Open Journal of Genetics
Vol.2 No.1(2012), Article ID:17764,24 pages DOI:10.4236/ojgen.2012.21002
QTL for floral stem lignin content and degradability in three recombinant inbred line (RIL) progenies of Arabidopsis thaliana and search for candidate genes involved in cell wall biosynthesis and degradability
![]()
1INRA, Unité de Génétique et d’Amélioration des Plantes, Lusignan, France
2CNRS, Université Paul Sabatier, Toulouse, France
3INRA, Institut Jean-Pierre Bourgin, Versailles, France
Email: yves.barriere@lusignan.inra.fr
Received 9 November 2011; revised 15 December 2011; accepted 24 January 2012
Keywords: Arabidopsis; QTL; Lignin; Secondary Wall; Digestibility; Candidate Gene
ABSTRACT
Deciphering the genetic determinants involved in cell wall assembly is a strategic issue for breeding programs that target both ruminant feeding and biofuel production. The Arabidopsis thaliana model system has great potentials to elucidate the genetic determinants involved in cell wall component biosynthesis and those involved in the regulation cascades allowing their coordinated assembly. QTL for biomass quality related traits (cell wall content, lignin content, and cell wall degradability) were mapped in the three Arabidopsis RIL progenies Bay0 × Shahdara, Bur0 × Col0, and Blh1 × Col0. Overall, 40 QTL were detected for these traits, explaining up to 33% and 12% of the observed phenotypic variation for lignin content and cell wall degradability respectively. Major QTL hotspots were mapped on chromosome 1 (position 5 Mbp), chromosome 4 (position 1 Mbp), and chromosome 5 (position 3 Mbp). A putative candidate gene set (82 genes) was considered including those previously described as involved in cell wall phenolic component biosynthesis, their regulation factors, and genes involved in lignified tissue patterning. Colocalisations observed (according to the reference sequence of Col0) between the detected QTL and these candidate genes did not prioritize any of the three gene groups (monolignol biosynthesis, transcription factors, lignified tissue patterning). Colocalizations were thus observed for 57% of monolignol biosynthesis related genes, 55% of the transcription factors considered, and 66% of genes considered to be involved in lignified tissue patterning and assembly. Colocalizations were observed for at least one member of all investigated gene families, except WRKY transcription factors. Colocalizations were also shown with several miRNA putatively involved in the regulation of lignifying tissue assembly. Taking into account the QTL shown in the Bur0 × Col0 progeny, allelic variations were shown in the MYB32, MYB58, MYB75, GRAS SCARECROW, AtC3H14 zinc finger, SHINE2, and IFL1 genes and in the AtMIR397a. Given that the list of candidate genes is not complete, and because the QTL support intervals encompassed genes of still unknown function, it is still not clear whether one of the selected candidates is responsible for the effect of a detected QTL. Mutant investigation and positional cloning steps are likely essential to clearly determine the causal mechanism involved in cell wall degradability variation.
1. INTRODUCTION
Lignocellulose biomass, which is the basis of herbivore nutrition, is also considered to be a major sustainable resource for second generation biofuel production. Lignocellulose biomass is made up of secondary walls and is mostly comprised of cellulose and hemicelluloses embedded in a phenolic component matrix. Lignification is the basic cause of reduced digestibility of forages in cattle digestive tracks [1,2] and of reduced fermentation of plant biomass into ethanol during green fuel production based on straw or stover materials [3-5]. In addition to lignin content, lignin structure and lignin associations with other cell wall components, including covalent linkages between phenolics and hemicelluloses, greatly influence cell wall properties [6,7]. Embedding between phenolics and carbohydrates thus prevent physical access of enzymes to cell wall carbohydrates and strongly limit their enzymatic hydrolysis. However, lignins and crosslinkages between cell wall components are essential as they contribute to the mechanical properties of tissues and impart hydrophobicity to vascular elements, allowing water and nutriment transportation.
Most forage and non-woody plants devoted to energy production for cattle feeding, biogas or biofuel production belong to the grass family. Nevertheless, the small mouse cress dicotyledonous Arabidopsis thaliana (abbreviated Arabidopsis) is a relevant model system to study the genetic basis of cell wall recalcitrance to saccharification. The monolignol biosynthesis pathway indeed has been extensively deciphered in Arabidopsis, with investigations most often based on mutant or deregulated plants. Another great interest of the Arabidopsis model system is related to the possible searches for regulation cascades of genes involved in cell wall component biosynthesis and genes controlling the cell wall assembly. Comprehensive investigations have thus described the transcription factor network regulating the organization of secondary cell wall biosynthesis in Arabidopsis [8,9].
The Arabidopsis lignified cell wall (type I cell wall as in all dicots and non grass monocots) is a composite material with phenolics, cellulose microfibrils, an amorphous matrix consisting predominantly of hemicelluloses (mainly xyloglucans and a few glucuronoxylans), and, to a lesser extent, pectins (rhamnogalacturonan I and II, and homogalacturonan). As in other dicotyledonous plants, Arabidopsis lignins are comprised of guaiacyl (G) units derived from coniferyl alcohol, syringyl (S) units derived from sinapyl alcohol, together with very low levels of p-hydroxyphenyl units (H) derived from p-coumaryl alcohol. The average relative frequencies of each monomeric units released by thioacidolysis of lignins of mature floral stems were shown ranging from 72% to 78% for G units and from 21% to 28% for S units, with less than 1% of H units [10-12]. Interfascicular fibers have a higher content in S units than vascular bundles [11]. Incorporation of ferulic acid in lignins has also been shown to occur in Arabidopsis, but in very low amount, nearly 100 times lower compared to maize [13,14]. Conversely to grasses, there is a quasi-lack of p-coumaric acid in Arabidopsis cell wall. Moreover, grass hemicelluloses mostly comprised glucurono-arabinoxylans, with very little pectins. Given the differences between type I and type II cell walls, it should be considered that different gene families will likely only be found in grass genomes, while several ortholog genes will have different functions in grass and dicot plants. According to Carpita and McCann [15] and to Bosch et al. [16], nearly two-third of cell wall related genes should be common in grasses and Arabidopsis.
A more complete understanding of the molecular networks involved in cell wall assembly is essential for further efficient breeding of plants with lignocellulosic biomass that can be more easily broken-down in animal digestive tracks or industrial bio-fermenters. QTL for cell wall related traits have been mostly detected in maize (reviews by Barrière et al. [6,17]) and to a lesser extent in woody species such as pine [18-20], poplar [21], and eucalyptus [22-24]. No studies have yet validated genes as responsible for the effect of the QTL, even if colocalizations between candidate genes and QTL were investigated in maize, poplar, and eucalyptus [17,24-26]. While many investigations related to cell wall assembly and cell wall component biosynthesis have been done in Arabidopsis based on mutant studies, no QTL analyses have been considered on this topic despite the early availability of the genomic reference sequence for this species, apart from two in the Bay0 × Shahdara progeny. One was devoted to primary wall carbohydrates [27] and one to floral stem lignification [28]. The objective of this work was thus to detect QTL involved in floral stem lignification in three Arabidopsis RIL progenies with a systematic search for candidate genes underlying each QTL. The list of candidate genes considered mainly focused on a limited set of genes which had been described with key-roles in constitutive phenolic component biosynthesis, in the regulation of phenolic component biosynthesis, or in the assembly of lignified tissues.
2. MATERIAL AND METHODS
2.1. RIL Progeny Description
The three recombinant inbred lines were derived at INRA Versailles by single seed descent (SSD) up until the F7 generation from crosses between Bay0 and Shahdara, Col0 and Bur0, and Blh1 and Col0 ( [29] , and VNAT database, http://dbsgap.versailles.inra.fr/vnat/). According to data available in the VNAT database, the Bay0 ecotype originates from a fallow-land habitat near Bayreuth (Germany) and the Shahdara ecotype was collected in the Pamiro-Alay mountains of Tadjikistan in the drainage basin of the Shakdara river. The Bur0 ecotype was collected from the roadside nearby Burren (Eire) and the sequenced ecotype Columbia descended from Landsberg which originates from Gorzow Wielkopolski (Landsberg/Warthe, Poland). Blh1 is an ecotype from Bulhary (Czechoslovakia) that was collected in a grassy place on the left riverside of the Thaya. The diversity in geographical and ecological origins of ecotypes is such that one would expect a significant allelic diversity that is especially well adapted to QTL investigations.
The genetic map of the Bay0 × Shahdara RIL progeny was originally based on 38 microsatellite markers [30], but it was further extended to 69 markers on the 420 RIL. The average genetic distance between two adjacent markers was thus 6.1 cM, corresponding on average to 2.0 Mb, with a global good allelic equilibrium (Bay0 51%, Shahdara 49%, VNAT database). This progeny was the basis of numerous QTL analyses including a first study of cell wall related QTL, which was based on the 38-marker map with a less elaborated NIRS calibration of cell wall traits [28]. The Bur0 × Col0 RIL progeny comprised 343 RIL genotyped with 87 markers. The average distance between markers was 4.4 cM, corresponding on average to 1.4 Mb, with a global good allelic equilibrium (Col0 51.3%, Bur0 48.7%, VNAT database). The Blh1 × Col0 progeny comprised 315 RIL successfully genotyped with 75 markers. The average distance between markers was 5.9 cM, on average equivalent to 1.65 Mb, with a convenient global allelic equilibrium (Col0 52.7%, Blh1 47.3%, VNAT database).
2.2. Plant Cropping
Plants were grown in a greenhouse at INRA Versailles (France), in a mix of sand and compost, and watered with a normal nutrient solution (harvested in 2001 for Bay0 × Shahdara, in 2005 for Bur0 × Col0 and Blh1 × Col0). The temperature was 23˚C during the day and 15˚C at night, with a 16 h light/8 h dark photoperiod. Additional light was added according to natural day length giving 105 mE/m2/s at the plant level. Floral stems were collected at seed maturity in successive harvests according to plant earliness. Only one replicate was cropped, but growth conditions ensured homogeneous materials allowing relevant phenotypic analysis.
2.3. Samples Analysis and Trait Variation
Floral stem samples cleared of siliques were dried in a ventilated oven (65˚C). Dry samples were then ground with a hammer mill to pass through a 1 mm screen for later analyses. Due to sample quantity available after grinding, investigations were carried out on 396 RIL out of 420 in Bay0 × Shahdara, 311 out of 344 in Bur0 × Col0, and 299 out of 315 in Blh1 × Col0. Plant cell wall estimated constituents included neutral detergent fiber (NDF, [31]) which is an estimate of cell wall content, Klason lignins (KL, [32]) which is an estimate of the whole lignins, acid detergent lignins (ADL, [31]) which is an estimate of the core acido-resistant fraction of lignins [33], and neutral detergent soluble fiber (NDSF, [34] ) which is an estimate of pectins. ADL and KL, which are constituents of the cell wall, were both expressed as percentage of NDF (ADL/NDF and KL/NDF). The in vitro dry matter degradability (IVDMD) was estimated based on enzymatic solubility of samples according to Aufrère and Michalet-Doreau [35]. Cell wall degradability was first considered according to Struik [36] and Dolstra and Medema [37] as the in vitro NDF degradability (IVNDFD), which is computed assuming that the non-NDF part of plant material is completely digestible [IVNDFD = 100 × (IVDMD – (100 – NDF))/NDF]. Cell wall degradability was also estimated according to the DINAGZ trait which has been proposed and used for maize plants by Argillier et al. [38] and Barrière et al. [39], considering that starch (St), soluble carbohydrates (SC), and crude protein (CP) are fully digestible [DINAGZ = 100 × (IVDMD – St – SC – CP)/(100 – St – SC – CP)]. Because no starch is present in Arabidopsis mature stems and because conversely to maize significant amounts of soluble pectins are present, starch content was replaced in the DINAGZ formula by NDSF content. For DINAGZ estimate, crude protein was estimated according to Kjeldahl nitrogen ´ 6.25, and soluble carbohydrates according to Lila [40]. All plant components and IVDMD were estimated using near infrared reflectance spectroscopy (NIRS, NIRS system 6500 spectrophotometer, FOSS technology). Calibration equations were developed at INRA Lusignan (Table 1), and calibration regressions were validated with laboratory analysis of nearly 40 samples per progeny.
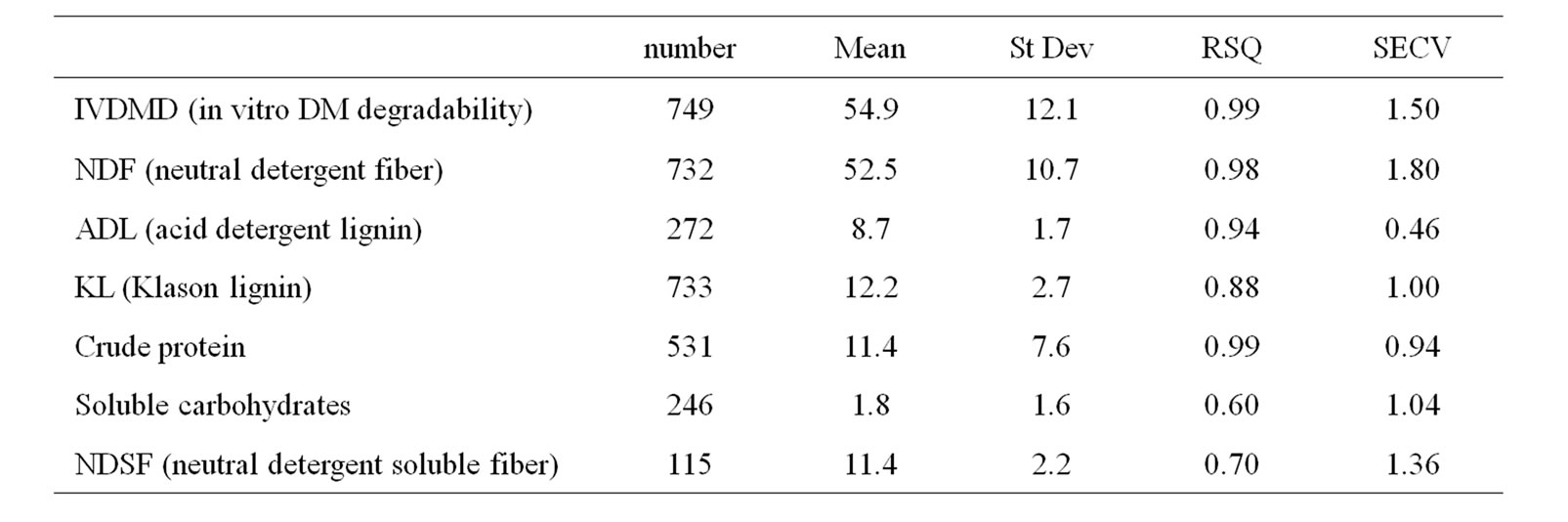
Table 1. Characteristics of NIRS calibrations developed for cell wall traits in Arabidopsis mature stems (for each trait, number is the numbers of analyzed samples with their mean and standard deviation (St Dev), RSQ is the coefficient of determination between laboratory analysis and NIRS prediction, and SECV is the standard error of cross validation prediction; DM = dry-matter).
Lower numbers of reference analyses were available for ADL, NDSF, and SC. However, fairly low coefficients of determination between laboratory analysis and NIRS prediction were only observed for NDSF and SC, two traits that were only involved in the estimate of the DINAGZ cell wall trait. Moreover, the average content in SC was very low in plant mature stems. Pearson’s phenotypic correlations between traits were computed for each progeny.
2.4. QTL Mapping and Candidate Gene Investigations
QTL mapping was based on individual values of each investigated RIL in each progeny, using the Composite Interval Mapping method (CIM) implemented in the PLABQTL computer package [41]. PLABQTL uses the regression method of Haley and Knott [42] in combination with selected markers as cofactors. Cofactors are selected by stepwise regression (option cov SELECT) with an “F-to-enter” and an “F-to-delete” value of 7, retaining markers significant at the 1% level. Lod thresholds are obtained by the permutation test method (1000 permutations, [43-45]). Percentages of phenoltypic variance ascribed to individual QTL are estimated with the approximate standard error of Kendall and Stuart [46]. LOD support intervals are constructed for each QTL according to Lander and Botstein [47], which could be underestimated in the case of CIM. The additive effects of QTL are estimated as half the difference between the phenotypic values of the respective homozygotes. Physical QTL positions (Mbp, Mega-base-pairs) were estimated based on physical positions of the two flanking markers assuming a constant and linear relationship between recombination and physical distances within this interval. Physical lengths of QTL support intervals were similarly estimated. A consensus map was drawn with MapChart, based on physical distances (http://www. biometris.wur.nl/uk/Software/MapChart/).
The list of putative candidate genes was established gathering available information on genes involved or putatively involved in the lignified cell wall assembly of Arabidopsis. This list thus comprised Arabidopsis genes belonging to the monolignol pathway, their transcription factors, and genes involved in lignified tissue assembly. The list of putative candidate genes also comprised orthologs of genes for which an involvement in cell wall lignification and deposition have been established in other dicotyledonous plants including poplar [48] and eucalyptus [49], but also maize [6]. The list took into account Arabidopsis expression data (http://genecat.mpg. de/; http://atted.jp, [50-53]) and only genes with expression in stems were considered. Gene physical positions were those given in “The Arabidopsis Information Resource” database (TAIR, http://www.arabidopsis.org).
