, the factors determining the selection and where is localized the origins which will be fired in a genome segment are still poorly understood, and a limited number of DNA replication origins had been studied in detail. The use of different methodologies for the analysis of replication intermediates indicates no consensus sequences for these regions [5] , as described only in S. cerevisiae and closely related species. Moreover, the selection process is influenced by the chromatin structure where the DNA replication origin is localized and the cell type [5] -[8] . The systems which are extensively used as a model for studying the replication origins of the multicellular eukaryotes are those where occurs the developmentally gene amplification. The best system studied in this research area is the Drosophila ovarian follicles cells and the genomic segments where the DNA puffs open in the larval Sciaridae salivary gland cells. The study of the DNA replication process is advantageous in both systems due to the high number of copies of DNA segments generated and the fact that the enzymatic machinery used is the same as described for regions not amplified [9] -[11] .
In Drosophila, transgenic constructs, mediated via P-element transformation in ectopic location, preserve the tissue and temporal pattern of gene amplification [12] . To avoid some problems in the chromosomal position, which can affect the process in some cases, vector elements insulators can be used as the suppressor of Hairywing insulator protein binding site [su (Hw) BS; 13]. The better characterized Drosophila amplified segment is located on the third chromosome (DAFC-66D), where were initially identified the preferential ori-β origin, an amplification control element (ACE3) located upstream of the chorionic gene S18 (CG6517), and four regions amplification enhancers, AER-a, b, c and d [14] . The elements ACE3 and ori-β are necessary and sufficient to promote amplification of transgenic constructs with insulator element, and these elements interact in cis [15] . To identify the sequences required for amplification of this region, was analyzed transgenic lines containing deletions in different regions of ACE3, on the gene S18 and ori-β DNA replication origin, resulting in quantitative effects on amplification levels [16] . Recently, were identified three intrinsically DNA bent sites on segment DAFC-66D. One of them was localized at ACE3 (b1) region and the others two in the β-ori origin (b2 and b3) [17] . However, the functional role of these DNA bent sites remains unclear. Thus, the aim of this work was to construct a vector transformation in Drosophila with a short deletion in ACE3 able to disrupt the b1 DNA bent curvature, in order to analysis the importance of this site in the gene amplification process.
2. Method
2.1. Computational Analysis
Map15a software was used for the analysis of ENDS ratio helical parameter (ratio of the length of the curve by the distances between the ends of a fragment) and the software 3D15m1 to visualize the two-dimensional projection of the three-dimensional fragment trajectory. Both the software is based on Eckdahl and Anderson algorithm and Bolshoy et al. [18] -[21] . The DAFC-66D 2348 bp fragment was analyzed, which includes the 320 bp from ACE3 region, a 1200 bp from S18 gene and 840 bp of the ori-β DNA replication origin segment.
2.2. Transformation Vector Construction Strategy
First, the 320 bp ACE3 segment was cloned into the vector pBluescript KS+ (Stratagene) in KpnI and BamHI restriction enzymes sites. The strategy to obtain a mutated sequence from ACE3 start with a special construction of complementary forward and reverse primers sequences: 5’-GTAATTTTATCTCATTTTCGTTTTGTATACAAATTC-3’ and 5’-GAATTTGTATACAAAAGCAAAATGAGATAAAATTAC-3’, Figure 1, underlined. The 5 bp deletion in ACE3 was designed in the positions 176 - 180 bp from the original ACE3 sequence, which is able to disrupt the intrinsically curvature b1 (Figure 1, bold).
