Advances in Biological Chemistry
Vol. 2 No. 1 (2012) , Article ID: 17272 , 11 pages DOI:10.4236/abc.2012.21002
Glycosylation and secretion of human α-amylases
![]()
1Laboratory of Glycobiology, The Noguchi Institute, Tokyo, Japan
2New Industry Creation Hatchery Center, Tohoku University, Sendai, Japan
Email: amano@noguchi.or.jp
Received 9 November 2011; revised 15 December 2011; accepted 22 December 2011
Keywords: Amylase; Gycosylation Efficiency; N-Glycan; Secretion
ABSTRACT
Three human α-amylases exist: Amy1 (salivary amylase), Amy2A (pancreatic amylase), and Amy2B (expressed in various tissues). These amylases share a 97% - 99% amino acid sequence identity, and two potential N-glycosylation sites (N427 and N476) are commonly found in the C-terminal region. In general, salivary amylase is more frequently glycosylated than pancreatic amylase, and it is still uncertain why differences in the glycosylation pattern among human amylase isozymes occur. In this study, we found that there was no significant change of ratio of glycosylated molecules among isozymes produced by the same cultured cells, indicating that glycosylation of amylase is influenced by the type of cell producing the enzyme rather than being an inherent property of the amylase isozymes. We analyzed the glycosylation efficiency of N-glycosylation sites in recombinant Amy2A mutants produced by HEK293 cells and found that glycosylation efficiencies of N427 and N476 were 3% - 18% and 40% - 52%, respectively, indicating that the major N-glycosylation site of glycosylated Amy2A produced by HEK293 cells is N476. The difference in the glycosylation efficiency of each N-glycosylation site also seemed to contribute in part to generate different glycosylation patterns of human amylases. We also confirmed that the C-terminal region of human amylase plays a critical role in secretion, although glycosylation does not play a part in this effect.
1. INTRODUCTION
There are three types of human α-amylases: Amy1 is the so-called salivary amylase and is mainly secreted in saliva; Amy2A is the so-called pancreatic amylase and is mainly secreted in pancreatic juice; Amy2B also is a pancreatic amylase but it is expressed in various tissues. These amylases share a 97% - 99% amino acid sequence identity and are composed of three domains: the signal peptide, the catalytic domain and the C-terminal region (Figure 1). The C-terminal region is not essential for their catalytic activity, but it has been suggested that it is indispensable for the correct post-translational folding of these enzymes that is required for their activity and that also allows them to be secreted properly [1]. Although there are two potential N-glycosylation sites (N427 and N476) in the C-terminal region of these amylases, glycosylation of the amylases seems to occur in a tissuespecific manner. The N-glycan structures of human amylase from the parotid are complex-type glycans with several glycoforms [2]. We studied the structures of N-glycans of human pancreatic amylase and found that they are different from those of salivary amylase (Oyama et al., submitted). The N-glycan structures of human amylase from tumors were similar but modified compared to those of parotid amylase [3,4]. In addition to the structural variety of N-glycans, there is a difference in the glycosylation pattern of human amylase isozymes [5,6]: both the N427 and N476 sites in human salivary amylase can be glycosylated to some extent, whereas those in human pancreatic amylase are either glycosylated to a lesser extent or not glycosylated. In the case of human amylase, glycosylation may not be important since the unglycosylated form of amylases is fully secreted and active. It is still uncertain, however, why differences in the glycosylation pattern among human amylase isozymes occur.
In this study, we evaluated the glycosylation state of human amylase isozymes and found that the different glycosylation patterns of human amylases were related to the types of amylase-producing cells rather than the types of amylase isozymes. We also found that the glycosylation efficiencies of the N427 and N476 sites were different and the N476 site was more efficiently glycosylated than the N427 site. These findings seem to be highly related to the different glycosylation pattern of human amylases. In addition, we discuss the importance of the C-terminal region of human amylase in secretion.