2.5. Allelic Variation Investigations
Alleles of the Bur0 and Col0 genomes were compared according to sequence data available in the public databases, with several validation based on direct sequencing. The sequence of the reference Col0 ecotype is available in the TAIR database (www.arabidopsis.org/) and the sequence of the Bur0 ecotype has been recently released [54,55] in the 1001genomes database (www.1001genomes.org/).
3. RESULTS AND DISCUSSION
3.1. Arabidopsis Genes Considered as Putative Candidates Underlying Cell Wall Trait QTL
3.1.1. Genes Involved in Monolignol Biosynthesis and Polymerization
The monolignol pathway has been extensively investigated in Arabidopsis, making it easy to list the genes involved [10,13,56-63]. After deamination of phenylalanine by phenylalanine ammonia lyases (PAL), monolignol biosynthesis is the result of successive steps of hydroxylation and methylation on the aromatic ring. Most of genes in the pathway belong to small multigene families, with different members likely involved in different metabolons. As far as information was available, the considered candidate gene list included only genes involved in constitutive lignification and excluded genes involved in lignification after non developmental signals such as abiotic or abiotic stresses (Table 2).
Class III peroxidases and laccases, which are both involved in monolignol polymerization, belong to larger multigene families. The considered members were chosen based on their expression in stem and/or lignified tissues [12,64-71]. There is an important redundancy of peroxidase genes and most often individual mutants did not clearly exhibit any lignin modified phenotype. Nevertheless, redundancy importance is likely reduced by the fact that many peroxidases have tissue specific expression patterns [72].
Before their polymerization, monolignols are transported from the cytosol to the cell wall, likely as monolignol glucosides [73,74]. Two uridine-diphosphateglucosyltransferases (UGT) have been shown with capacity to glucosylate coniferyl and sinapyl alcohols [75, 76]. The release of monolignol aglycone from its glucosidic form at the cell wall for subsequent lignin polymerization is thought to be mediated by specific glucosedases. Arabidopsis β-Glu45 and β-Glu46 β-glucosidases (UGT), which are strongly expressed in lignifying organs, encode proteins with narrow specificity towards the three monolignol glucosides [74]. In addition, and even if this
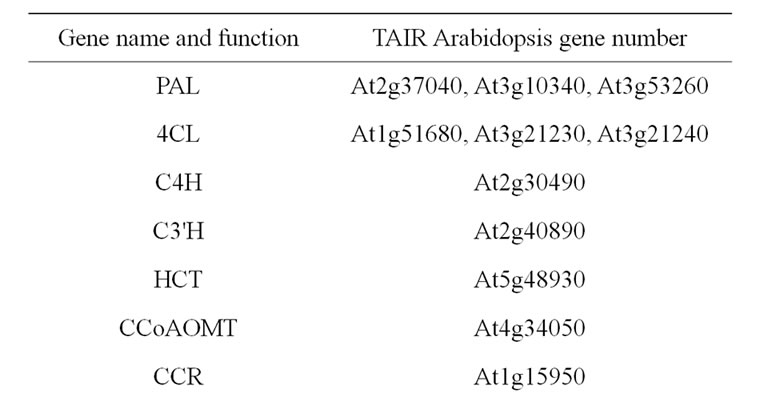
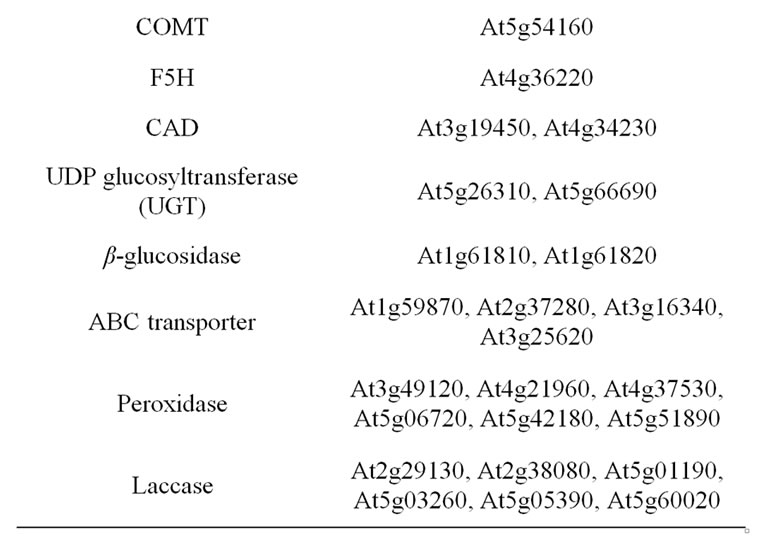
Table 2. List of considered candidate gene underlying cell wall related QTL. Genes involved in monolignol biosynthesis and polymerization.
step is not fully elucidated, ABC transporters are very likely involved in the transport of monolignols across membranes [66,77-80].
3.1.2. Transcription Factors Involved in Regulation of Lignified Cell Wall Biosynthesis Genes
Comprehensive investigations have used the Arabidopsis model system in order to document the transcription factors regulating the secondary wall biosynthetic program (Table 3).
Different Arabidopsis MYB and NAC proteins regulate the expression of several transcription factors and/or genes involved in secondary cell wall biosynthesis [8,9, 81-85]. NAC NST1 and NST2 (secondary wall thickening promoting factor), SND1 (secondary wall associated NAC domain protein 1), and VND6 and VND7 (vascular-related NAC domain) are thus cell-type specific and partly functionally redundant “master” genes activating the entire secondary wall biosynthetic pathways [81,82]. In addition, VNI2 (VND-INTERACTING2, and to a lower extent VNI1), is a transcriptional repressor of vessel-specific genes regulated by VND7 [86]. Interactions mostly occur between VNI2 and VND7, but also exist to a lower extent with other VND proteins and possibly other NAC factors. Downstream these genes, direct target transcription factors such as AtMYB46 and At
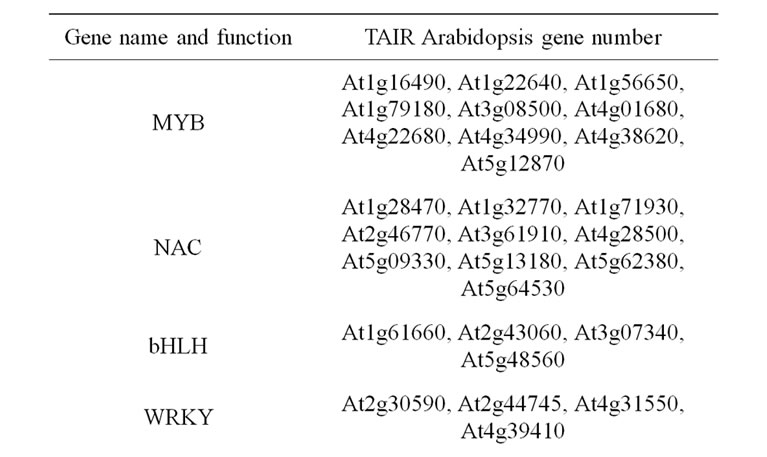
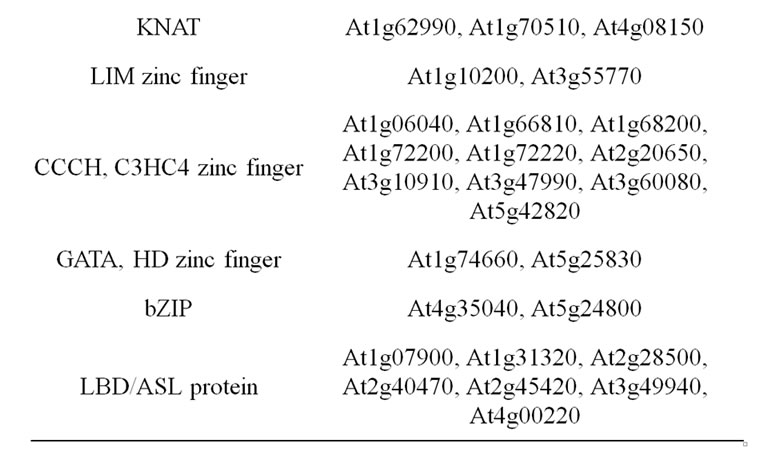
Table 3. List of considered candidate gene underlying cell wall related QTL. Transcription factors involved in secondary wall biosynthesis and assembly.
MYB83, and several secondary targets regulating monolignol biosynthesis genes were also considered to be relevant candidates [9,84,86-90].
Complementarily, different LIM and WRKY transcription factors have to be considered as candidate genes. The tobacco NtLIM1 gene was shown to be a positive regulator of the lignin pathway [91]. LIM proteins are characterized by zinc-binding domains that ligate two zinc ions. Unlike the classical zinc fingers, these domains do not bind DNA, but mediate interactions with other proteins [92]. WRKY proteins are involved in many processes, especially including defense against biotic stresses and their role in cell wall constitutive lignification is not yet established [93,94]. However, several AtWRKY genes were highly expressed in lignifying stems, and the mutation of the AtWRKY12 gene induced secondary wall formation of pith cell [53]. Several WRKY genes expressed in healthy lignifying stems were thus considered as candidates.
Transcription factors belonging to other families have been related to lignified tissue biosynthesis, and could also consequently be considered as candidates underlying lignified cell wall related QTL. Zinc finger proteins, which constitute one of the largest families of transcripttion factor regulatory proteins, are involved in numerous regulations of plant development, including lignified tissue deposition. Zinc-finger C2H2 genes were the most frequently represented transcription factors in eucalyptus secondary xylem libraries [49]. The CCCH zinc finger protein (AtC3H14) has been shown to activate all of the secondary wall phenolics and carbohydrate related genes tested [95]. Both SND1 and MYB46 proteins bound to the AtC3H14 promoter, and AtC3H14 may function as master regulator of secondary wall biosynthesis, located downstream of MYB46.
Basic region/leucine zipper motif (bZIP) transcription factors regulate several processes during plant development and are involved in biotic and abiotic stress responses. More especially, studies of group I bZIP genes have given converging evidence that members of this family might regulate vascular development [96].
LBD or ASL (LATERAL ORGAN BOUNDARIES DOMAIN or ASYMMETRIC LEAVES) proteins are a family of plant-specific transcription factors with 43 members in Arabidopsis which regulate a variety of developmental processes [97]. ASL19/LBD30 and ASL20/ LBD18 are expressed under the control of VND6 and VND7, and LBD18 or LBD30 over-expression induced trans-differentiation of cells from non-vascular tissues into tracheary-like cells [98]. These LBD proteins appear to be involved in a feedback loop with VND7 that regulates genes involved in tracheary element differentiation [98,99]. In addition, Yordanov et al. [100] showed similar results in poplar based on investigations in a LBD1 mutant which displayed an increased secondary phloem. The LBD1 and LBD11 Arabidopsis genes are the closest orthologs of poplar PtaLBD1.
Basic helix-loop-helix (bHLH) transcription factors belong to a family of transcriptional regulators for which many different functions have first been identified in animals, including the control of cell proliferation and development of specific cell lineages. Their mechanism for controlling gene transcription often involves homodimerization or heterodimerization. At least 133 bHLH genes have been shown in Arabidopsis and the AtbHLH gene family also constitutes one of the largest families of transcription factors in this species [101]. Genes considered in the candidate gene list were those co-expressed with lignin related genes and significantly expressed in stems.
3.1.3. Candidate Genes Putatively Involved in Lignified Tissue Patterning
The COV1 (continuous vascular ring) gene encodes an integral membrane protein of unknown function which is supposed to be involved in a mechanism that negatively regulates the differentiation of stem vascular tissue by a mechanism independent of auxin [102]. The Arabidopsis HCA (HIGH CAMBIAL ACTIVITY) mutant also exhibits an increase in vascular tissue development characterized by a continuous ring of xylem and phloem with an unusual cambial activity. Conversely to COV1, HCA was shown to control the arrangement of vascular bundles by regulating the auxin-cytokinin sensitivity of vascular cambial cells [103]. Another mutant with high cambial activity (HCA2) has been described [104]. Its phenotype was shown to result from an elevated expression of a DOF transcription factor (DOF5.6, DNA binding with one finger), preferentially expressed in the cambium, phloem, and interfascicular parenchyma cells of inflorescence stems. Ectopic lignification was also related to variation in DOF gene expression in pom1, eli1 (ectopic lignification 1) and det3 (de-etiolated 3) Arabidopsis mutants, in addition to expression variation of MYB genes [105] (Table 4).
The GRAS SCARECROW and SCARECROW-like proteins belong to a plant specific transcription factor family which contains basic leucine zipper regions [106, 107]. These proteins are involved in complex regulatory pathways regulating tissue patterning and differentiation [107]. The SCARECROW protein is thus involved in bidirectional cell signaling mediated by miRNA165/6 and interfering with the transcription factor SHORT ROOT (SHR, equally expressed in stem and root) and class III homeodomain leucine zipper proteins towards the control of xylem patterning [108].
Members of a small class III homeodomain-leucine
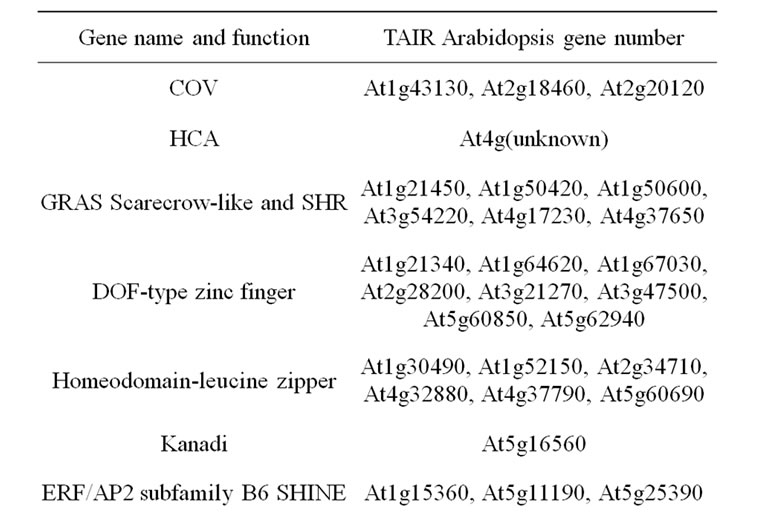
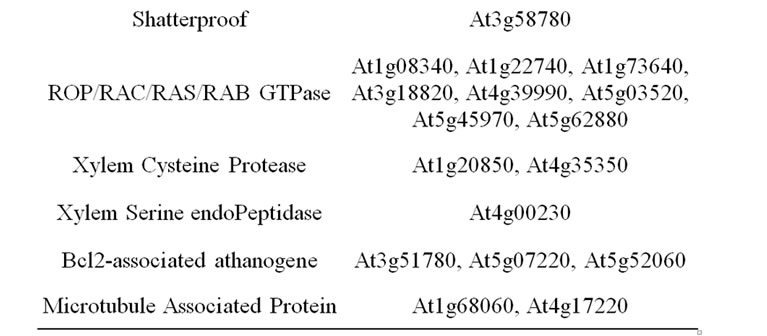
Table 4. List of considered candidate gene underlying cell wall related QTL. Genes involved in lignified tissue patterning.
zipper family out of which AtHB8, AtHB9 (PHAVOLUTA), AtHB14 (PHABULOSA), AtHB15 (CORONA), and IFL1 (REVOLUTA), are expressed in vascular tissues and have been shown to play regulatory roles in vascular differentiation [109-114]. The IFL1 Arabidopsis gene has two maize orthologs of which mutants have rolled leaf phenotypes (RLD1 and RLD2). The maize RLD1 gene is regulated by the ZmmiR166 miRNA [115]. The expression of the aspen PtaHB1 gene, which is also orthologous to IFL1, is also inversely correlated with the level of miR166 miRNA [116]. Similarly, the AtmiR166g over-expression in Arabidopsis jabba-1D (jba-1D) mutant plants affects the transcripts of several class III AtHDZIP family target genes. PHABULOSA, PHAVOLUTA, and CORONA genes have thus a significantly reduced expression in the jba-1D background, while REVOLUTA expression is increased and ATHB8 expression is unchanged [117]. In addition, interactions between HDZIP III and KANADI gene family members were shown to be involved in the establishment of the spatial arrangement of phloem, cambium and xylem. It was considered that HDZIP III and KANADI transcripttion factors control cambium activity, with KANADI proteins acting on auxin transport, and HDZIP III proteins promoting axial cell elongation and xylem differentiation [114]. These genes could therefore be considered as candidates even if their involvement occurs upstream in the pathway.
The SHP1 (SHATTERPROOF MADS-box) gene, which has been shown to specify with SHP2 the lignified valve margin of mature Arabidopsis siliques [118], likely has other roles in tissue lignification as it is also expressed in stems and down-regulated at the maturing stage [51]. Moreover, an ortholog exist in maize (ZmZAG5), the function of which is not known [6,119,120].
Two microtubule-associated proteins (AtMAP70-5 and AtMAP70) were shown to be essential for defining where secondary cell wall polymers are applied at the cell cortex in wood-forming cells [121]. These two genes were consequently added to the candidate gene list.