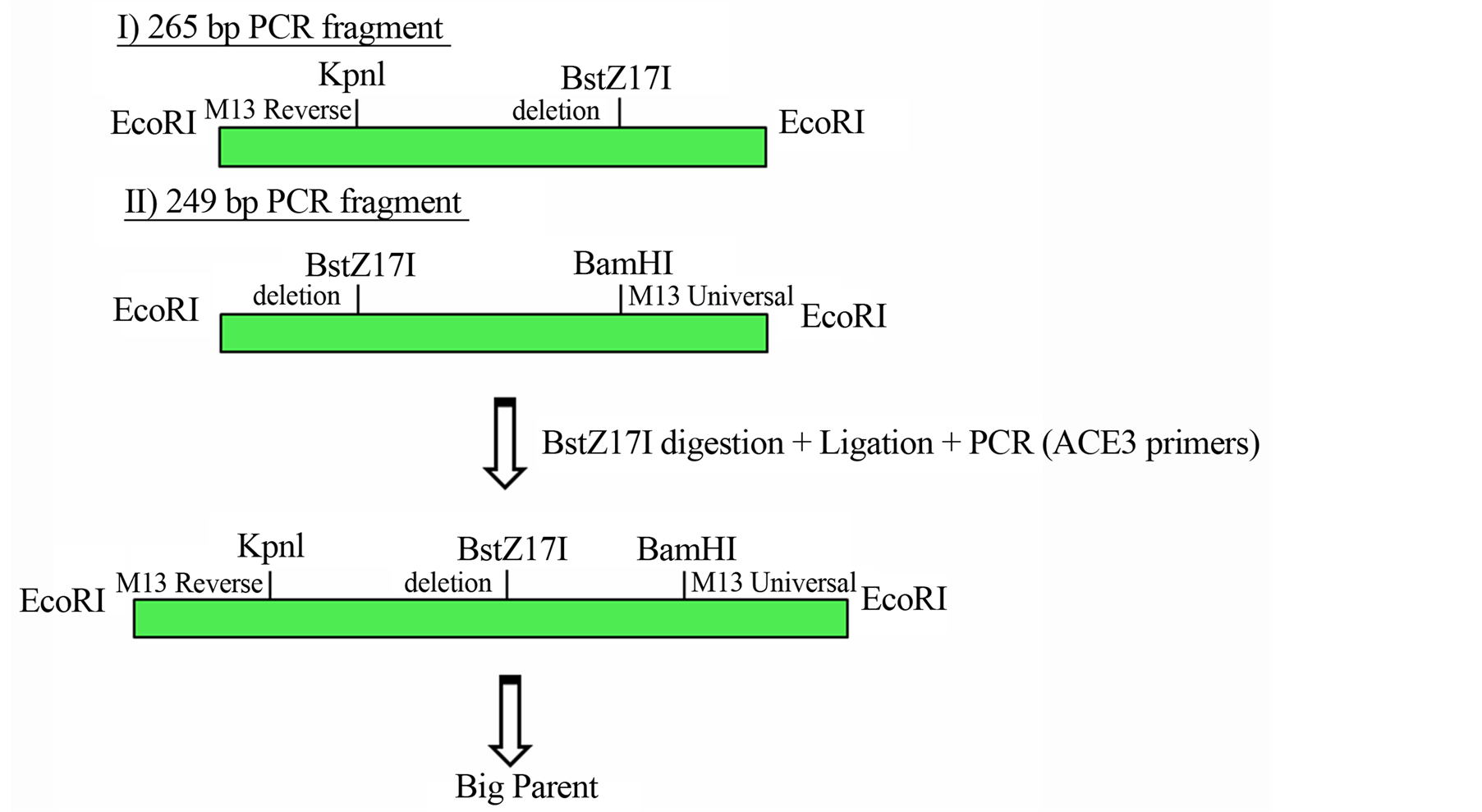
The substitution of a C nucleotide base for a T nucleotide in the 196 bp position was capable to promote the sequence of a new enzyme site, the BstZ171. For this construction, was necessary two individually PCR reactions, one using the plasmid M13 universal primer and the primer containing the deletion and another using the plasmid reverse primer plasmid and the reverse primer from the plasmid deletion. Figure 2(a) shows the details from these reactions.

Figure 1. Drosophila ACE3 vector cloning strategy. This figure display the 320 pb from Drosophila ACE3 and the annealing primers region are underlined. The five nucleotides in bold (ATTTT) at the positions 176 - 180 bp was deleted to abolished the intrinsically DNA bent site b1. The nucleotide T is replaced by C (196 bp position), to created the restriction site enzyme BstZ17I, is shadowed in gray.
