Open Journal of Genetics
Vol.1 No.3(2011), Article ID:8888,4 pages DOI:10.4236/ojgen.2011.13007
No correlation between X chromosome inactivation pattern and autistic spectrum disorders in an Italian cohort of patients
1Institute of Medical Genetics, Catholic University School of Medicine, Rome, Italy;
2Institute of Clinical Pediatrics, Catholic University School of Medicine, Rome, Italy.
Email: fgurrieri@rm.unicatt.it
Received 15 September 2011; revised 28 October 2011; accepted 13 November 2011.
Keywords: Autism; X Chromosome Inactivation
ABSTRACT
Autistic spectrum disorders (ASD) occur more frequently in males, suggesting a major pathogenic role for genes located on the X-chromosome. The analysis of X chromosome inactivation (XCI) pattern may help to identify XCI skewing in those families in which such genes are involved, even without identifying the specific genetic mutation. In order to identify such families, we determined the XCI pattern in 40 females with ASD and 58 mothers of children with ASD, as well as in 80 matched control females. The X inactivation assay was carried out on genomic DNA extracted from peripheral blood. XCI was calculated for informative heterozygous individuals as the ratio of the peak area of two alleles of the highly polymorphic CAG repeat of the androgen receptor (AR) gene (Xq11-12). Our results indicate that there is no difference in XCI pattern both in ASD females and in the mothers of ASD patients when compared with the appropriate controls. These findings suggest that the contribution of X-linked genes to the etiology of ASD is still likely but it is not supported by X-inactivation patterns on peripheral blood cells.
1. INTRODUCTION
Autistic spectrum disorders (ASD) are a group of childhood neurodevelopmental disorders characterized by difficulties in socialization and communication and stereotypic behaviors. Although the role of genetic factors in the etiology of ASD is under intense scrutiny [1-3], little is known concerning the relationship of genetic, epigenetic and environmental factors with respect to the core clinical features and the underlying neuropathological substrate. The prevalence of ASD is about 0.6:100, with an overall excess of males with autism in a proportion of about 4:1 in general and of 9:1 in the cases of Asperger syndrome [4], suggesting a major involvement of genes on the X chromosome. Mutations in two X-linked genes encoding neuroligins, NLGN3 and NLGN4 respectively, have been identified in males with ASD, including Asperger syndrome [5]. In addition, ASD is frequently associated with two X-linked conditions, i.e. fragile X and Rett syndrome [6,7]. X chromosome inactivation (XCI) occurs early in embryonic development of somatic cells in human females to achieve gene dosage compensation with males [8]. Therefore, one of the two X chromosomes is inactivated in each female cell at random which then results in an equal number of active X chromosome genes in both male and female cells. The X inactivation is a complex process and requires three main steps: initiation, spreading, and maintenance [9,10]. Skewed XCI is a rare event in the normal female population [11]. In contrast, an extremely skewed XCI can be observed in heterozygous females carrying gene mutations involved in X-linked conditions, such as X-linked intellectual disability, Barth syndrome and X-linked sideroblastic anemia [12-14].
In ASD, Talebizadeh et al. 2005 reported an excess of XCI skewing in affected girls (33%, 10/30) compared to controls (11%, 4/35), suggesting a major contribution of X-linked genes to ASD in this cohort. A subsequent study investigated the XCI pattern on a larger cohort, including affected females and mothers of ASD children [15], but failed to identify an altered X inactivation pattern, with respect to a control sample. Since contrasting results are shown in these reports, we decided to investtigate the XCI pattern in an Italian cohort of 40 autistic females and in a sample of 58 mothers of children with ASD.
2. METHODS
A total of 98 females, with the same ethnic background, were analyzed in our study, consisting of 40 females with ASD and 58 mothers of children with ASD. All the patients present intellectual disabilities and classical autistic stereotypies, but not other clinical features like seizures or hypotonia. DNA from 80 females, without a history of autism or mental retardation was used as control. All were of the same ethnic background as the ASD group. Of these, 40 were DNA from healthy girls of the same age as the ASD girls, and 40 were DNA from unaffected women with an average age of 45 years, matched with the age of the ASD mothers. The diagnosis of autism was established in the affected females with the use of the Autism Diagnostic Interview-Revised (ADI-R) [16] and clinical evaluation. Chromosome analysis, as well as molecular tests for fragile X testing and Rett syndrome, were reported normal. Informed, written consent to this study was obtained from all participating subjects. All the experiments have been performed with the approval of the Catholic University of Rome ethic committee.