A member of the plant ROP family (EgROP1) was shown to be preferentially expressed in the cambial zone and differentiating xylem of eucalyptus. Its over-expression in Arabidopsis altered vessel formation and fibre growth in secondary xylem [122,123]. AtROP/AtRAC/ AtRAB genes encode geranyl-geranylated GTP-binding proteins (GTPases) involved in the auxin proteolysis pathway. The latter are thought to provide a universal mechanism in the control of extracellular signal transmission to intracellular metabolic pathways related to growth, differentiation, development and defense responses [124,125]. Several of them are involved in autophagy and xylem development [126]. Eight Arabidopsis RAS/RAC/RAB GTPases expressed in stems were thus considered as candidate genes.
Candidate genes were also considered in the ERF/AP2 (ethylene responsive factor/APETALA2) SHINE1 family. Based on over-expression investigations in rice [127], an ERF/AP2 gene was indeed considered as an upstream transcriptional regulator of both master and secondary target genes involved in the biosynthesis of cell wall phenolic and carbohydrate components. It was suggested that this ERF/AP2 transcription factor directly binds to promoter regions of NAC and MYB genes involved in regulation of cell wall assembly. Rice plants over-expressing Arabidopsis SHINE2 gene had lower lignin and higher cellulose and hemicellulose contents, without changes in plant strength and overall performances.
Finally, candidate genes were considered in families involved in proteolysis and apoptosis, which are keysteps in the lignification process. Plant BAG (Bcl-2-associated athanogene) proteins are multi-functional and they regulate apoptotic-like processes in plant development and responses to pathogen attack or abiotic stress [128] . Two cysteine protease genes (XCP1, XCP2), highly up-regulated in maturing stems, and one xylem serine peptidase (XSP1) were considered as putative candidates underlying cell wall QTL [51,88,129].
3.1.4. miRNA Involved in Regulation of Cell Wall Related Genes as Candidates Underlying QTL
Several AtMIR were considered as possible candidate following the description of their involvement in ligninrelated gene regulation (Table 5). AtMIR165 and AtMIR166 have been shown to target and regulate the transcription class III AtHD-ZIP genes, including those involved in vascular tissue differentiation [115,130-133]. AtMIR397a and b, AMIR408, and AtMIR857 control laccase genes, some of them potentially involved in monolignol polymerization in the cell wall through the regulation of copper homeostasis [134]. In addition, AtMIR858 targets the MYB83 gene which interacts with VND7 and negatively regulates xylem vessel formation [132].
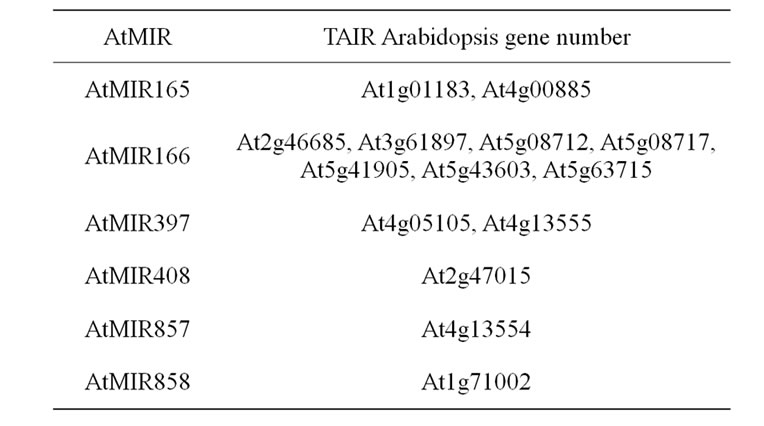
Table 5. List of considered candidate gene underlying cell wall related QTL. miRNA regulating lignified tissue biosynthesis and assembly.
3.1.5. The Final Candidate Gene List
Ultimately, 133 putative candidate genes were considered (Table 2), out of which 39, 18, 22, 23, and 31 were located on chromosomes 1, 2, 3, 4, and 5, respectively, while 12, 5, 1, 11, and 11 QTL were located on the same chromosomes, respectively. Out of the 16 putative genes involved in constitutive monolignol biosynthesis, 2, 3, 5, 4, and 2 are located on chromosomes 1, 2, 3, 4, and 5, respectively without any clustering in the monolignol pathway or between members of multigene families. Six peroxidase genes out of the 73 described in the Arabidopsis genome [64], and six laccase genes out of the 17 described in the Arabidopsis genome [65], were considered. These were mostly located on chromosomes 4 and 5. Transcription factors considered as putatively involved in regulation of cell wall phenolic component biosynthesis were mostly located on chromosome 1 (19 out of 53), while the genes involved in lignified tissue patterning were mostly located on chromosomes 1 and 5 (15 and 11 out of 44, respectively). This candidate gene list, which focused on genes involved in cell wall phenolic component biosynthesis and regulation, cannot be considered as exhaustive. Several other genes, whose roles in cell wall lignification and deposition have not yet been defined, should be added to the list of candidates underlying cell trait related QTL. Moreover, a few genes encoding proteins of unknown function, which were expressed in stems, colocalized with QTL. However, the latter were not taken into account as their possible involvement in lignified tissue biosynthesis and deposition is still unknown.
3.2. Trait Phenotypic Variation and Correlations in RIL Progenies
Observed means and ranges of variation were nearly similar for cell wall traits in the three RIL progenies (Table 6), with nevertheless some distinctive characteristics between families. NDF content was lower in the Bay0 × Shahdara progeny. Average ADL lignin content was nearly 3 percent higher in Bur0 × Col0, with correlatively lower cell wall degradabilities. Unlike with ADL lignins, no differences between progenies were observed for average Klason lignin content. Lignin polymers in the different progenies were thus characterized by the loss of a variable acid-soluble part during the first step of the ADL procedure [33,135].
Correlations were high between cell wall content and other traits, except with Klason lignins (Table 7, correlations significantly different from zero at P = 0.01 for absolute values higher than 0.15). These results highlighted an important difference between Arabidopsis and maize. In Arabidopsis cell wall degradability was highly related to cell wall content, with an average correlation value equal to –0.85, while it was only 0.13 in maize, as an average of two RIL progenies in per se value and topcross experiments ([17,136] and unpublished data). However, the cell wall degradability was similarly related to ADL lignin content in maize (average r = –0.80) and Arabidopsis (average r = –0.70). The weak or null relationship between Klason lignin and cell wall degradability in both maize and Arabidopsis strengthened the role of only the core lignin part (estimated as ADL/NDF) in the inhibition of cell wall degradability. The lack of a secondary cambium and the different tissue organization between grasses and dicotyledonous plants partly limit gene discovery when using Arabidopsis as a model system to investigate crop plants such as maize. However, major determinants of lignification and correlative negative effects on cell wall degradability can likely be considered in Arabidopsis to identify candidate genes in maize.
3.3. QTL Analyses in the Three RIL Progenies
Lod thresholds equal to 2.3 and 3.0, 2.5 and 3.3, and 2.5 and 3.5 yielded experiment-wise error rates equal to 5% and 1% for Bay0 × Shahdara, Bur0 × Col0, and Blh1 × Col0, respectively. QTL were therefore only considered significant in each progeny for error equal to or lower than 5%, and overall 40 QTL were mapped (Figure 1). Because data were available from only one replicate per RIL, the total percentages of variance explained by all QTL for each trait in each progeny were moderate and
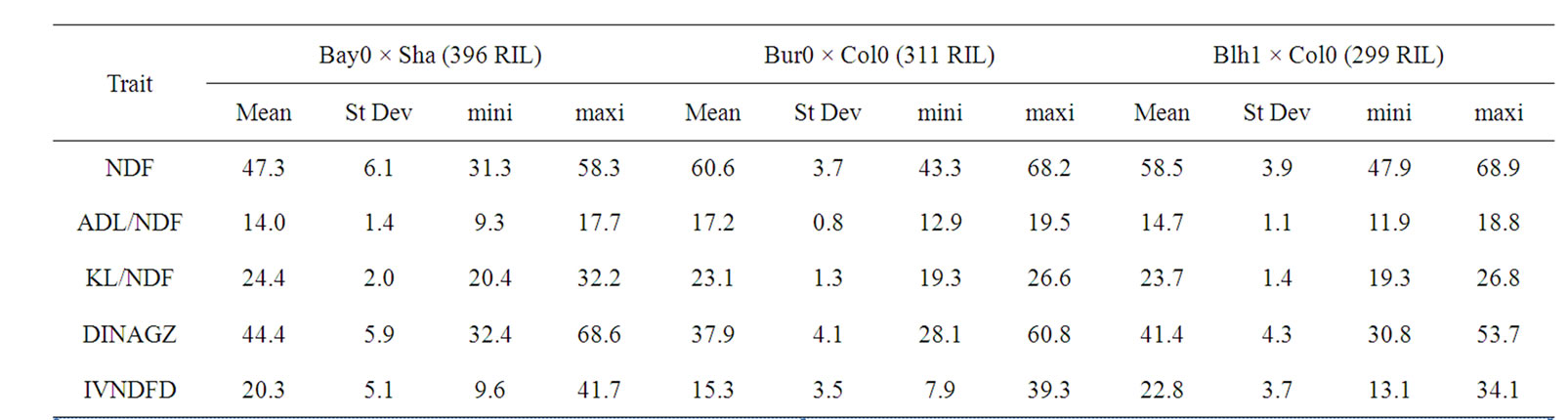
Table 6. Means, standard deviations, and extreme values for investigated traits in the three RIL progenies.
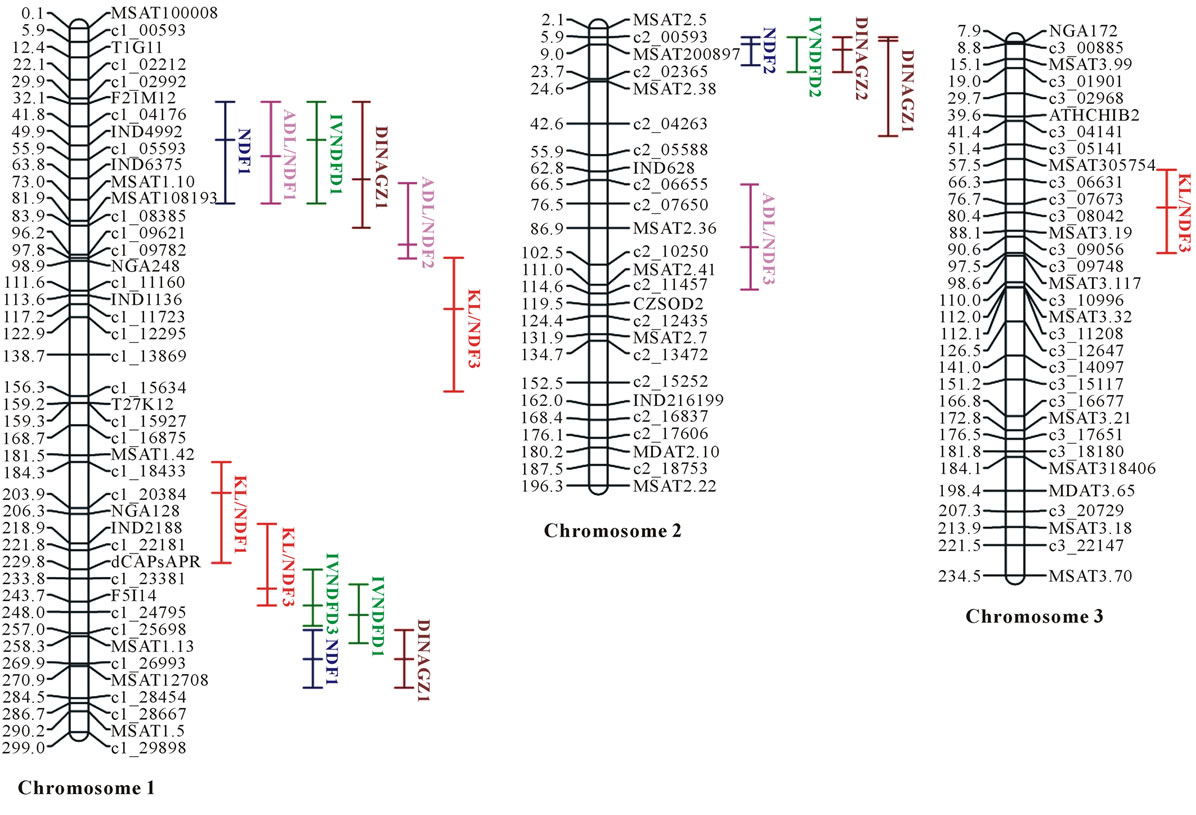
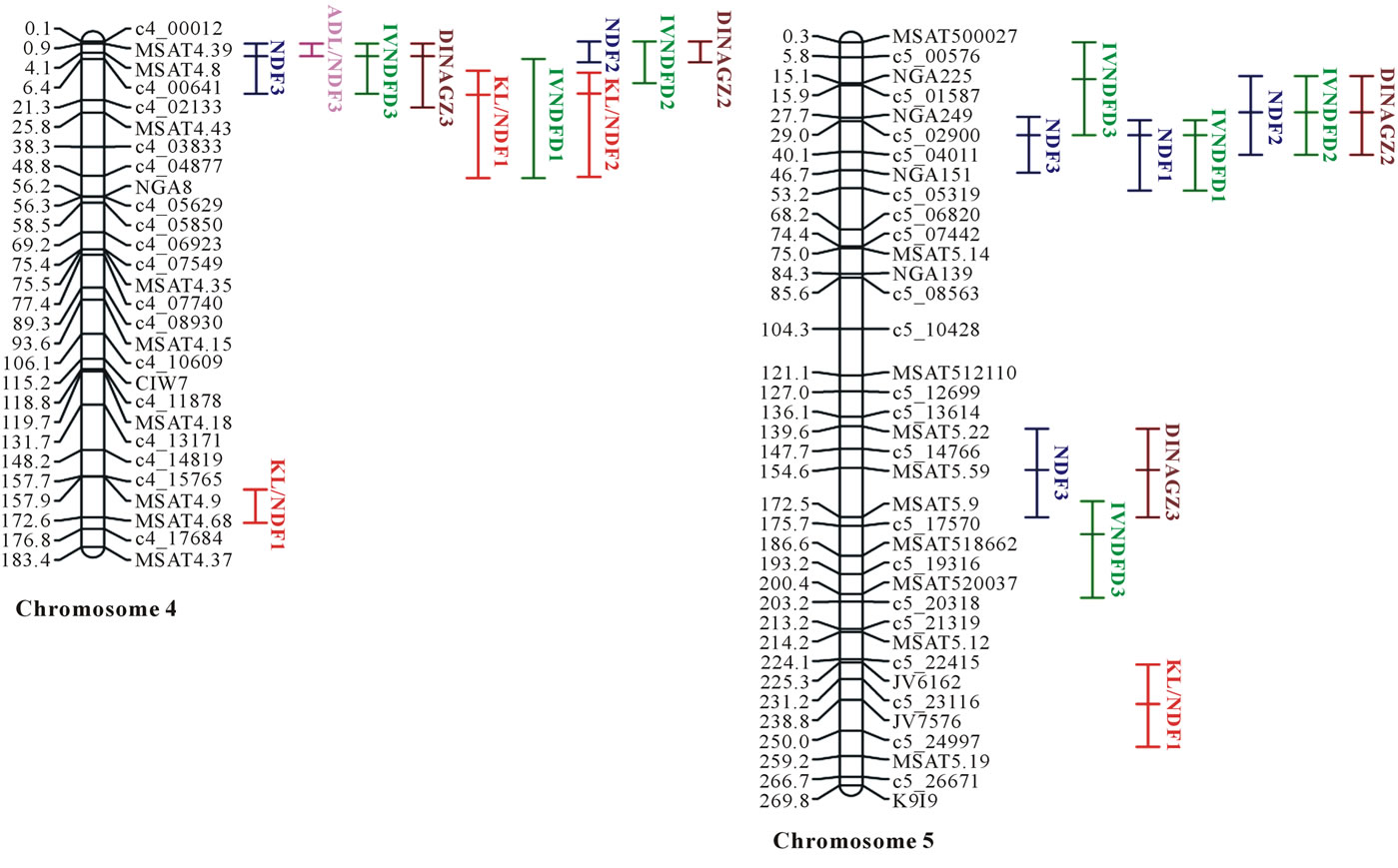
Figure 1. Consensus map of the three RIL progenies based on physical distances (distances were given as 101 Mbp; numbers after QTL trait names indicated the respective progenies with 1 = Bur0 × Col0, 2 = Blh1 × Col0, 3 = Bay0 × Shahdara).

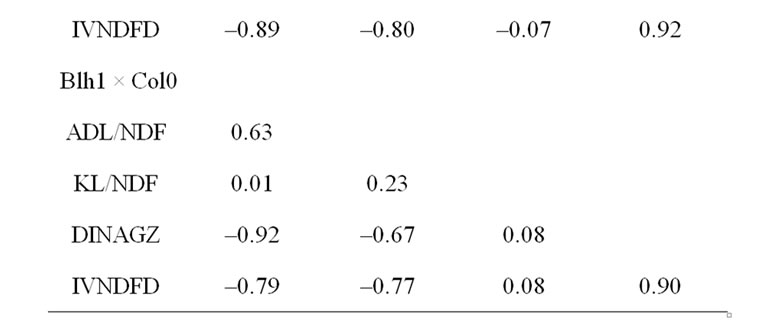
Table 7. Correlations between investigated traits in the three RIL progenies.
nearly equal to 20%, 30%, and 30% in the Bay0 × Shahdara, Bur0 × Col0, and Blh1 × Col0 progenies, respectively. The lack of experimental replicates thus induces some inaccuracies in the phenotypic values of RIL with consequently a lowering of QTL Lod values and lack of detection of QTL of weaker effects. Eight QTL explaining more or at least 10% of the observed phenoltypic variation were nevertheless shown, with corresponding Lod values equal or greater than 6.0. In addition, no significant interactions between any QTL were observed.