 (a)
(a)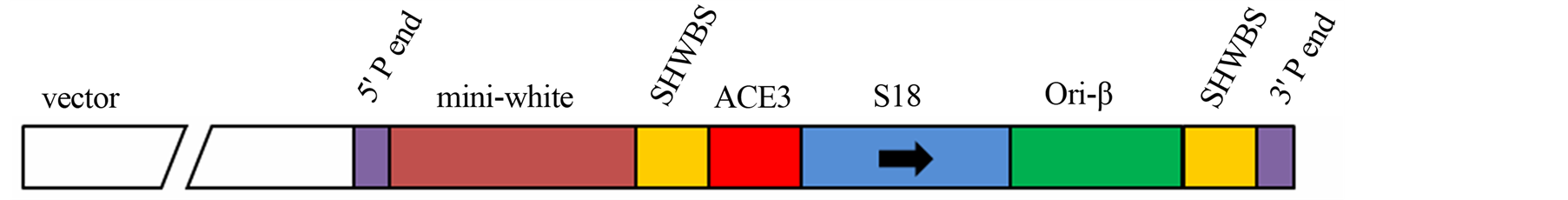 (b)
(b)
Figure 2. (a) Cloning strategy to ensure a final vector with intrinsically DNA bent site b1 abolished and a creation of a new restriction site enzyme BstZ17I; (b) Scheme of the Big Parent vector, showing ACE3, S18 gene and ori-β flanked by the insulator element SHWBS.
The products were cloned separately into the vector pGEM (Promega). Both clones were cleaved with the enzyme and BstZ17I and ligated together. The product was subjected to a PCR reaction with ACE3 primers (forward 5’-GGTACCCTGAGCCTGGCCAAC-3’ and reverse 5’-GGATCCGCATAGTTTCGATC-3’) with the objective of generating a fragment of 320 containing the 5bp deletion and a new BstZ171 restriction site. This product was cloned into the pGEM vector, and then was moved to the pBluescript KS+ vector containing a fragment of 2.8 kb vector Big Parent via the KpnI and BamHI sites. Then, the original 2.8 kb segment of Big Parent ACE3 without the deletion was replaced by the fragment containing the deletion via XhoI and NotI sites.
The products resulting from each step of the construction were subjected to sequencing reactions with DYEnamicTM ET Terminator kit (Amersham Biosciences) and the products were analyzed on MegaBACE 1000 sequencing system, in order to confirm the b1 site deletion and the new site BstZ17I.
The Drosophila Big Parent transforming vector was kindly provided by Dr. John Tower (University of Southern California, USA). This vector contains the ACE3, the S18 gene and ori-β element flanked by insulator SHWBS (Figure 2(b)).
3. Results
The computational analysis of the 2348 bp fragment from Drosophila DAFC-66D with the software Map15a and 3D15m1 revealed that deletion of the five nucleotides, ATTTT, from positions 176 - 180 bp within ACE3 region cause a significant reduction in the curvature of the intrinsically DNA bent site b1 (Figures 3 and 4).
Thus, the first step in the strategy to generate a transforming vector taking this unstructured intrinsically bent DNA site b1, was to construct a primer pair complementary introducing a deletion of 5 bp. The last step of the construction was to replace the original fragment of 2.8 kb within the Big Parent vector for a similar fragment containing the deletion. At the end, it has been isolated 12 positive clones and for all of them was extracted the plasmid DNA, which were subjected to restriction analysis, which selected six clones for sequencing.
The results of the sequencing reaction confirmed the presence of deletion and a new site for BstZ17I restriction enzyme in all clones (Figure 5). Thus, one of the clones will be used to obtain transgenic Drosophila lines for subsequent functional analysis of intrinsically bent DNA site b1 curvature on amplification processes in the segment analyzed.

Figure 3. ACE3 curvature analysis. The ENDS ratio analysis by Map15a software from the 2348 pb sequence that contains the 320 pb ACE3, ~1200 pb from S18 corionic gene and 840 pb from the ori-β replication origin. (A) original sequence; (B) deleted sequence, that shows a significative reduction from the intrinsically DNA bent site b1 ENDS ratio value (arrow).