X Inactivation Analysis
The X inactivation assay was carried out on genomic DNA extracted from peripheral blood. XCI was calculated for informative heterozygous individuals as the ratio of the peak area of two alleles of the highly polymorphic CAG repeat of the androgen receptor (AR) gene (Xq11-12) after digestion with the methylation sensitive enzyme HpaII, corrected with the ratio of the peak area of the two alleles before digestion for preferential amplification of one of the alleles [11].
Briefly, 500 ng of genomic DNA was digested with 2U DdeI plus 10 U HpaII, and with 2U DdeI alone at 37˚C overnight. Both mixtures were PCR-amplified with forward primer 50-TCCAGA ATC TGT TCC AGA GCG TGC-30, fluorescently labelled with 6-FAM (6-carboxyfluorescein), and reverse primer 50-GCT GTG AAG GTT GCT GTT CCT CAT-30. The resulting PCR products were run on ABI 3130 Genetic Analyzer DNA sequencer and the peak area was measured by Gene Mapper v4 software (Applied Biosystems, Foster City, CA).
The range of XCI ratio can vary from 50:50 (random XCI) to 100:0 (completely skewed XCI). One female sample with known XCI ratio (95:5) and one male sample were used as controls in each batch of samples to verify the complete digestion and replication. The frequency of XCI skewing was analyzed with Student test.
3. RESULTS
35 of the 40 females with ASD and 50 of the 58 mothers of affected children were informative for the AR gene polymorphism. Likewise, 37 of the 40 unaffected women and 36 of the 40 unaffected girls, used as controls, were informative. In the mothers group, ASD females group, and respective controls, the subjects were classified into three subgroups representing random, moderately and highly skewed X inactivation pattern (see Figures 1-2). The mothers group was further divided in female mothers and males mothers. A highly skewed XCI was observed in 4% of mothers of ASD males, in 7% of mothers of ASD females and 8% of controls (Figure 1). This difference did not reach statistical significance.
Again, we observed no significant difference between ASD females (6%) and the young females control group (9%) with a highly skewed XCI (see Figures 2-3).
4. DISCUSSION
Our results indicate that there is no difference in XCI pattern both in ASD females and in the mothers of ASD patients when compared with the appropriate controls. Even though our analysis is based on a small sample, the results are in agreement with those obtained by Gong et al. [15] on a large cohort of ASD individuals and controls. None of two previous studies [15,17] analyzed separately ASD-female’s mothers and ASD-male’s mothers: if X-linked genes were involved, a more consistent XCI skewing should have been observed in the latter group. However, this was not the case in our study as we
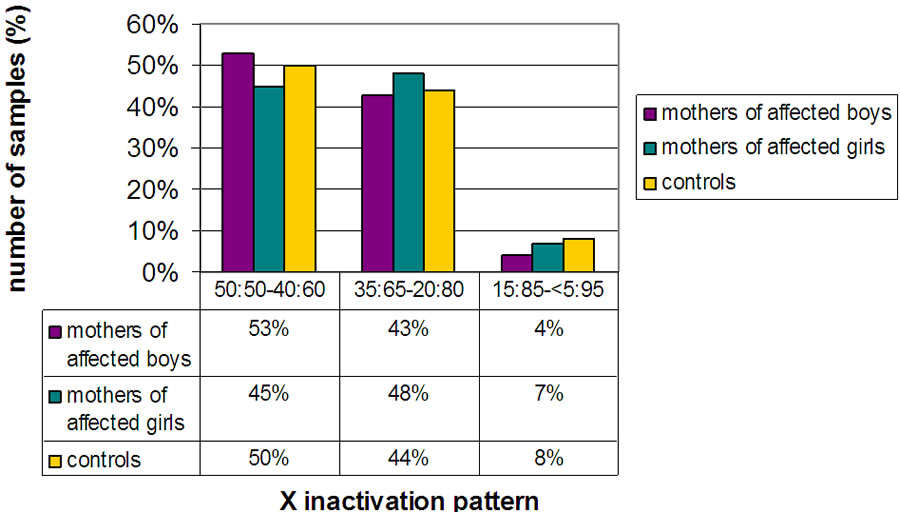
Figure 1. Histogram showing the distribution of ASD mothers and controls in three subgroups including non skewed (50:50% - 60:40%), mildly skewed (35:65% - 20:80%) and highly skewed (15:85% - 5:95%).