Only 5 QTL out of 40 were mapped in isolated position, including one ADL/NDF QTL in the Bay0 × Shahdara progeny, and four KL/NDF QTL in Bur0 × Col0 and Bay0 × Shahdara progenies (Table 8). Half of KL/NDF QTL was thus located in isolated positions, strengthening the relative independence between Klason lignins, ADL lignins or cell wall degradability. Two locations on chromosomes 4 and 5 brought together 15 QTL for the different cell wall related traits observed in the three RIL progenies. Four locations including three on chromosome 1 and one on chromosome 2 brought together 15 QTL for the different traits and two RIL progenies, while three QTL were only observed for the Bay0 × Shahdara progeny on chromosome 5.
Most QTL with the highest Lod values were observed in the Bur0 × Col0 progeny, probably reflecting larger differences in cell traits between these two ecotypes. However, a reverse situation was shown on the upstream part of chromosome 4, with QTL of high Lod values for Bay0 × Shahdara and Blh1 × Col0 RIL, and lower values for Bur0 × Col0 RIL. In addition, a Klason lignin QTL with a Lod value equal to 26.4 and explaining 33.4% of the phenotypic variation for this trait was shown at position 1.9 Mbp in the Blh1 × Col0 progeny. The existence of two genetic traits underlying QTL in positions 0.3 and 1.9 Mbp was strengthened by the fact that alleles increasing Klason lignin content and cell wall degradability originated from the same parent (Col0, based on additive values) in the Blh1 × Col0 progeny.
Plant resistance to pests and diseases included both specific and general mechanisms, out of which increased lignin contents and tissue stiffness. Hence, an effort was made to identify relationships between cell wall QTL shown in the three investigated progenies and QTL previously shown and involved in pathogen resistance. On chromosome 5 in position 50 cM, a QTL of resistance to Botrytis cinerea was shown in the Bay0 × Shahdara progeny [137]. It colocalized with a QTL of cell wall degradability with Bay0 alleles increasing both disease susceptibility and cell wall degradability. In the Bur0 × Col0 progeny, a QTL controlling partial resistance to clubroot (Plasmodiophora brassicae) was shown on chromosome 5 in position 11 cM [138]. It similarly colocalized with a QTL of cell wall degradability with Col0 alleles increasing both disease susceptibility and cell wall degradability. Another QTL was shown on chromosome 1 in position 12 cM which colocalized with QTL of ADL lignin and cell wall degradability, the Col0 allele increasing disease susceptibility and cell wall degradability and the Bur0 allele increasing lignin content. Whether these colocalizations correspond to the same genetic determinant or to different genes in close positions is still unknown. However, a genetic mechanism inducing simultaneously greater cell wall degradability and greater disease susceptibility is a plausible hypothesis. The MYB58 and SHINE2 genes could be involved in these capacities
3.4. Colocalisation between QTL in the Three RIL Progenies and Putative Candidate Genes
3.4.1. Chromosome 1, QTL Mapped at 5.5 Mbp
Several highly relevant candidate genes colocalized with QTL of large effects detected at 5.0 Mbp on chromosome 1 (Table 9). AtCCR1 is an obvious candidate, owing to its important role in the lignin pathway as shown in studies of mutants or transgenic approaches in Arabidopsis [13,56] and in other species including tobacco [139], eucalyptus [140], poplar [14,141], spruce [142], and maize [143]. The second candidate to be considered
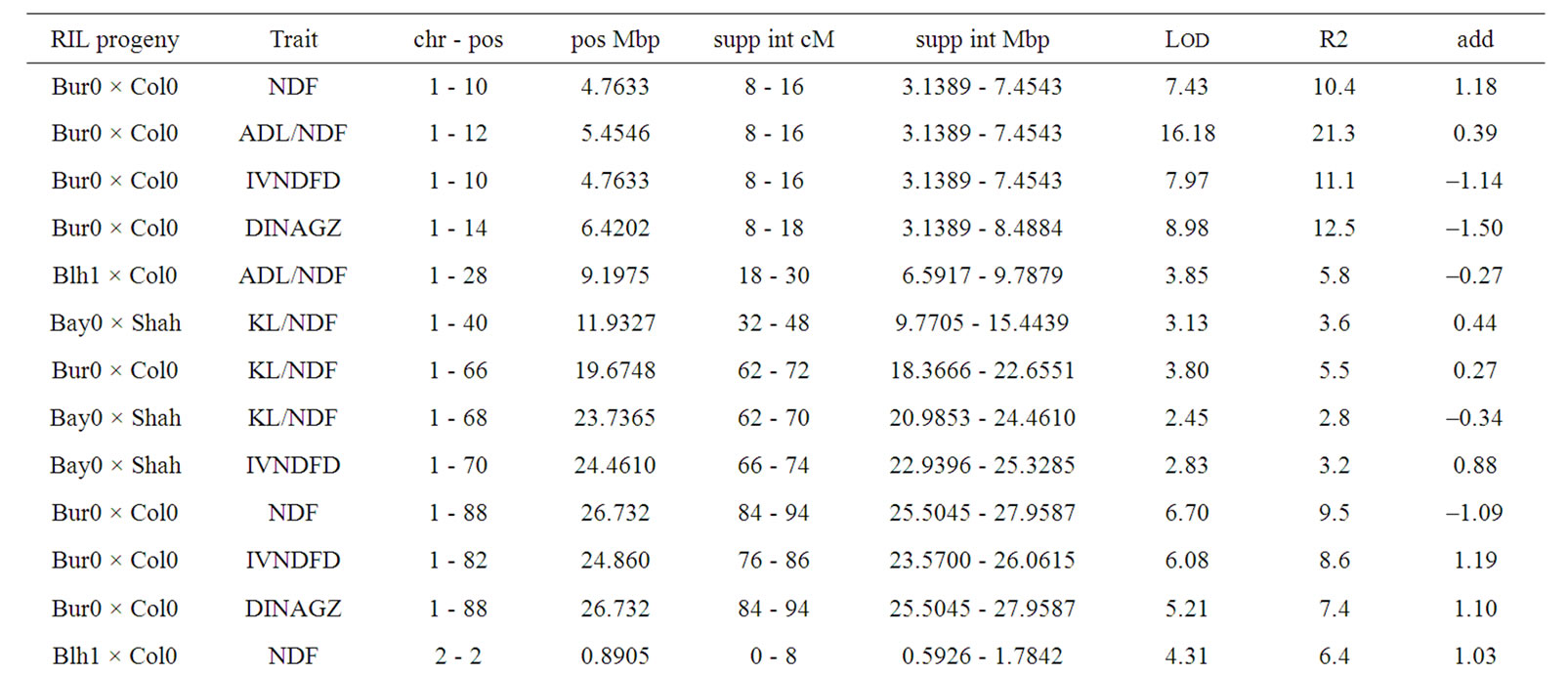
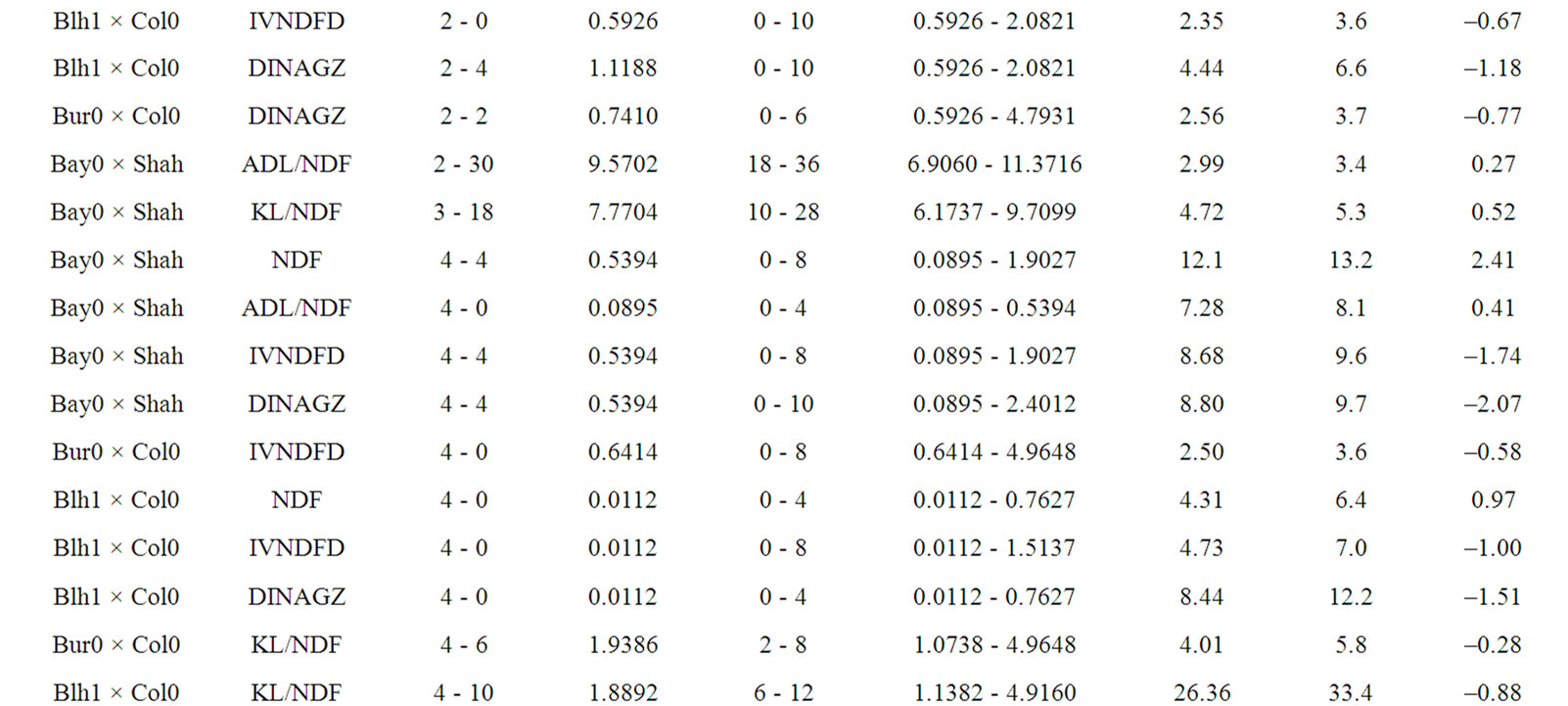
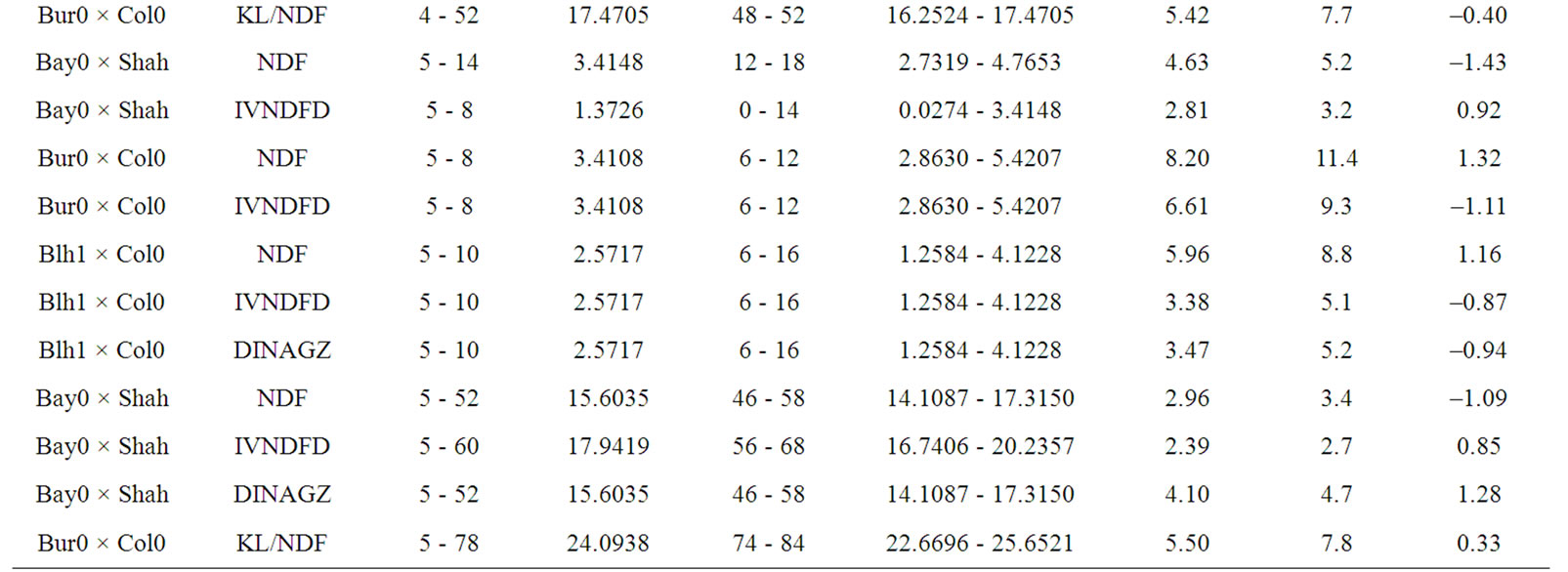
Table 8. Putative QTL observed for lignin contents and degradability traits the three RIL progenies. Additive value of traits is positive when allele increasing trait value originated from Shahdara, Bur0, and Blh1, and negative when allele increasing trait value originated from Bay0, and Col0.
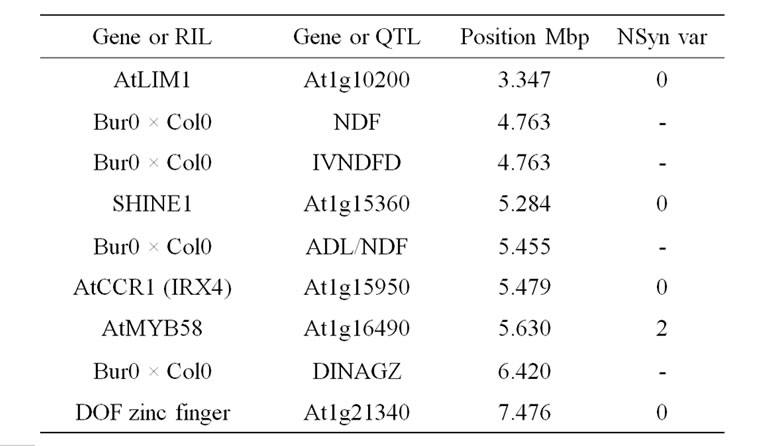
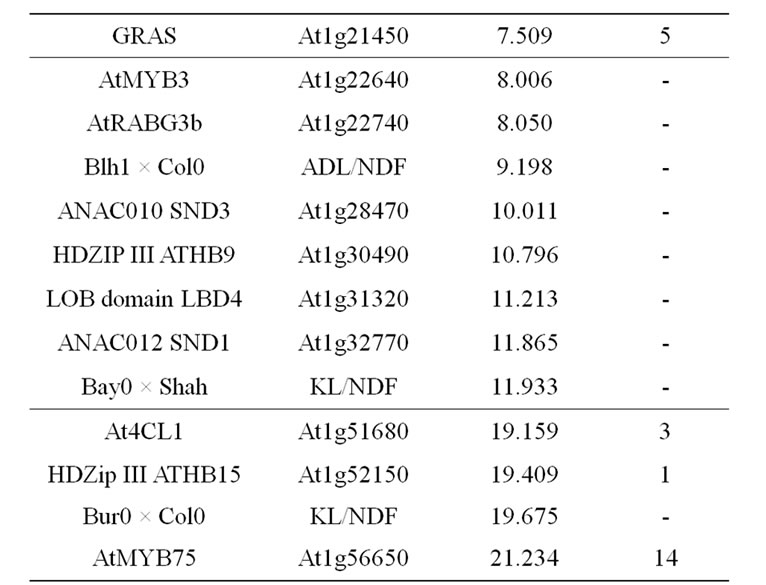
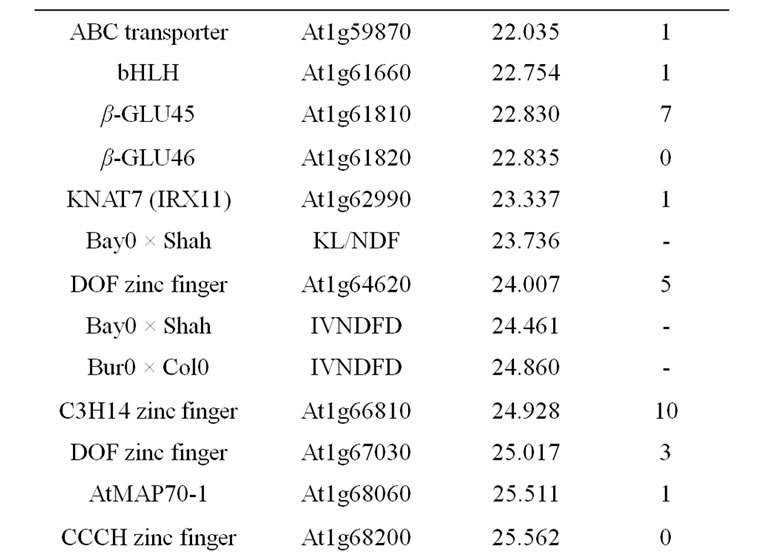
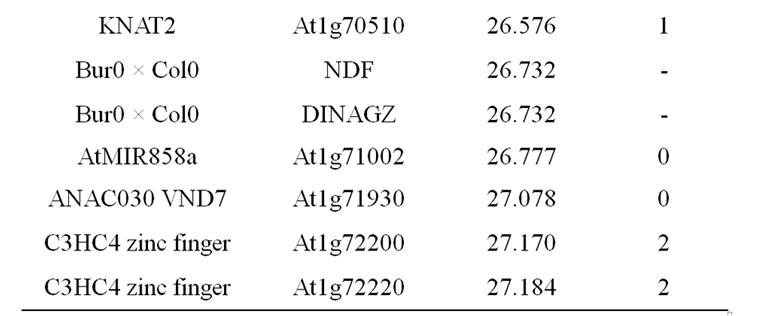
Table 9. Colocalizations between QTL positions and putative candidate genes on chromosome 1. Allelic variation of candidate genes (CDS) between Bur0 and Col0 (NSyn var = non synonymous variation including SNP and Indel).