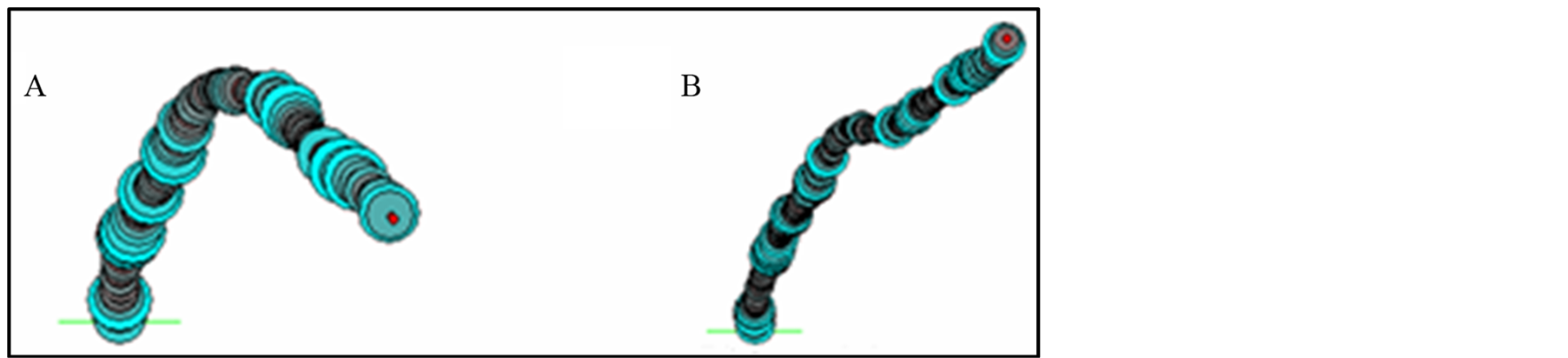
Figure 4. Two-dimensional (2D) projection of the 3D trajectories from ACE3 320 bp sequence by 3D15m1 software. (A) ACE3 bending curved structure; (B) straight ACE3 sequence with 176 - 180 bp deletion.

Figure 5. ACE3 sequence alignment. The sequence of the six modified ACE3 clones (C1-6), with the original ACE3 sequence is showed. The 5 bp deletion is underlined and the new restriction site BstZ17I is shaded.
4. Discussion
Intrinsic curvature of DNA can be found in promoter regions of prokaryotes and eukaryotes replication origins and transcription regions (reviewed in [22] ). Bramhill and Kornberg [23] proposed a model involving both discrete sites of intrinsically DNA bent in replication origins of prokaryotes, which can act at the opening of double-stranded DNA mediated by protein DnaA. Saccharomyces cerevisiae intrinsically DNA bent sites, occurs naturally in the initiation of replication sites (ARS), and can be bent functionally replaced by synthetic DNA [24] . A study in the amplified dihydrofolate reductase (DHFR) locus in Chinese hamster cultured cells identified a site of intrinsically bent DNA as one of the four elements required for the activity of ori-β [25] .
Such sites can make easier or help the unwinding of the double helix target and the interaction of proteins in the processes of transcription and DNA replication, since DNA usually intrinsically curvatures are formed in regions rich in A/T [26] , and some proteins such as HMGs, which bind, preferentially, an unusual DNA structures [27] . However, the function of these sites is still unclear, and the analysis of bends present in the amplified locus on Drosophila melanogaster chromosome 3 using the methodology of P-element mediated transformation, may contribute to a better understanding of the functional role of intrinsic curvatures DNA.
The vector constructed can be used to obtain a transgenic line of Drosophila in order to assess the level of amplification of the element containing the deletion in ACE3 and determine whether the curvature present in this region shows any role in the activation of preferential ori-β origin. The absence of this region drastically reduces element ACE3 amplification levels [28] , and it was observed that this element interacts in cis with this DNA replication origin [15] . Austin et al. [29] observed that the ACE3 is sufficient for localization of ORC in vivo, making compelling analysis in the role of the b1 curvature in the association of this region with the ORC from the methodology for chromatin immunoprecipitation. Remus et al. [30] showed an increased affinity to DmORC ACE3 and ori-β segments with negative supercoiling in vitro, demonstrating the possible influence of topological properties in the process of DNA replication initiation.
In addition, our work could be also evaluated the b1 binding curvature with Myb complex proteins, which presents role in the nucleation of pre-RC complex in Drosophila. Members of this complex specifically bind to the ori-β and ACE3 [31] [32] , and consensus sequences for the association of proteins Dm-myb and Mip120 are present at the ends of curvature of ACE3. It is possible to formulate the hypothesis that a conformational change, resultant from the interaction between the complex and the Myb site, may increase the affinity of the complex pre-RC for the region. It may also be investigated the role of proteins that exhibit preferential binding to unusual DNA structures like the HMGs, since it was observed that HMGB1 protein may have a regulatory role in the process of DNA replication [33] .
The construction of a Drosophila transformation vector containing a region of an unstructured intrinsically DNA curvature at an amplified locus during development to allow analysis of the functional role of this curvature in the process of gene amplification in order to contribute to a better understanding of the process initiation of replication in multicellular eukaryotes and the functional role of intrinsic DNA bends.
Acknowledgements
This research was supported by CNPq, CAPES, Fundação Araucária. The authors thank the Complexo de Centrais de Apoio à Pesquisa, COMCAP from Universidade Estadual de Maringá, UEM, PR, Brasil.