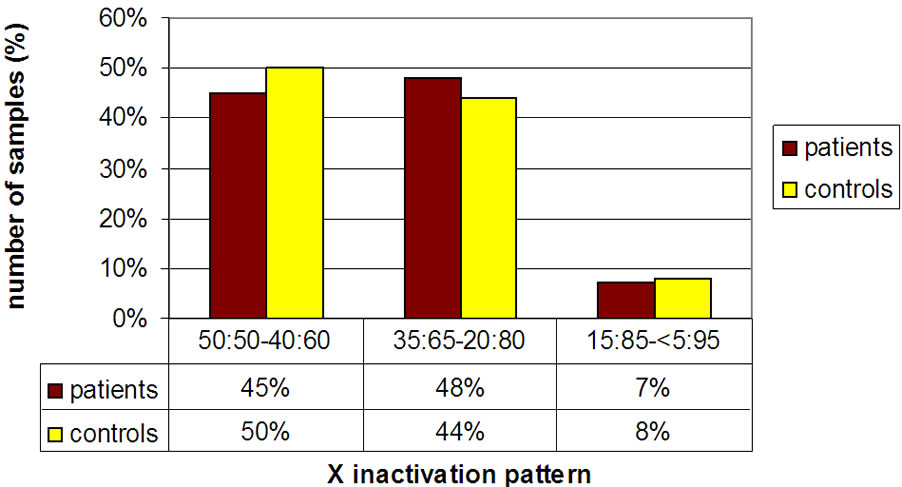
Figure 2. Histogram showing the distribution of affected ASD females and controls in three subgroups including non skewed (50:50% - 60:40%), mildly skewed (35:65% - 20:80%) and highly skewed (15:85% - 5:95%).
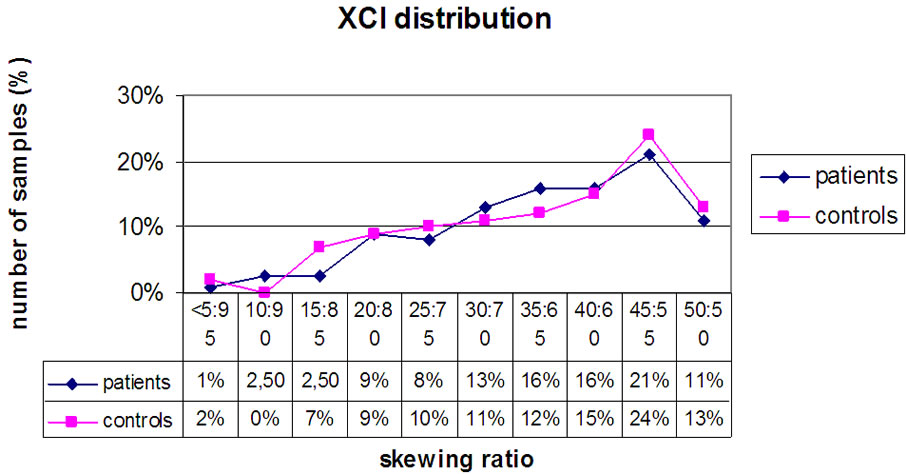
Figure 3. Diagram showing the distribution of affected ASD females (diamonds) and controls (squares) with respect to the XCI pattern.
observed a similar distribution of XCI in the two groups. Therefore, it is likely that the excess of ASD males is related to X-linked genes, whose function may be altered by some unknown factors. For instance, the male offspring inherits the maternal X chromosome, which is, because of meiotic crossing over, a composite of regions derived from the mother’s nonmethylated X chromosome and of others from the methylated X chromosome. Jones et al. [18] hypothesize that the male’s demethylating capacity may be ineffective if the male inherits a predominantly methylated X chromosome. In this case, uncomplete erasing of methylation in early postmeiotic cells might happen, causing a mosaic state in which a number of crucial genes remain methylated and therefore inactive in target cells. In any case, it should be noted that, since we could not analyse the XCI pattern in the target tissue and at different loci on the X-chromosome, we could not rule out that skewing may occur for instance in the brain or at definite loci on the X chromosome.
Besides, it has been proposed that the paternal X chromosome, inherited only by females, preserves them from ASD, explaining why females are less frequently affected than males (Skuse). If this were the case, ASD females should have a preferential inactivation of the paternal X chromosome. In our study, only three ASD females had a 10:90 skewing pattern, but we could not determine the parental origin of the inactive chromosome for lack of informativeness. The mothers of ASD patients with NLGN3 or MECP2 mutations show highly skewed XCI, suggesting that ascertainment of XCI could reveal families with X-linked mutations [15]. Other ASD causes not related to X chromosome gene mutations /alterations seems to not have skewed XCI. Nagarajan et al. 2008 [16] described an aberrant methylation status into the MECP2 promoter in male autism brain DNA. Besides, autistic female brain DNA showed evidence for aberrant MECP2 promoter methylation but no skewing in XCI. These data suggests the presence of locus-specific methylation changes rather than global chromosome methylation modifications.
Possible epigenetic mechanisms cannot be excluded to explain the major prevalence in males. Further studies are needed to elucidate the pathogenesis of ASD.