in this locus is the AtMYB58 gene, which is a transcripttional activator of monolignol biosynthesis genes in the SND1-mediated network [84]. AtMYB58 binds to an AC element of monolignol biosynthesis genes (PAL1, 4CL1, HCT, C3H1, CCoAOMT1, CCR1). Its over-expression especially greatly increased expression of PAL1, 4CL1, and CCoAOMT1, but a significant, even lesser, increase was also observed for the COMT gene in which AC elements have not been shown [84]. AtMYB58 underor over-expression have no effect on F5H1 activity, corroborating the fact that this F5H gene is directly regulated by the master genes NST1/SND1 [144]. AtMYB58 also regulated LAC4, but not LAC17 expression [84]. Moreover, a potential natural antisens gene At1g16489 overlaps with the AtMYB58 (At1g16490) gene and it could also be the underlying determinant of the observed QTL. The AtLIM1 gene is orthologous to NtLIM1 [91] and also to ZmLIM1. The SHINE1 gene located close to the QTL position, the DOF-type zinc finger and the GRAS SCARECROW-like1 genes located at the basal part of the QTL support intervals are also possible underlying candidate genes.
3.4.2. Chromosome 1, QTL Mapped at 9 and 12 Mbp
The ADL/NDF QTL detected at 9.2 Mbp in the Blh1 × Col0 progeny and the KL/NDF QTL detected at 11.9 Mbp in the Bay0 × Shahdara progeny, were in close positions with the two master NAC transcription factors SND3 and SND1, respectively. Moreover, the AtMYB3 transcription factor that represses phenylpropanoid biosynthesis gene expression was also located in the upstream area of QTL support intervals. In the QTL support intervals were also located the LBD4 gene preferentially expressed in phloem, the ATHB9 leucine zipper gene, and the AtRABG3b (or RAB7) gene which were likely less probable candidates at these loci than the MYB and the two NAC transcription factors.
3.4.3. Chromosome 1, QTL Mapped at 20 and 25 Mbp
The Bur0 × Col0 and Bay0 × Shahdara QTL that mapped from 19.7 Mbp to 26.7 Mbp had partly overlapping support intervals. Even if it was not possible to definitely affirm the presence of only a single QTL or two distinct QTL in this area, this second hypothesis is the most probable given the structure of QTL positions and support intervals. Several major transcription factors involved in cell wall lignification (AtMYB75, and KNAT7), two DOF-type zinc fingers, the ATHB15 leucine zipper, and one gene involved in monolignol biosynthesis (At4CL1) were in the support intervals of the upstream part of this QTL location. Moreover, KNAT7 and AtMYB75 physically interacts, forming functional complexes that integrate the metabolic flux through the lignin, flavonoid and polysaccharide pathways in inflorescence stems [89]. KNAT7 could thus have a simultaneous effect on lignin content and cell wall degradability. The Bur0 × Col0 QTL located in the downstream part of this QTL position first colocalized with the finger transcription factor AtC3H14, a CCCH zinc finger protein which has been shown to activate a lot of the secondary wall phenolics and carbohydrate related genes [95], three other zinc fingers, AtMAP70-1, and with KNAT2. This QTL also colocalized with the master NAC gene VND7 Moreover, the AtMIR858, which targets the master gene AtMYB83, also colocalized with this set of QTL and could be considered a relevant candidate.
3.4.4. Chromosome 2, QTL Mapped 0.8 Mbp
Based on the proposed gene list, which is not exhaustive, no candidate gene was located in the support intervals of these QTL (Table 10). Given that no QTL for lignin traits were observed in this position, the underlying determinant should therefore be related to carbohydrate structure. The determinants of the latter were not included in the candidate gene list. However, none of the three cellulose synthase expressed in stems, and no COBRA or FRAGILE FIBER genes were located in QTL support intervals. A survey of genes located under these QTL support intervals shows the presence, between At2g02260 and At2g05620, of 384 protein-coding genes out of which 64 encode proteins of unknown function.
3.4.5. Chromosome 2, QTL Mapped at 9.5 Mbp
Three putative candidate genes with established involvement in lignified tissue assembly colocalized with the ADL/NDF QTL detected in the Bay0 × Shahdara progeny. The continuous vascular ring (COV1) defective mutant displays an increase in vascular tissue development in the stem interfascicular regions [102]. The LCV3 (like-COV-3), which is located in close upstream position to COV1, is also supposed to be involved in stem vascular tissue pattern formation. The C3HC4 zinc finger was the third possible candidate gene underlying this QTL.
3.4.6. Chromosome 3, QTL Mapped at 7.8 Mbp
Only one QTL for KL/NDF was observed on chromosome 3, in the Bay0 × Shahdara progeny. The AtCAD-C and two 4CL genes (possibly) involved in the constitutive monolignol biosynthesis pathway were located in the upstream part of the QTL, together with an ABC transporter in the downstream part (Table 11). In addition, a DOF-type zinc finger and the AtBAB7b GTPase genes were also located in the QTL support interval.
3.4.7. Chromosome 4, QTL Mapped at 0.3 Mbp
Eight QTL originating from the three progenies and re
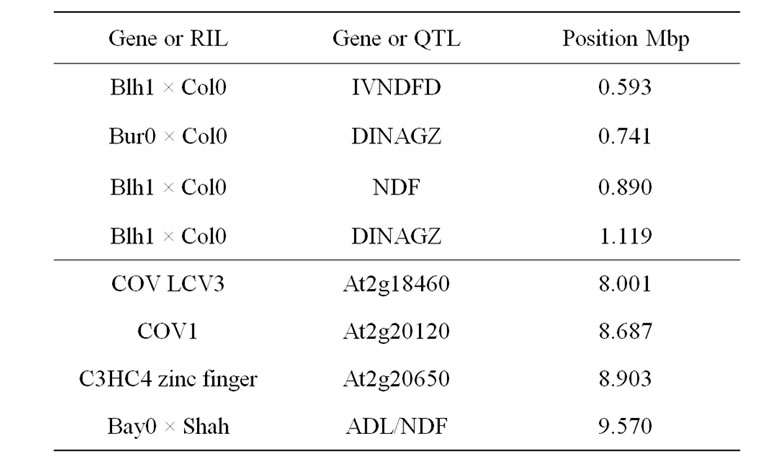
Table 10. Colocalizations between QTL positions and putative candidate genes on chromosome 2.

Table 11. Colocalizations between QTL positions and putative candidate genes on chromosome 3.
lated to cell wall content, lignin content and cell wall digestibility colocalized in the upstream part of chromosome 4, centred on the position 0.3 Mbp (Table 12). Only two cell wall related candidate genes were shown for the upstream QTL, with the LBD30 and XSP1 genes. In addition, the AtMIR165b miRNA, which was also located in the central part of the QTL support interval, targets HDZIP genes involved in vascular tissue differentiation, including PHV, PHB, REV, ATHB-8, and ATHB-15. Variation in AtMIR165b expression of efficiency could therefore explain differences in lignin content and cell wall degradability. However, another hypothesis could be considered in the search for the underlying determinant of these QTL. The FRIGIDA (flowering locus A, FRI, At4g00650) gene, which encodes a major determinant of natural variation in Arabidopsis flowering time, is located in the central part of the QTL support intervals at position 0.269 Mbp. It could therefore be assumed that the differences observed in lignin traits might result from differences in earliness, duration of stem growth, and consequential different lignification between parental ecotypes. Many early flowering accessions carry loss-of-function mutation in the FRI alleles, including the reference ecotype Col0 [145], and Bay-0
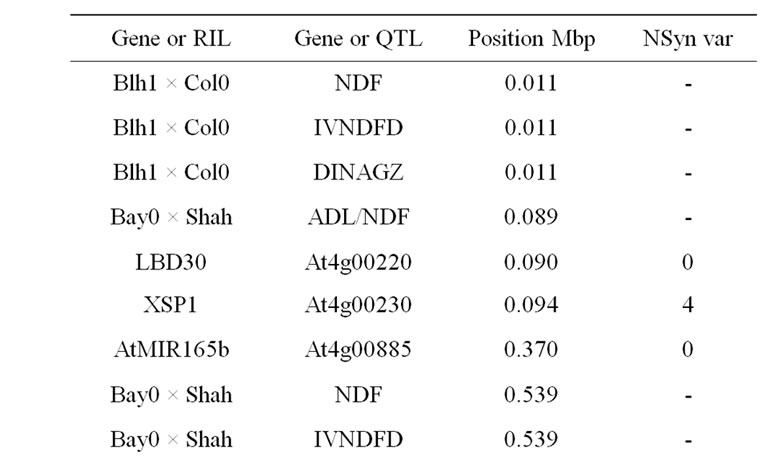
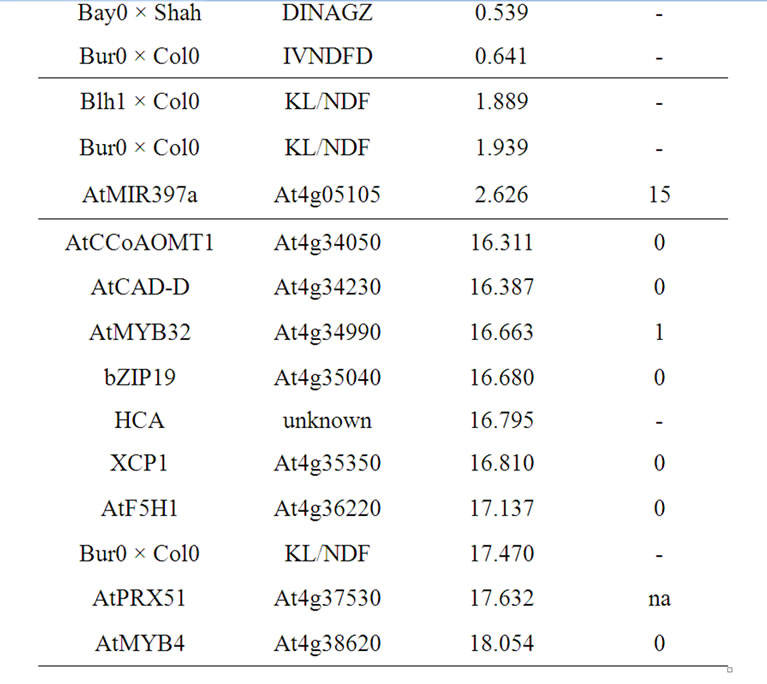
Table 12. Colocalizations between QTL positions and putative candidate genes on chromosome 4. Allelic variation of candidate genes (CDS) between Bur0 and Col0 (NSyn var = non synonymous variation including SNP and Indel, na = missing data).
which carry the same recessive allele as Col0. Conversely, Shahdara FRI allele was considered fully functional [30,146]. The two ecotypes Bur0 and Blh1 also have a functional FRI allele ([147]; Camilleri et al., unpublished data). In all cases, alleles increasing cell wall degradability were shown in ecotypes with loss-of-function FRI allele (Bay0 and Col0). A candidate gene or miRNA effect strengthened by differences in growth duration could also be hypothesized.
3.4.8. Chromosome 4, QTL Mapped at 1.9 Mbp
Even if the two KL/NDF QTL mapped at position 1.9 Mbp on chromosome 4 had partly overlapping positions with several QTL at the 0.3 Mbp position, they likely corresponded to a different underlying determinant. Moreover, the KL/NDF QTL observed in the Blh1 × Col0 progeny had the highest Lod (26.4) and R2 (33.4%) val-
ues out of all observed QTL. No candidate genes were shown colocalizing with these two QTL. However, the AtMIR397a was located at the basal part of the QTL support intervals. The AtMIR397a encodes a microRNA that targets several laccase family members. In Arabidopsis, two double mutants AtLAC4-AtLAC17 had a reduced Klason lignin content by 20% and 40%, respecttively [12] .
3.4.9. Chromosome 4, QTL Mapped at 17.5 Mbp
The isolated KL/NDF mapped at this position on chromosome 4 colocalized with several lignin-related genes, including genes involved in monolignol biosynthesis (AtCCoAOMT1, AtF5H1, AtCAD-D) and the AtPRX51 peroxidase. Colocalization was also shown with one bZIP, the XCP1 serine protease, and the HCA gene of which the position is only approximate. The latter were all putatively involved in tissue assembly and patterning.
3.4.10. Chromosome 5, QTL Mapped at 2.5 and 3.4 Mbp
On the upstream part of chromosome 5, all seven detected QTL have overlapping support intervals. It could not definitely be concluded whether one or two genetic determinants corresponded to the most likely situation. This QTL area is typified by the fact that colocalizations brought together QTL for cell wall content and degradeability, but no QTL for lignin content. The AtMYB46 gene located at the downstream part of QTL support intervals (Table 13), is, along with AtMYB83, the closest ortholog of EgMYB2, a positive regulator of lignification [148]. This AtMYB46 gene could have been a relevant candidate. However, variations driven by such a determinant might have also induced differences and QTL for lignin content. The two NAC genes VNI1 and VNI2 are repressors of xylem vessel development in roots and aerial organs [86]. The ERF/AP2 SHINE2, AtRAB8c, and AtBAG3 genes could also be considered as inducing variations in the assembly of the different lignified tissues. The laccases and peroxidase were less probable candidates as they also might have induced differences in lignin content. In addition, the two AtMIR166, which were located in central part of QTL support intervals, could also be the underlying determinants.