References
- Messer, W. (2002) The Bacterial Replication Initiator DnaA. DnaA and oriC, the Bacterial Mode to Initiate DNA Replication. FEMS Microbiology Reviews, 26, 355-374.
- Robinson, N.P. and Bell, S.D. (2005) Origins of DNA Replication in the Three Domains of Life. FEBS Journal, 272, 3757-3766. http://dx.doi.org/10.1111/j.1742-4658.2005.04768.x
- Bell, S.P. and Dutta, A. (2002) DNA Replication in Eukaryotic Cells. Annual Review of Biochemistry, 71, 333-374.
- Stillman, B. (2005) Origin Recognition and the Chromosome Cycle. FEBS Letters, 579, 877-884. http://dx.doi.org/10.1016/j.febslet.2004.12.011
- Biamonti, G., Paixão, S., Montecucco, A., Peverali, F.A., Riva, S. and Falaschi, A. (2003) Is DNA Sequence Sufficient to Specify DNA Replication Origins in Metazoan Cells? Chromosome Research, 11, 403-412. http://dx.doi.org/10.1023/A:1024910307162
- Debatisse, M., Toledo, F. and Anglana, M. (2004) Replication Initiation in Mammalian Cells: Changing Preferences. Cell Cycle, 3, 19-21. http://dx.doi.org/10.4161/cc.3.1.628
- Cvetic, C. and Walter, J.C. (2005) Eukaryotic Origins of DNA Replication: Could You Please Be More Specific? Seminars Cell Developmental Biology, 16, 343-353. http://dx.doi.org/10.1016/j.semcdb.2005.02.009
- Aladjem, M.I. (2007) Replication in Context: Dynamic Regulation of DNA Replication Patterns in Metazoans. Nature Review Genetics, 8, 588-600. http://dx.doi.org/10.1038/nrg2143
- Spradling, A.C. (1999) ORC Binding, Gene Amplification, and the Nature of Metazoan Replication Origins. Genes Development, 13, 2619-2623. http://dx.doi.org/10.1101/gad.13.20.2619
- Tower, J. (2004) Developmental Gene Amplification and Origin Regulation. Annual Review of Genetics, 38, 273-304. http://dx.doi.org/10.1146/annurev.genet.37.110801.143851
- Claycomb, J.M. and Orr-Weaver, T.L. (2005) Developmental Gene Amplification: Insights into DNA Replication and Gene Expression. Trends Genetics, 21, 149-162. http://dx.doi.org/10.1016/j.tig.2005.01.009
- de Cicco, D.V. and Spradling, A.C. (1984) Localization of a Cis-Acting Element Responsible for the Developmentally Regulated Amplification of Drosophila Chorion Genes. Cell, 38, 45-54. http://dx.doi.org/10.1016/0092-8674(84)90525-7
- Lu, L. and Tower, J. (1997) A Transcriptional Insulator Element, the su(Hw) Binding Site, Protects a Chromosomal DNA Replication Origin from Position Effects. Molecular Cell Biology, 17, 2202-2206.
- Orr-Weaver, T.L., Johnston, C.G. and Spradling, A.C. (1989) The Role of ACE3 in Drosophila Chorion Gene Amplification. EMBO Journal, 8, 4153-4162.