REFERENCES
- Abrahams, B.S. and Geschwind, D.H. (2008) Advances in autism genetics: On the threshold of a new neurobiology. Nature, Reviews, Genetics, 9, 341-355. doi:10.1038/nrg2346
- Morrow, E.M, Yoo, S.Y., Flavell, S.W., Kim,T.K., Lin, Y., Hill, R.S., et al. (2008). Identifying autism loci and genes by tracing recent shared ancestry. Science, 321, 218-223. doi:10.1126/science.1157657
- Sebat, J., Lakshmi, B., Malhotra, D., Troge, J., LeseMartin, C., Walsh, T., et al., (2007) Strong association of de novo copy number mutations with autism. Science, 316, 445-449. doi:10.1126/science.1138659
- Volkmar, F.R., Szatmari, P. and Sparrow, S.S. (1993) Sex differences in pervasive developmental disorders. Journal of Autism Developmental Disorders, 23, 579-591. doi:10.1007/BF01046103
- Jamain, S., Quach, H., Betancur, C., Råstam, M., Colineaux, C., Gillberg, I.C. et al. (2003) Mutations of the X-linked genes encoding neuroligins NLGN3and NLGN4 are associated with autism. Nature Genetics, 34, 27-29. doi:10.1038/ng1136
- Carney, R.M., Wolpert, C.M., Ravan, S.A. Shahbazian, M., Ashley-Koch, A., Cuccaro, M.L.M., et al., (2003) Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatric Neurology, 28, 205-211. doi:10.1016/S0887-8994(02)00624-0
- Reddy, K.S. (2005) Cytogenetic abnormalities and fragile-X Syndrome in autism spectrum disorder. BMC Medical Genetics, 6, 3. doi:10.1186/1471-2350-6-3
- Lyon, M.F. (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature, 190, 372-373. doi:10.1038/190372a0
- Penny, G.D., Kay, G.F., Sheardown, S.A., Rastan, S., Brockdorff, N. (1996) Requirement for Xist in X chromosome inactivation. Nature, 379, 131-137. doi:10.1038/379131a0
- Willard, H.F. (1995) The sex chromosome and X-chromosome inactivation. In: Scriver, C.R., Beaudet, A.L., Sly, W.S. and Valle, D., Eds., The Metabolic and Molecular Bases of Inherited Disease, McGraw-Hill, New York, 717-737.
- Amos-Landgraf, J.M., Cottle, A., Plenge, R.M., Friez, M., Schwartz, C.E., Longshore, J. and Willard, H.F. (2006) X chromosome-inactivation patterns of 1005 phenotypically unaffected females. American Journal of Human Genetics, 79, 493-499. doi:10.1086/507565
- Orstavik, K.H., Orstavik, R.E., Naumova, A.K., D’Adamo, P., Gedeon, A., Bolhuis, P.A., et al. (1998) X chromosome inactivation in carriers of Barth syndrome. American Journal of Human Genetics, 63, 1457-1463. doi:10.1086/302095
- Cazzola, M., May, A., Bergamaschi, G., Cerani, P., Rosti, V. and Bishop, D.F. (2000) Familial-skewed X-chromosome inactivation as a predisposing factor for late-onset X-linked sideroblastic anemia in carrier females. Blood, 96, 4363-4365.
- Plenge, R.M., Stevenson, R.A., Lubs, H.A., Schwartz, C.E. and Willard, H.F. (2002) Skewed X-chromosome inactivation is a common feature of X-linked mental retardation disorders. American Journal of Human Genetics, 71, 168-173. doi:10.1086/341123
- Gong, X., Bacchelli, E., Blasi, F., Toma, C., Betancur, C., Chaste, P., et al. (2008) Analysis of X chromosome inactivation in autism spectrum disorders. America Journal of medical Gentics, 147B, 830-835.
- Nagarajan, R.P., Patzel, K.A., Martin, M., Yasui, D.H., Swanberg, S.E., Hertz-Picciotto, I., et al. (2008) MECP2 promoter, methylation and X chromosome inactivation in autism. Autism Research, 1, 169-178. doi:10.1002/aur.24
- Talebizadeh, Z., Bittel, D.C., Veatch, O.J., Kibiryeva, N., Butler, M.G. (2005) Brief report: Non-random X chromosome inactivation in females with autism. Journal of Autism Developmental Disorders, 35, 675-681. doi:10.1007/s10803-005-0011-z
- Jones J.R., Skinner C., Friez M.J., Schwartz, C.E. and Stevenson, R.E. (2008) Hypothesis: Dysregulation of methylation of brain-expressed genes on the X chromosome and autism spectrum disorders. American Journal of Medical Genetics Part A, 146A, 2213-2220. doi:10.1002/ajmg.a.32396