3.4.11. Chromosome 5, QTL Mapped at 17.0 Mbp
Considering only one genetic determinant for the three QTL only detected in Bay0 × Shahdara progeny at the basal part of chromosome 5, one NAC, one CCCH zinc finger, one bHLH gene, and one member of the ROP GTPase family, could be assumed as the main candidates. In addition the two AtMIR166e and AtMIR166f also colocalized with the QTL position. The AtHCT gene was
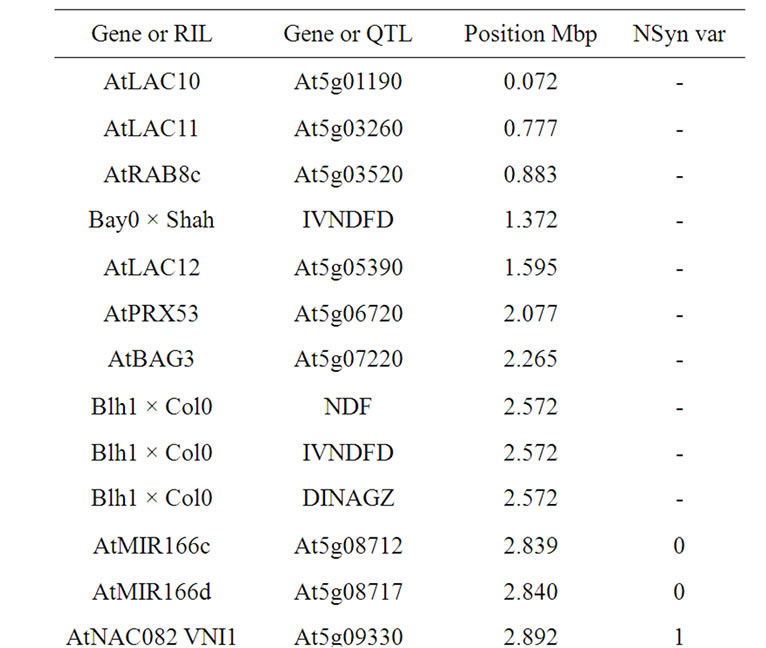
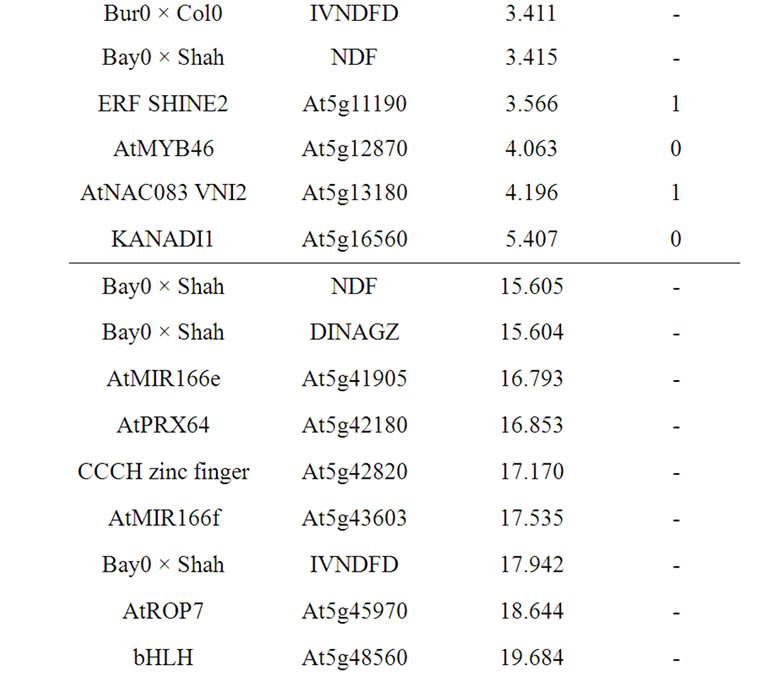
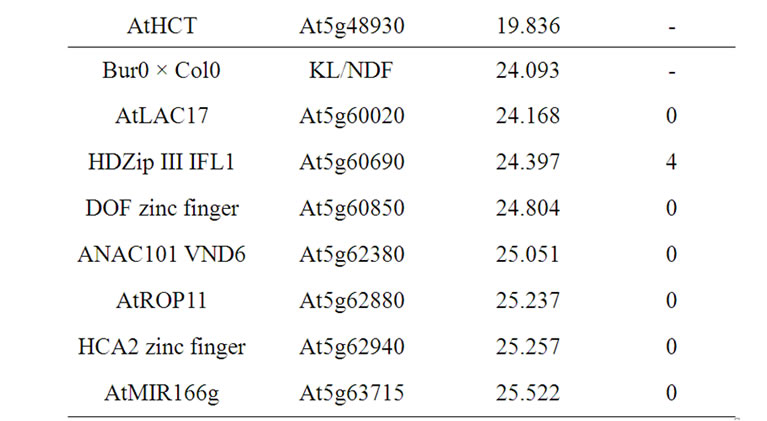
Table 13. Colocalizations between QTL positions and putative candidate genes on chromosome 5. Allelic variation of candidate genes (CDS) between Bur0 and Col0 (NSyn var = non synonymous variation including SNP and Indel).
a less possible candidate as it was located at the downstream part of the IVNDFD QTL support interval.
3.4.12. Chromosome 5, QTL Mapped at 24.1 Mbp
Several candidate genes were closely located to the position of the isolated KL/NDF QTL on chromosome 5, including the master NAC VND6, the HDZip IFL1, the HCA2 DOF-type zing finger and another DOF-type zinc finger, and the AtROP11 RAC GTPase gene. In addition, the AtMIR166g was located at the basal part of the QTL support interval.
3.5. Allelic Variation of Candidate Genes between Bur0 and Col0 Ecotypes
Allelic variation has been investigated in the coding sequence (CDS) of 43 candidate genes located in the support intervals of seven QTL shown in the Bur0 × Col0 progeny on chromosomes 1 (23 genes), 4 (9 genes), and 5 (11 genes). Among these genes, no SNP or Indel allelic variation was shown in 14 genes. In the 29 genes with allelic variation, 93 synonymous SNP, 56 non synonymous SNP and 6 Indels were shown. However, three genes (GRAS SCARECROW-like At1g21450, AtMYB75 At1g56650, and AtC3H14 zinc-finger At1g66810) exhibited most of the observed variation with 41 synonymous SNP, 27 non synonymous SNP, and 2 Indels (Tables 9, 12 and 13).
3.5.1. Chromosome 1, Bur0 × Col0 QTL Mapped at 5.5 Mbp
Among several candidate genes located in the support interval of lignin-related QTL, the CCR1 gene was a priori the most probable. However, no SNP or Indel were shown in the two CDS between Bur0 and Col0 (Table 9), nor in the 5’ areas (data not shown). Conversely, a non synonymous SNP (A/C substitution in position 836) and two glutamine deletions (ACAACA deletion in position 912) were shown in the MYB58 gene sequences. The QQ deletion was just flanking the right side of the R2R3 motif WFKHLESELGLEExDNQQ [149] of the MYB58 gene. Five non synonymous SNP were also shown in the GRAS SCARECROW-like gene in positions 1111, 1133, 1433, 1434, and 1633. In addition, the GRAS SCARECROW-like genes in Bur0 and Col0 exhibited large sequence extra variation with 25 synonymous SNP in the CDS.
3.5.2. Chromosome 1, Bur0 × Col0 QTL Mapped at 20 and 25 Mbp
Seventeen candidate genes were located in the support intervals of lignin-related QTL, with at least two and possibly three QTL positions. In the upstream part of the region, three non synonymous SNP were shown in the 4CL1 CDS (T/C, A/G, and C/A in positions 159, 517, and 1484, respectively). The AtMYB75 gene was extremely different between Col0 and Bur0 (Table 9). More especially, a G insertion in position 200 induced a modified ORF and a stop codon with a protein reduced to 68 amino acids in comparison with the 248 amino acids of the normal protein. The AtMYB75 was shown to act as a repressor of the lignin pathway [89]. The AtMYB75 inactivation in Bur0 would be in agreement with the increasing allelic effect of the KL/NDF lignin QTL located in close position to the latter gene. While the CDS of the β-glucosidase β-GLU46 were very similar in Bur0 and Col0 with only one synonymous SNP, the opposite situation was observed in the β-GLU45 β-glucosidase with two synonymous and seven non synonymous SNP between Bur0 and Col0 likely inducing different conformations and efficiencies of the two proteins. In a close position to IVNDFD and NDF QTL, the gene encoding the AtC3H14 zinc finger protein was also very different in the two ecotypes with nine non synonymous SNP and one Indel inducing a K/EF change in the protein (AAT insertion in position 172). Similarly to the β-GLU45 gene, these changes likely induced different conformations and efficiencies in the Bur0 and Col0 AtC3H14 zinc finger proteins. Because the AtC3H14 has been shown to activate a lot of the secondary wall phenolics and carbohydrate related genes [95], and because alleles from Bur0 increased IVNDFD and NDF content, it should be considered that the AtC3H14 has a lower efficiency resulting in less NDF content of likely higher degradability.
3.5.3. Chromosome 4, Bur0 × Col0 QTL Mapped at 0.3 Mbp
Only two candidate genes were shown in the support interval of QTL located in the upstream part of chromosome 4. Only the XSP1 exhibited CDS variation with two non synonymous SNP in positions 545 and 821 (A/G and T/A changes) inducing N/I and V/D amino acid changes. These changes were likely to induce changes in protein conformations and efficiencies. In addition, no changes were shown in the AtMIR165b sequence between Col0 and Bur0 (Table 12).
3.5.4. Chromosome 4, Bur0 × Col0 QTL Mapped at 1.9 Mbp
In this position corresponding to a KL/NDF QTL, only the AtMIR397a was considered as a candidate gene. AtMIR397a is a microRNA that targets several laccase family members. The two corresponding sequences were highly different between Col0 and Bur0 with 14 base substitutions in a 109 bp long cDNA. These substitutions did not affect the 21 bp long mature MIR sequence, but likely affected the stem-loop secondary structure and thus the subsequent processing of the hairpin loop.
3.5.5. Chromosome 4, Bur0 × Col0 QTL Mapped at 17.5 Mbp
Only one candidate gene located in the support interval of the KL/NDF QTL shown in the basal part of chromosome 4 exhibited non synonymous SNP variation. In position 729 of the AtMYB32 gene, a T/A CDS substitution induced a Y/H amino acid change, with possible effects on protein conformations and efficiencies.
3.5.6. Chromosome 5, Bur0 × Col0 QTL Mapped at 3.4 Mbp
The AtMYB46 gene, whose role in Arabidopsis stem lignification has been clearly established, was not considered to be the most probable candidate for the cell wall degradability QTL located in the upstream part of chromosome 5 because the lack of colocalizations with lignin QTL (Table 13). No SNP or Indel were indeed shown for AtMYB46 CDS between Col0 and Bur0. Moreover, the two alleles were also identical in their 5’ and 3’ sequences (data not shown). The cell wall degradability QTL colocalized in this location with cell wall (NDF) content QTL. Observed variation in cell wall degradability would be free of variation in lignification intensity, but negatively related to cell wall content. Among the candidate genes located in this area, CDS allelic variation were only shown for the SHINE2 gene with an AAG deletion in Bur0 (position 483) inducing a change from KE to K amino acids, with the loss of a glutamic acid. The SHINE2 ERF/AP2 gene was considered to be an upstream transcriptional regulator of both master and secondary target genes involved in the biosynthesis of cell wall phenolic and carbohydrate components. Its higher expression induced higher cellulose and hemicellulose contents [127] . Because allele increasing NDF content originated from Bur0, a higher efficiency of the protein with the E deletion should be considered.
3.5.7. Chromosome 5, Bur0 × Col0 QTL Mapped at 24.1 Mbp
Five putative candidate genes out of the six considered did not exhibit any SNP or Indel in their corresponding CDS, and similarly for the colocalizing AtmiRNA166g. Only one non synonymous SNP in the CDS of the HDZIPIII IFL1 REVOLUTA gene was shown with a T/C substitution in position 1874 inducing an F/S amino acid change. Little variation was observed for considered genes between Bur0 and Col0 in this area of chromosome 5.
4. CONCLUSIONS
While QTL investigations have been reported in numerous plant species, the underlying candidate genes have rarely been identified. Moreover, it is not known whether a QTL most often corresponds to one or several physiccally linked genes, and in case of QTL colocalization for different related traits, whether there are correlated or pleiotropic actions of only one gene, or several genes with close physical positions, each affecting one or a subset of trait variation.
Based on the set of 133 putative candidates considered, colocalizations with QTL were observed for 70 genes in the three RIL progenies, thus corresponding to 52% of colocalizing genes while the sum of QTL support intervals corresponded to 30% of the Arabidopsis genome (35 out of 115 Mbp, excluding centromer areas). Out of the 16 genes of the monolignol pathway, 9 were found colocalizing with QTL, including 4CL1/2/5, HCT, CCoAOMT1, F5H, CCR1, AtCAD-C/D, but no PAL which are the entry genes of the monolignol pathway. Considering genes involved in monolignol polymerization, three peroxidases out of six, and four laccases out of six, were shown colocalizing with QTL. Colocalizations between genes and QTL were also shown for transcription factors of the NAC and MYB families with six NAC out of ten and three MYB out of eight in colocalizing positions. Similarly, six CCCH/C3HC4 zinc finger genes out of ten and two KNAT genes out of three were in colocalizing positions. For genes putatively involved in tissue patterning, five DOF-type zinc fingers out of six and three HDZip out of six colocalized with QTL. DOF zinc finger, NAC, C3H zinc finger, HDZip, and MYB were in decreasing orders the families with members more frequently associated with QTL positions. Similarly, eight AtMIR out of the 14 considered also colocalized with QTL, including five miRNA166 out of seven, one miRNA165, one miRNA397, and the miRNA858a. Colocalizations were also shown for genes in families with one or a few members (LIM, HCA, COV, ERF SHINE, MAP, GRAS SCARECROW-like and KANADI1 genes). No colocalization was observed for SHATTERPROOF gene likely corroborating its major involvement in silique lignification. Moreover, no WRKY transcription factors colocalized with QTL, and only two ROP out of seven, and two LBD out of seven were in colocalizing positions This likely indicates that these families are possibly not or little involved in the regulation of lignified tissue assembly resulting in degradability differences.
As a tentative conclusion regarding putative determinants and based on observed colocalizations in the three RIL progenies and allelic variation between Bur0 and Col0 CDS, genes underlying the cell wall related QTL are more probably transcription factors regulating cell wall phenolic component biosynthesis. In addition, they also are more likely to be genes involved in lignified tissue patterning than genes involved in monolignol biosynthesis and polymerization. The MYB32, MYB58, MYB75, GRAS SCARECROW, AtC3H14 zinc finger, SHINE2, IFL1, and the AtMIR397a were shown to be very plausible candidates. However, this does not obscure the fact that the true candidates are possibly also outside the considered list, especially because QTL support intervals encompassed genes of still unknown function. Moreover, for application to grasses, the lack of secondary cambium in grasses and the role of p-hydroxycinnamic acid in grass cell walls might be taken into account. Many cell wall genes are specific to grasses, and orthologs of genes involved in secondary cambium assembly of dicotyledonous plants might have an originnal function in grasses or do not exist.
The search for candidate gene and allele CDS variation is a first step towards functional validation of the highlighted candidates underlying QTL. In addition to whole allele sequencing, it is worth investigating mutants of the considered candidates that have not yet been studied. Positional cloning, which is a long and tedious approach, will probably be inescapable for final validations. In any case, investigating QTL for cell wall degradability and their underlying determinants is an essential key approach when it comes to improving forage feeding value and saccharification yield in bio-ethanol production.
5. ACKNOWLEDGEMENTS
The authors thank Christine Camilleri and Matthieu Simon (INRA, Versailles, France, Genetics and Plant Breeding Laboratory, Arabidopsis thaliana Resource Centre) for providing floral stems of RIL. They thank David Sabourin for his contribution to the establishment of the Arabidopsis NIRS calibration equations. They also thank Christiane Minault, Dominique Denoue, and Pascal Vernoux for their technical assistance. Finally, the authors thank Corinne Melin for the management of the maize bibliographical database.
REFERENCES
- Argillier, O., Barrière, Y., Dardenne, P., Emile, J. and Hébert, Y. (1998) Genotypic variation for in vitro criteria and relationships with in vivo digestibility in forage maize hybrids. Plant Breeding, 117, 437-441. doi:10.1111/j.1439-0523.1998.tb01969.x
- Barrière, Y., Surault, F. and Emile, J.C. (2003) Genetic variation of silage maize ingestibility in dairy cattle. Animal Research, 52, 489-500. doi:10.1051/animres:2003042
- Carroll, A. and Somerville, C. (2009) Cellulosic biofuels. Annual Review of Plant Biology, 60, 165-182. doi:10.1146/annurev.arplant.043008.092125
- Harris, D. and DeBolt, S. (2010) Synthesis, regulation and utilization of lignocellulosic biomass. Plant Biotechnology Journal, 8, 244-262. doi:10.1111/j.1467-7652.2009.00481.x
- Li, X., Ximenes, E., Kim, Y., Slininger, M., Meilan, R., Ladisch, M. and Chapple, C. (2010) Lignin monomer composition affects Arabidopsis cell-wall degradability after liquid hot water pretreatment. Biotechnology for Biofuels, 3, 27. doi:10.1186/1754-6834-3-27
- Barriere, Y., Méchin, V., Lafarguette, F., Manicacci, D., Guillon, F., Wang, H., Lauressergues, D., Pichon, M., Bosio, D. and Tatout, C. (2009) Toward the discovery of maize cell wall genes involved in silage quality and capacity to biofuel production. Maydica, 54, 161-198.
- Grabber, J.H., Mertens, D.R., Kim, H., Funk, C., Lu, F.C. and Ralph, J. (2009) Cell wall fermentation kinetics are impacted more by lignin content and ferulate cross-linking than by lignin composition. Journal of the Science of Food and Agriculture, 89, 122-129. doi:10.1002/jsfa.3418
- Zhong, R.Q. and Ye, Z.H. (2007) Regulation of cell wall biosynthesis. Current Opinion in Plant Biology, 10, 564- 572. doi:10.1016/j.pbi.2007.09.001
- Zhong, R.Q. and Ye, Z.H. (2009) Transcriptional regulation of lignin biosynthesis. Plant Signaling and Behavior, 4, 1-7. doi:10.4161/psb.4.11.9875
- Goujon, T., Sibout, R., Eudes, A., MacKay, J. and Jouanin, L. (2003) Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiology and Biochemistry, 41, 677-687. doi:10.1016/S0981-9428(03)00095-0
- Ruel, K., Berrio-Sierra, J., Derikvand, M.M., Pollet, B., Thevenin, J., Lapierre, C., Jouanin, L. and Joseleau, J.P. (2009) Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytologist, 184, 99-113. doi:10.1111/j.1469-8137.2009.02951.x
- Berthet, S., Demont-Caulet, N., Pollet, B., Bidzinski, P., Cezard, L., Le Bris, P., Borrega, N., Herve, J., Blondet, E., Balzergue, S., Lapierre, C. and Jouanin, L. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell, 23, 1124-1137. doi:10.1105/tpc.110.082792
- Mir Derikvand, M., Sierra, J.B., Ruel, K., Pollet, B., Do, C.T., Thevenin, J., Buffard, D., Jouanin, L. and Lapierre, C. (2008) Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta, 227, 943-956. doi:10.1007/s00425-007-0669-x
- Ralph, J., Kim, H., Lu, F., Grabber, J.H., Leplé, J.C., Berrio-Sierra, J., Derikvand, M.M., Jouanin, L., Boerjan, W. and Lapierre, C. (2008) Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant Journal, 53, 368-379. doi:10.1111/j.1365-313X.2007.03345.x
- Carpita, N.C. and Mccann, M.C. (2008) Maize and sorghum: genetic resources for bioenergy grasses. Trends in Plant Science, 13, 415-420. doi:10.1016/j.tplants.2008.06.002
- Bosch, M., Mayer, C.D., Cookson, A. and Donnison, I.S. (2011) Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. Journal of Experimental Botany, 62, 3545-3561. doi:10.1093/jxb/err045
- Barriere, Y., Mechin, V., Denoue, D., Bauland, C. and Laborde, J. (2010) QTL for Yield, earliness, and cell wall quality traits in topcross experiments of the F838 × F286 early maize RIL progeny. Crop Science, 50, 1761-1772. doi:10.2135/cropsci2009.11.0671
- Sewell, M.M., Davis, M.F., Tuskan, G.A., Wheeler, N.C., Elam, C.C., Bassoni, D.L. and Neale, D.B. (2002) Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). II. Chemical wood properties. Theoretical and Applied Genetics, 104, 214-222. doi:10.1007/s001220100697
- Markussen, T., Fladung, M., Achere, V., Favre, J.M., Faivre-Rampant, P., Aragones, A., Perez, D.D., Harvengt, L., Espinel, S. and Ritter, E. (2003) Identification of QTLs controlling growth, chemical and physical wood property traits in Pinus pinaster (Ait.). Silvae Genetica, 52, 8-15.