- Lu, L., Zhang, H. and Tower, J. (2001) Functionally Distinct, Sequence-Specific Replicator and Origin Elements Are Required for Drosophila Chorion Gene Amplification. Genes Development, 15, 134-146. http://dx.doi.org/10.1101/gad.822101
- Zhang, H. and Tower, J. (2004) Sequence Requirements for Function of the Drosophila Chorion Gene Locus ACE3 Replicator and Ori-Beta Origin Elements. Development, 131, 2089-2099. http://dx.doi.org/10.1242/dev.01064
- Gimenes, F., Assis, M.A., Fiorini, A., Mareze, V.A., Monesi, N. and Fernandez, M.A. (2009) Intrinsically Bent DNA Sites in the Drosophila melanogaster Third Chromosome Amplified Domain. Molecular Genetics Genomics, 281, 539-549. http://dx.doi.org/10.1007/s00438-009-0430-1
- Eckdahl, T.T. and Anderson, J.N. (1987) Computer Modelling of DNA Structures Involved in Chromosome Maintenance. Nucleic Acids Research, 15, 8531-8545. http://dx.doi.org/10.1093/nar/15.20.8531
- Bolshoy, A., McNamara, P., Harrington, R.E. and Trifonov, E.N. (1991) Curved DNA without A-A: Experimental Estimation of All 16 DNA Wedge Angles. Proceedings of the National Academy of Sciences of the United States of America, 88, 2312-2316. http://dx.doi.org/10.1073/pnas.88.6.2312
- Pasero, P., Sjakste, N., Blettry, C., Got, C. and Marilley, M. (1993) Long-Range Organization and Sequence-Directed Curvature of Xenopus laevis Satellite 1 DNA. Nucleic Acids Research, 21, 4703-4710. http://dx.doi.org/10.1093/nar/21.20.4703
- Marilley, M. and Pasero, P. (1996) Common DNA Structural Features Exhibited by Eukaryotic Ribosomal Gene Promoters. Nucleic Acids Research, 24, 2204-2211. http://dx.doi.org/10.1093/nar/24.12.2204
- Fiorini, A., Gimenes, F., Lima Neto, Q.A., Rosado, F.R. and Fernandez, M.A. (2011) Sequence-Directed DNA Curvature in Replication Origins Segments. In: Croatia, R., Ed., Fundamental Aspects of DNA Replication, InTech Open, Rijeka Croatia, 145-160.
- Bramhill, D. and Kornberg, A. (1988) Duplex Opening by dnaA Protein at Novel Sequences in Initiation of Replication at the Origin of the E. coli Chromosome. Cell, 52, 743-755. http://dx.doi.org/10.1016/0092-8674(88)90412-6
- Williams, J.S., Eckdahl, T.T. and Anderson, J.N. (1988) Bent DNA Functions as a Replication Enhancer in Saccharomyces cerevisiae. Molecular Cell Biology, 8, 2763-2769.
- Altman, A.L. and Fanning, E. (2004) Defined Sequence Modules and an Architectural Element Cooperate to Promote Initiation at an Ectopic Mammalian Chromosomal Replication Origin. Molecular Cell Biology, 24, 4138-4150. http://dx.doi.org/10.1128/MCB.24.10.4138-4150.2004
- Anderson, J.N. (1986) Detection, Sequence Patterns and Function of Unusual DNA Structures. Nucleic Acids Research, 14, 8513-8533. http://dx.doi.org/10.1093/nar/14.21.8513
- Stros, M. (2010) HMGB Proteins: Interactions with DNA and Chromatin. Biochimica et Biophysica Acta, 1799, 101-113. http://dx.doi.org/10.1016/j.bbagrm.2009.09.008
- Austin, R.J., Orr-Weaver, T.L. and Bell, S.P. (1999) Drosophila ORC Specifically Binds to ACE3, an Origin of DNA Replication Control Element. Genes Development, 13, 2639-2649. http://dx.doi.org/10.1101/gad.13.20.2639
- Swimmer, C., Delidakis, C. and Kafatos, F.C. (1989) Amplification-Control Element ACE-3 Is Important but Not Essential for Autosomal Chorion Gene Amplification. Proceedings of the National Academy of Sciences of the United States of America, 86, 8823-8827. http://dx.doi.org/10.1073/pnas.86.22.8823
- Remus, D., Beall, E.L. and Botchan, M.R. (2004) DNA Topology, Not DNA Sequence, Is a Critical Determinant for Drosophila ORC-DNA Binding. EMBO Journal, 23, 897-907. http://dx.doi.org/10.1038/sj.emboj.7600077
- Beall, E.L., Bell, M., Georlette, D. and Botchan, M.R. (2004) Dm-myb Mutant Lethality in Drosophila Is Dependent upon mip130: Positive and Negative Regulation of DNA Replication. Genes Development, 18, 1667-1680. http://dx.doi.org/10.1101/gad.1206604
- Beall, E.L., Manak, J.R., Zhou, S., Bell, M., Lipsick, J.S. and Botchan, M.R. (2002) Role for a Drosophila Myb-Containing Protein Complex in Site-Specific DNA Replication. Nature, 420, 833-837. http://dx.doi.org/10.1038/nature01228
- Topalova, D., Ugrinova, I., Pashev, I.G. and Pasheva, E.A. (2008) HMGB1 Protein Inhibits DNA Replication in Vitro: A Role of the Acetylation and the Acidic Tail. The International Journal of Biochemistry and Cell Biology, 40, 1536-1542. http://dx.doi.org/10.1016/j.biocel.2007.11.014