- Pot, D., Rodrigues, J.C., Rozenberg, P., Chantre, G., Tibbits, J., Cahalan, C., Pichavant, F. and Plomion, C. (2006) QTLs and candidate genes for wood properties in maritime pine (Pinus pinaster Ait.). Tree Genetics and Genomes, 2, 10-24. doi:10.1007/s11295-005-0026-9
- Yin, T., Zhang, X., Gunter, L., Priya, R., Sykes, R., Davis, M., Wullschleger, S.D. and Tuskan, G.A. (2010) Differential detection of genetic loci underlying stem and root lignin content in Populus. Plos One, 5, 11. doi:10.1371/journal.pone.0014021
- Freeman, J.S., Whittock, S.P., Potts, B.M. and Vaillancourt, R.E. (2009) QTL influencing growth and wood properties in Eucalyptus globulus. Tree Genetics and Genomes, 5, 713-722. doi:10.1007/s11295-009-0222-0
- Thumma, B.R., Southerton, S.G., Bell, J.C., Owen, J.V., Henery, M.L. and Moran, G.F. (2010) Quantitative trait locus (QTL) analysis of wood quality traits in Eucalyptus nitens. Tree Genetics and Genomes, 6, 305-317. doi:10.1007/s11295-009-0250-9
- Gion, J.M., Carouche, A., Deweer, S., Bedon, F., Pichavant, F., Charpentier, J.P., Bailleres, H., Rozenberg, P., Carocha, V., Ognouabi, N., Verhaegen, D., Grima-Pettenati, J., Vigneron, P. and Plomion, C. (2011) Comprehensive genetic dissection of wood properties in a widelygrown tropical tree: Eucalyptus. BMC Genomics, 12, 301.
- Ranjan, P., Yin, T., Zhang, X., Kalluri, U.C., Yang, X., Jawdy, S. and Tuskan, G.A. (2010) Bioinformatics-based identification of candidate genes from QTLs associated with cell wall traits in Populus. Bioenergy Research, 3, 172-182. doi:10.1007/s12155-009-9060-z
- Thomas, J., Guillaumie, S., Verdu, C., Denoue, D., Pichon, M. and Barriere, Y. (2010) Cell wall phenylpropanoid-related gene expression in early maize recombinant inbred lines differing in parental alleles at a major lignin QTL position. Molecular Breeding, 25, 105-124. doi:10.1007/s11032-009-9311-x
- Mouille, G., Witucka-Wall, H., Bruyant, M.P., Loudet, O., Pelletier, S., Rihouey, C., Lerouxel, O., Lerouge, P., Hofte, H. and Pauly, M. (2006) Quantitative trait loci analysis of primary cell wall composition in Arabidopsis. Plant Physiology, 141, 1035-1044. doi:10.1104/pp.106.079384
- Barrière, Y., Laperche, A., Barrot, L., Aurel, G., Briand, M. and Jouanin, L. (2005) QTL analysis of lignification and cell wall digestibility in the Bay-0 × Shahdara RIL progeny of Arabidopsis thaliana as a model system for forage plant. Plant Science, 168, 1235-1245. doi:10.1016/j.plantsci.2005.01.001
- Simon, M., Loudet, O., Durand, S., Berard, A., Brunel, D., Sennesal, F.X., Durand-Tardif, M., Pelletier, G. and Camilleri, C. (2008) Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus singlenucleotide polymorphism markers. Genetics, 178, 2253- 2264. doi:10.1534/genetics.107.083899
- Loudet, O., Chaillou, S., Camilleri, C., Bouchez, D. and Daniel-Vedele, F. (2002) Bay-0 × Shahdara recombinant inbred line population: A powerful tool for the genetic dissection of complex traits in Arabidopsis. Theoretical and Applied Genetics, 104, 1173-1184. doi:10.1007/s00122-001-0825-9
- Goering, H.K. and Van Soest, P.J. (1970) Forage fiber analysis (Apparatus, reagents, procedures and some applications). US Agricultural Research Service, Washington DC, 33-61.
- Dence, C.W. and Lin, S.Y. (1992) The determination of lignin. In: Methods in lignin chemistry. Springer-Verlag, Berlin, 33-61. doi:10.1007/978-3-642-74065-7_3
- Hatfield, R.D., Jung, H.J.G., Ralph, J., Buxton, D.R. and Weimer, P.J. (1994) A comparison of the insoluble residues produced by the klason lignin and acid detergent lignin procedures. Journal of the Science of Food and Agriculture, 65, 51-58. doi:10.1002/jsfa.2740650109
- Hall, M.B., Lewis, B.A., VanSoest, P.J. and Chase, L.E. (1997) A simple method for estimation of neutral detergent-soluble fibre. Journal of the Science of Food and Agriculture, 74, 441-449. doi:10.1002/(SICI)1097-0010(199708)74:4<441::AID-JSFA813>3.0.CO;2-C
- Aufrère, J. and Michalet-Doreau, B. (1983) In vivo digestibility and prediction of digestibility of some byproducts. Proceedings of an EEC Seminar, Melle Gontrode, 26-29 September 1983, 25-33.
- Struik, P. (1983) Physiology of forage maize (Zea mays L.) in relation to its production and quality. Ph. Dissertation, Agricultural University, Wageningen, 1-252.
- Dolstra, O. and Medema, J.H. (1990) An effective screening method for genetic improvement of cell-wall digestibility in forage maize. Proceedings 15th Congress Maize and Sorghum Section of Eucarpia. Baden, 258- 270.
- Argillier, O., Barrière, Y. and Hébert, Y. (1995) Genetic variation and selection criterion for digestibility traits of forage maize. Euphytica, 82, 175-184. doi:10.1007/BF00027064
- Barrière, Y., Guillet, C., Goffner, D. and Pichon, M. (2003) Genetic variation and breeding strategies for improved cell wall digestibility in annual forage crops. A review. Animal Research, 52, 193-228. doi:10.1051/animres:2003018
- Lila, M. (1977) Influence of drying conditions on evaluation of soluble nitrogen and soluble carbohydrate content of forages—Consequences for trial harvesting. Annales de l’Amélioration des Plantes, 27, 465-475.
- Utz, H. and Melchinger, A. (1996) PLABQTL: A program for composite interval mapping of QTL. Journal of Agricultural Genomics, 2, 1-6.
- Haley, C.S. and Knott, S.A. (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity, 69, 315-324. doi:10.1038/hdy.1992.131
- Churchill, G.A. and Doerge, R.W. (1994) Empirical threshold values for quantitative trait mapping. Genetics, 138, 963-971.
- Melchinger, A.E., Utz, H.F. and Schön, C.C. (2004) QTL analyses of complex traits with cross validation, bootstrapping and other biometric methods. Euphytica, 137, 1-11. doi:10.1023/B:EUPH.0000040498.48379.68
- Schön, C., Utz, H., Groh, S., Truberg, B., Openshaw, S. and Melchinger, A. (2004) Quantitative trait locus mapping based on resampling in a vast maize testcross experiment and its relevance to quantitative genetics for complex traits. Genetics, 167, 485-498. doi:10.1534/genetics.167.1.485
- Kendall, M.G. and Stuart, A. (1961) The advanced theory of statistics. Inference and relationship. 3rd Edition, Griffin, London.
- Lander, E.S. and Botstein, D. (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics, 121, 185-199.
- Ohtani, M., Nishikubo, N., Xu, B., Yamaguchi, M., Mitsuda, N., Goue, N., Shi, F., Ohme-Takagi, M. and Demura, T. (2011) A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant Journal, 67, 499-512. doi:10.1111/j.1365-313X.2011.04614.x
- Rengel, D., Clemente, H.S., Servant, F., Ladouce, N., Paux, E., Wincker, P., Couloux, A., Sivadon, P. and Grima-Pettenati, J. (2009) A new genomic resource dedicated to wood formation in Eucalyptus. BMC Plant Biology, 9, 36.
- Obayashi, T., Hayashi, S., Saeki, M., Ohta, H. and Kinoshita, K. (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Research, 37, D987-D991. doi:10.1093/nar/gkn807
- Ko, J.H. and Han, K.H. (2004) Arabidopsis whole-transcriptome profiling defines the features of coordinated regulations that occur during secondary growth. Plant Molecular Biology, 55, 433-453. doi:10.1007/s11103-004-1051-z
- Minic, Z., Jamet, E., San-Clemente, H., Pelletier, S., Renou, J.P., Rihouey, C., Okinyo, D.P., Proux, C., Lerouge, P. and Jouanin, L. (2009) Transcriptomic analysis of Arabidopsis developing stems: A close-up on cell wall genes. BMC Plant Biology, 9, 6. doi:10.1186/1471-2229-9-6
- Wang, H., Avci, U., Nakashima, J., Hahn, M.G., Chen, F. and Dixon, R.A. (2010) Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proceedings of the National Academy of Sciences of the United States of America, 107, 22338-22343. doi:10.1073/pnas.1016436107
- Schneeberger, K., Ossowski, S., Ott, F., Klein, J.D., Wang, X., Lanz, C., Smith, L.M., Cao, J., Fitz, J., Warthmann, N., Henz, S.R., Huson, D.H. and Weigel, D. (2011) Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proceedings of the National Academy of Sciences of the United States of America, 108, 10249-10254. doi:10.1073/pnas.1107739108
- Cao, J., Schneerman, M.C., Ossowski, S., gunther, T., Bender, S., Fitz, J., Koenig, D., Lao, N.T., Stegle, O., lippert, C., wang.xi, Ott, F., Muller, J., Alonso-Blanco, C., Borgwardt, K., Schmid, K. and Weigel, D. (2011) Wholegenome sequencing of multiple Arabidopsis thaliana populations. Nature, 43, 956-963. doi:10.1038/ng.911
- Jones, L., Ennos, A.R. and Turner, S.R. (2001) Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant Journal, 26, 205-216. doi:10.1046/j.1365-313x.2001.01021.x
- Schöch, G., Goepfert, S., Morant, M., Hehn, A., Meyer, D., Ullmann, P. and Werck-Reichhart, D. (2001) CYP98A3 from Arabidopsis thaliana is a 3’-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. Journal of Biological Chemistry, 276, 36566-36574. doi:10.1074/jbc.M104047200
- Costa, M.A., Collins, R.E., Anterola, A.M., Cochrane, F.C., Davin, L.B. and Lewis, N.G. (2003) An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry, 64, 1097-1112. doi:10.1016/S0031-9422(03)00517-X
- Raes, J., Rohde, A., Christensen, J.H., Van de Peer, Y. and Boerjan, W. (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiology, 133, 1051-1071. doi:10.1104/pp.103.026484
- Sibout, R., Eudes, A., Pollet, B., Goujon, T., Mila, I., Granier, F., Seguin, A., Lapierre, C. and Jouanin, L. (2003) Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiology, 132, 848-860. doi:10.1104/pp.103.021048
- Sibout, R., Eudes, A., Mouille, G., Pollet, B., Lapierre, C., Jouanin, L. and Seguin, A. (2005) Cinnamyl alcohol dehydrogenase-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell, 17, 2059-2076. doi:10.1105/tpc.105.030767
- Eudes, A., Pollet, B., Sibout, R., Do, C.T., Seguin, A., Lapierre, C. and Jouanin, L. (2006) Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta, 225, 23-39. doi:10.1007/s00425-006-0326-9
- Bonawitz, N.D. and Chapple, C. (2010) The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annual Review of Genetics, 44, 337-363. doi:10.1146/annurev-genet-102209-163508
- Valerio, L., De Meyer, M., Penel, C. and Dunand, C. (2004) Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry, 65, 1331-1342. doi:10.1016/j.phytochem.2004.04.017
- Mccaig, B.C., Meagher, R.B. and Dean, J.F.D. (2005) Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta, 221, 619-636. doi:10.1007/s00425-004-1472-6
- Ehlting, J., Mattheus, N., Aeschliman, D.S., Li, E.Y., Hamberger, B., Cullis, I.F., Zhuang, J., Kaneda, M., Mansfield, S.D., Samuels, L., Ritland, K., Ellis, B.E., Bohlmann, J. and Douglas, C.J. (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant Journal, 42, 618-640. doi:10.1111/j.1365-313X.2005.02403.x
- Cai, X., Davis, E.J., Ballif, J., Liang, M., Bushman, E., Haroldsen, V., Torabinejad, J. and Wu, Y. (2006) Mutant identification and characterization of the laccase gene family in Arabidopsis. Journal of Experimental Botany, 57, 2563-2569. doi:10.1093/jxb/erl022
- Cosio, C. and Dunand, C. (2009) Specific functions of individual class III peroxidase genes. Journal of Experimental Botany, 60, 391-408. doi:10.1093/jxb/ern318
- Koizumi, K., Yokoyama, R. and Nishitani, K. (2009) Mechanical load induces upregulation of transcripts for a set of genes implicated in secondary wall formation in the supporting tissue of Arabidopsis thaliana. Journal of Plant Research, 122, 651-659. doi:10.1007/s10265-009-0251-7
- Tokunaga, N., Kaneta, T., Sato, S. and Sato, Y. (2009) Analysis of expression profiles of three peroxidase genes associated with lignification in Arabidopsis thaliana. Physiologia Plantarum, 136, 237-249. doi:10.1111/j.1399-3054.2009.01233.x
- Pedreira, J., Teresa Herrera, M., Zarra, I. and Revilla, G. (2011) The overexpression of AtPrx37, an apoplastic peroxidase, reduces growth in Arabidopsis. Physiologia Plantarum, 141, 177-187. doi:10.1111/j.1399-3054.2010.01427.x
- Fagerstedt, K.V., Kukkola, E.M., Koistinen, V.V., Takahashi, J. and Marjamaa, K. (2010) Cell wall lignin is polymerised by class III secretable plant peroxidases in Norway spruce. Journal of Integrative Plant Biology, 52, 186-194. doi:10.1111/j.1744-7909.2010.00928.x
- Lim, E.K., Jackson, R.G. and Bowles, D.J. (2005) Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Letters, 579, 2802-2806. doi:10.1016/j.febslet.2005.04.016
- Escamilla-Trevino, L.L., Chen, W., Card, M.L., Shih, M.C., Cheng, C.L. and Poulton, J.E. (2006) Arabidopsis thaliana beta-glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides. Phytochemistry, 67, 1651- 1660. doi:10.1016/j.phytochem.2006.05.022
- Lim, E., Jackson, R. and Bowles, D. (2005) Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Letters, 579, 2802-2806. doi:10.1016/j.febslet.2005.04.016
- Lanot, A., Hodge, D., Jackson, R., George, G., Elias, L., Lim, E., Vaistij, F. and Bowles, D. (2006) The glucosyltransferase UGT72E2 is responsible for monolignol 4- O-glucoside production in Arabidopsis thaliana. Plant Journal, 48, 286-295. doi:10.1111/j.1365-313X.2006.02872.x
- Sanchez-Fernandez, R., Davies, T.G.E., Coleman, J.O.D. and Rea, P.A. (2001) The Arabidopsis thaliana ABC protein superfamily, a complete inventory. Journal of Biological Chemistry, 276, 30231-30244. doi:10.1074/jbc.M103104200
- Samuels, A.L., Rensing, K.H., Douglas, C.J., Mansfield, S.D., Dharmawardhana, D.P. and Ellis, B.E. (2002) Cellular machinery of wood production: Differentiation of secondary xylem in Pinus contorta var. latifolia. Planta, 216, 72-82. doi:10.1007/s00425-002-0884-4
- Miao, Y.C. and Liu, C.J. (2010) ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proceedings of the National Academy of Sciences of the United States of America, 107, 22728-22733. doi:10.1073/pnas.1007747108
- Kaneda, M., Schuetz, M., Lin, B., Chanis, C., Hamberger, B., Western, T., Ehlting, J. and Samuels, A. (2011) ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. Journal of Experimental Botany, 62, 2063-2077. doi:10.1093/jxb/erq416
- Yamaguchi, M., Kubo, M., Fukuda, H. and Demura, T. (2008) Vascular-related nac-domain7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant Journal, 55, 652-664. doi:10.1111/j.1365-313X.2008.03533.x
- Zhong, R.Q., Lee, C., McCarty, R.L. and Ye, Z.H. (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell, 20, 2763-2782. doi:10.1105/tpc.108.061325
- Sonbol, F.M., Fornale, S., Capellades, M., Encina, A., Tourino, S., Torres, J.L., Rovira, P., Ruel, K., Puigdomenech, P., Rigau, J. and Caparros-Ruiz, D. (2009) The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Molecular Biology, 70, 283-296. doi:10.1007/s11103-009-9473-2
- Zhou, J., Lee, C., Zhong, R.Q. and Ye, Z.H. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell, 21, 248-266. doi:10.1105/tpc.108.063321
- Zhong, R., Lee, C. and Ye, Z.H. (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Molecular Plant, 3, 1087-1103. doi:10.1093/mp/ssq062
- Yamaguchi, M., Ohtani, M., Mitsuda, N., Kubo, M., Ohme-Takagi, M., Fukuda, H. and Demura, T. (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell, 22, 1249-1263. doi:10.1105/tpc.108.064048
- Bonke, M., Thitamadee, S., Mahonen, A.P., Hauser, M.T. and Helariutta, Y. (2003) APL regulates vascular tissue identity in Arabidopsis. Nature, 426, 181-186. doi:10.1038/nature02100
- Zhao, C.S., Avci, U., Grant, E.H., Haigler, C.H. and Beers, E.P. (2008) XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant Journal, 53, 425-436. doi:10.1111/j.1365-313X.2007.03350.x
- Bhargava, A., Mansfield, S.D., Hall, H.C., Douglas, C.J. and Ellis, B.E. (2010) MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiology, 154, 1428-1438. doi:10.1104/pp.110.162735
- Grant, E.H., Fujino, T., Beers, E.P. and Brunner, A.M. (2010) Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta, 232, 337-352. doi:10.1007/s00425-010-1181-2
- Kawaoka, A. and Ebinuma, H. (2001) Transcriptional control of lignin biosynthesis by tobacco LIM protein. Phytochemistry, 57, 1149-1157. doi:10.1016/S0031-9422(01)00054-1
- Matthews, J., Bhati, M., Lehtomaki, E., Mansfield, R., Cubeddu, L. and MacKay, J. (2009) It takes two to tango: the structure and function of LIM, RING, PHD and MYND domains. Current Pharmaceutical Design, 15, 3681-3696. doi:10.2174/138161209789271861
- Wu, K.L., Guo, Z.J., Wang, H.H. and Li, J. (2005) The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research, 12, 9-26. doi:10.2174/138161209789271861
- Rushton, P.J., Somssich, I.E., Ringler, P. and Shen, Q.J. (2010) WRKY transcription factors. Trends in Plant Science, 15, 247-258. doi:10.1016/j.tplants.2010.02.006
- Ko, J.H., Kim, W.C. and Han, K.H. (2009) Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant Journal, 60, 649-665. doi:10.1111/j.1365-313X.2009.03989.x
- Jakoby, M., Weisshaar, B., Droge-Laser, W., VicenteCarbajosa, J., Tiedemann, J., Kroj, T. and Parcy, F. (2002) bZIP transcription factors in Arabidopsis. Trends in Plant Science, 7, 106-111. doi:10.1016/S1360-1385(01)02223-3
- Husbands, A., Bell, E.M., Shuai, B., Smith, H.M. and Springer, P.S. (2007) Lateral organ boundaries defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Research, 35, 6663-6671. doi:10.1093/nar/gkm775
- Soyano, T., Thitamadee, S., Machida, Y. and Chua, N.H. (2008) Asymmetric leaves2-like19/lateral organ boundaries domain30 and Asl20/Lbd18 regulate tracheary element differentiation in Arabidopsis. Plant Cell, 20, 3359- 3373. doi:10.1105/tpc.108.061796
- Yamaguchi, M., Mitsuda, N., Ohtani, M., Ohme-Takagi, M., Kato, K. and Demura, T. (2011) Vascular-related NACdomain 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant Journal, 66, 579-590. doi:10.1111/j.1365-313X.2011.04514.x
- Yordanov, Y.S., Regan, S. and Busov, V. (2010) Members of the lateral organ boundaries domain transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell, 22, 3662-3677. doi:10.1105/tpc.110.078634
- Heim, M.A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B. and Bailey, P.C. (2003) The basic helix-loophelix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Molecular Biology and Evolution, 20, 735-747. doi:10.1093/molbev/msg088
- Parker, G., Schofield, R., Sundberg, B. and Turner, S. (2003) Isolation of COV1, a gene involved in the regulation of vascular patterning in the stem of Arabidopsis. Development, 130, 2139-2148. doi:10.1242/dev.00441
- Pineau, C., Freydier, A., Ranocha, P., Jauneau, A., Turner, S., Lemonnier, G., Renou, J.P., Tarkowski, P., Sandberg, G., Jouanin, L., Sundberg, B., Boudet, A.M., Goffner, D. and Pichon, M. (2005) HCA: An Arabidopsis mutant exhibiting unusual cambial activity and altered vascular patterning. Plant Journal, 44, 271-289. doi:10.1111/j.1365-313X.2005.02526.x
- Guo, Y., Qin, G., Gu, H. and Qu, L.J. (2009) Dof5.6/ HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell, 21, 3518-3534. doi:10.1105/tpc.108.064139
- Rogers, L.A., Dubos, C., Surman, C., Willment, J., Cullis, I.F., Mansfield, S.D. and Campbell, M.M. (2005) Comparison of lignin deposition in three ectopic lignification mutants. New Phytologist, 168, 123-140. doi:10.1111/j.1469-8137.2005.01496.x
- DiLaurenzio, L., WysockaDiller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A. and Benfey, P.N. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell, 86, 423-433. doi:10.1016/S0092-8674(00)80115-4
- Lee, H., Kim, B., Song, S.K., Heo, J.O., Yu, N.I., Lee, S.A., Kim, M., Kim, D.G., Sohn, S.O., Lim, C.E., Chang, K.S., Lee, M.M. and Lim, J. (2008) Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Molecular Biology, 67, 659-670. doi:10.1007/s11103-008-9345-1
- Carlsbecker, A., Lee, J.Y., Roberts, C.J., Dettmer, J., Lehesranta, S., Zhou, J., Lindgren, O., Moreno-Risueno, M.A., Vaten, A., Thitamadee, S., Campilho, A., Sebastian, J., Bowman, J.L., Helariutta, Y. and Benfey, P.N. (2010) Cell signalling by microRNA165/6 directs gene dosedependent root cell fate. Nature, 465, 316-321. doi:10.1038/nature08977
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I. and Morelli, G. (2001) The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiology, 126, 643-655. doi:10.1104/pp.126.2.643
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J. and Barton, M.K. (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature, 411, 709-713. doi:10.1038/35079635
- Talbert, P.B., Adler, H.T., Parks, D.W. and Comai, L. (1995) The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development, 121, 2723-2735.
- Ratcliffe, O.J., Riechmann, J.L. and Zhang, J.Z. (2000) Interfascicular fiberless1 is the same gene as revoluta. Plant Cell, 12, 315-317.
- Green, K.A., Prigge, M.J., Katzman, R.B. and Clark, S.E. (2005) CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell, 17, 691-704. doi:10.1105/tpc.104.026179
- Ilegems, M., Douet, V., Meylan-Bettex, M., Uyttewaal, M., Brand, L., Bowman, J.L. and Stieger, P.A. (2010) Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development, 137, 975-984. doi:10.1242/dev.047662
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A. and Timmermans, M.C.P. (2004) MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature, 428, 84-88. doi:10.1038/nature02363
- Ko, J.H., Prassinos, C. and Han, K.H. (2006) Developmental and seasonal expression of PtaHB1, a Populus gene encoding a class IIIHD-Zip protein, is closely associated with secondary growth and inversely correlated with the level of microRNA (miR166). New Phytologist, 169, 469-478. doi:10.1111/j.1469-8137.2005.01623.x
- Williams, L., Grigg, S.P., Xie, M.T., Christensen, S. and Fletcher, J.C. (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development, 132, 3657-3668. doi:10.1242/dev.01942
- Liljegren, S.J., Ditta, G.S., Eshed, H.Y., Savidge, B., Bowman, J.L. and Yanofsky, M.F. (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature, 404, 766-770. doi:10.1038/35008089
- Guillaumie, S., Pichon, M., Martinant, J.P., Bosio, M., Goffner, D. and Barrière, Y. (2007) Differential expression of phenylpropanoid and related genes in brownmidrib bm1, bm2, bm3, and bm4 young near-isogenic maize plants. Planta, 226, 235-250. doi:10.1007/s00425-006-0468-9
- Guillaumie, S., Goffner, D., Barbier, O., Martinant, J.P., Pichon, M. and Barrière, Y. (2008) Expression of cell wall related genes in basal and ear internodes of silking brown-midrib-3, caffeic acid O-methyltransferase (COMT) down-regulated, and normal maize plants. BMC Plant Biology, 8, 71. doi:10.1186/1471-2229-8-71
- Pesquet, E., Korolev, A.V., Calder, G. and Lloyd, C.W. (2010) The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Current Biology, 20, 744-749. doi:10.1016/j.cub.2010.02.057
- Rengel, D., San-Clemente, H., Servant, F., Ladouce, N., Paux, E., Wincker, P., Couloux, A., Sivadon, P. and GrimaPettenati, J. (2009) A new genomic resource dedicated to wood formation in Eucalyptus. BMC Plant Biology, 27, 9-36.
- Foucart, C., Jauneau, A., Gion, J.M., Amelot, N., Martinez, Y., Panegos, P., Grima-Pettenati, J. and Sivadon, P. (2009) Overexpression of EgROP1, a Eucalyptus vascular-expressed Rac-like small GTPase, affects secondary xylem formation in Arabidopsis thaliana. New Phytologist, 183, 1014-1029. doi:10.1111/j.1469-8137.2009.02910.x
- Gu, Y., Wang, Z. and Yang, Z. (2004) ROP/RAC GTPase: An old new master regulator for plant signaling. Current Opinion in Plant Biology, 7, 527-536. doi:10.1016/j.pbi.2004.07.006
- Nibau, C., Wu, H.M. and Cheung, A.Y. (2006) RAC/ROP GTPases: “Hubs” for signal integration and diversification in plants. Trends in Plant Science, 11, 309-315. doi:10.1016/j.tplants.2006.04.003
- Kwon, S.I., Cho, H.J., Jung, J.H., Yoshimoto, K., Shirasu, K. and Park, O.K. (2010) The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant Journal, 64, 151-164.
- Ambavaram, M.M., Krishnan, A., Trijatmiko, K.R. and Pereira, A. (2011) Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiology, 155, 916-931. doi:10.1104/pp.110.168641
- Kabbage, M. and Dickman, M. (2008) The BAG proteins: A ubiquitous family of chaperone regulators. Cellular and Molecular Life Sciences, 65, 1390-1402. doi:10.1007/s00018-008-7535-2
- Zhao, C.S., Johnson, B.J., Kositsup, B. and Beers, E.P. (2000) Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiology, 123, 1185-1196. doi:10.1104/pp.123.3.1185
- Zhong, R.Q. and Ye, Z.H. (2004) amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant and Cell Physiology, 45, 369-385. doi:10.1093/pcp/pch051
- Kim, J., Jung, J.H., Reyes, J.L., Kim, Y.S., Kim, S.Y., Chung, K.S., Kim, J.A., Lee, M., Lee, Y., Kim, V.N., Chua, N.H. and Park, C.M. (2005) microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant Journal, 42, 84-94. doi:10.1111/j.1365-313X.2005.02354.x
- Fahlgren, N., Howell, M.D., Kasschau, K.D., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., Law, T.F., Grant, S.R., Dangl, J.L. and Carrington, J.C. (2007) Highthroughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of miRNA genes. Plos One, 2, 219. doi:10.1371/journal.pone.0000219
- Yang, T.W., Xue, L.G. and An, L.Z. (2007) Functional diversity of miRNA in plants. Plant Science, 172, 423- 432. doi:10.1016/j.plantsci.2006.10.009
- Abdel-Ghany, S.E. and Pilon, M. (2008) MicroRNAmediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. Journal of Biological Chemistry, 283, 15932- 15945. doi:10.1074/jbc.M801406200
- Jung, H., Mertens, D. and Payne, A. (1997) Correlation of acid detergent lignin and Klason lignin with digestibility of forage dry matter and neutral detergent fiber. Journal of Dairy Science, 80, 1622-1628. doi:10.3168/jds.S0022-0302(97)76093-4
- Barrière, Y., Thomas, J. and Denoue, D. (2008) QTL mapping for lignin content, lignin monomeric composition, p-hydroxycinnamate content, and cell wall digestibility in the maize recombinant inbred line progeny F838 × F286. Plant Science, 175, 585-595. doi:10.1016/j.plantsci.2008.06.009
- Rowe, H.C. and Kliebenstein, D.J. (2008) Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics, 180, 2237-2250. doi:10.1534/genetics.108.091439
- Jubault, M., Lariagon, C., Simon, M., Delourme, R. and Manzanares-Dauleux, M.J. (2008) Identification of quantitative trait loci controlling partial clubroot resistance in new mapping populations of Arabidopsis thaliana. Theoretical and Applied Genetics, 117, 191-202. doi:10.1007/s00122-008-0765-8
- Chabannes, M., Barakate, A., Lapierre, C., Marita, J.M., Ralph, J., Pean, M., Danoun, S., Halpin, C., Grima-Pettenati, J. and Boudet, A.M. (2001) Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant Journal, 28, 257-270. doi:10.1046/j.1365-313X.2001.01140.x
- Thumma, B.R., Nolan, M.R., Evans, R. and Moran, G.F. (2005) Polymorphisms in cinnamoyl CoA reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics, 171, 1257-1265. doi:10.1534/genetics.105.042028
- Leplé, J.C., Dauwe, R., Morreel, K., Storme, V., Lapierre, C., Pollet, B., Naumann, A., Kang, K.Y., Kim, H., Ruel, K., Lefebvre, A., Joseleau, J.P., Grima-Pettenati, J., De Rycke, R., ndersson-Gunneras, S., Erban, A., Fehrle, I., Petit-Conil, M., Kopka, J., Polle, A., Messens, E., Sundberg, B., Mansfield, S.D., Ralph, J., Pilate, G. and Boerjan, W. (2007) Downregulation of cinnamoyl-coenzyme a reductase in poplar: Multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell, 19, 3669-3691. doi:10.1105/tpc.107.054148
- Wadenbäck, J., von Arnold, S., Egertsdotter, U., Walter, M.H., Grima-Pettenati, J., Goffner, D., Gellerstedt, G., Gullion, T. and Clapham, D. (2008) Lignin biosynthesis in transgenic Norway spruce plants harboring an antisense construct for cinnamoyl CoA reductase (CCR). Transgenic Research, 17, 379-392. doi:10.1007/s11248-007-9113-z
- Tamasloukht, B., Lam, M.S.-J.W.Q., Martinez, Y., Tozo, K., Barbier, O., Jourda, C., Jauneau, A., Borderies, G., Balzergue, S., Renou, J.P., Huguet, S., Martinant, J.P., Tatout, C., Lapierre, C., Barriere, Y., Goffner, D. and Pichon, M. (2011) Characterization of a cinnamoyl-CoA reductase 1 (CCR1) mutant in maize: Effects on lignification, fibre development and global gene expression. Journal of Experimental Botany, 62, 3837-3848. doi:10.1093/jxb/err077
- Zhao, Q., Wang, H., Yin, Y., Xu, Y., Chen, F. and Dixon, R.A. (2010) Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch. Proceedings of the National Academy of Sciences of the United States of America, 107, 14496-14501. doi:10.1073/pnas.1009170107
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R. and Dean, C. (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science, 290, 344-347. doi:10.1126/science.290.5490.344
- Gazzani, S., Gendall, A.R., Lister, C. and Dean, C. (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiology, 132, 1107-1114. doi:10.1104/pp.103.021212
- Shindo, C., Aranzana, M.J., Lister, C., Baxter, C., Nicholls, C., Nordborg, M. and Dean, C. (2005) Role of frigida and flowering Locus C in determining variation in flowering time of Arabidopsis. Plant Physiology, 138, 1163-1173. doi:10.1104/pp.105.061309
- Goicoechea, M., Lacombe, E., Legay, S., Mihaljevic, S., Rech, P., Jauneau, A., Lapierre, C., Pollet, B., Verhaegen, D., Chaubet-Gigot, N. and Grima-Pettenati, J. (2005) EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant Journal, 43, 553-567. doi:10.1111/j.1365-313X.2005.02480.x
- Stracke, R., Werber, M. and Weisshaar, B. (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology, 4, 447-456. doi:10.1016/S1369-5266(00)00199-0