2. MATERIALS AND METHODS
2.1. Materials
Human saliva was obtained from a healthy male. Human pancreatic amylase was purchased from Meridian Life Science, Inc. (Saco, ME). Peptide: N-glycosidase (PNGase) F was purchased from Takara (Shiga, Japan). Tunicamycin was purchased from Sigma (St. Louis, MO). Z-VADfmk was purchased from BioVision (Mountain View, CA). 2-Acetamido-1,2-dideoxynojirimycin (2-ADN) was purchased from Tronto Research Chemicals Inc. (Ontario, Canada).
2.2. Preparation of De-N-Glycosylated Amylase
For preparation of de-N-glycosylated amylase, the sample was treated with recombinant PNGase F, which is free of protease activity, according to the manufacturer’s instructions. The sample buffer for the SDS-poly-acrylamide gel electrophoresis (SDS-PAGE) was added to the reaction products and boiled. The boiled samples were subjected to SDS-PAGE.
2.3. Cell Culture
HEK293, COS-7 and CHO cells were obtained from the RIKEN Bioresource Center (Tsukuba, Japan), and KLM- 1 cells were obtained from the Institute of Development, Aging, and Cancer, Tohoku University. HEK293 cells were cultured and maintained in minimum essential medium supplemented with 10% fetal calf serum. COS-7 cells were cultured and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. CHO cells were cultured and maintained in minimum essential medium α supplemented with 10% fetal calf serum. KLM-1 cells were cultured and maintained in RPMI 1640 medium supplemented with 10% fetal calf serum. The cultivation was performed at 37˚C and under 5% CO2 with media change with every 4 - 7 days.
2.4. cDNA Cloning of Human Amylases
Total RNA was extracted from KLM-1 cells by using the SV total RNA Isolation System (Promega, Madison, WI), and first-strand cDNA was synthesized using the ImProm-II Reverse Transcription System (Promega) according to the manufacturer’s instructions. Total RNA of human salivary gland was purchased from Clontech (Mountain View, CA), and first-strand cDNA was synthesized as described above. First-strand cDNA of human pancreas was purchased from Clontech (Human multiple tissue cDNA panel). To obtain the entire coding region of human amylases, polymerase chain reactions (PCRs) were performed as follows using PrimeSTAR HS DNA polymerase (Takara): 98˚C for 5 s; 45 cycles of 98˚C for 10 s55˚C for 15 s, and 72˚C for 150 s; and 72˚C for 60 s. PCR primers used in this study are summarized in Table 1. For the amplification of Amy1 cDNA, primers NG5+ and NG6– were used with the above first-strand cDNA of salivary gland as a template. For the amplification of Amy2A cDNA, primers NG5+ and NG6– were used with the above first-strand cDNA of pancreas as a template. For the amplification of Amy2B cDNA, primers NG5+ and NG6– were used with the above first-strand cDNA of KLM-1 cell as a template. The PCR products were cloned into the EcoRV site of the pBluescript II SK(+) vector. The nucleotide sequences were confirmed by DNA sequencing.
2.5. Determination of the Expressing Amylase Genes
To determine the amylase genes expressed in tissues, PCRs were performed as described above except using primers Pr1+ and NG4–. The PCR products were digested with PstI, BsaAI, and Aor51HI, respectively electrophoresed on a 2% agarose gel, stained with ethidium bromide, and then visualized under UV light. The expressing amylase genes were determined by the restriction enzyme digestion patterns of the PCR products.
2.6. Construction of Epitope-Tagged Expression Vectors
The DNA fragment containing HindIII and BamHI sites and the FLAG-epitope (DYKDDDDK) tag-encoding sequence with the stop codon was synthesized by annealing the following primers: 5’-AGCTTGACTACAAGGACG ACGATGACAAGTGAG-3’ and 5’-GATCCTCACTTG TCATCGTCGTCCTTG-TAGTCA-3’. The annealed fragment was cloned into the HindIII-BamHI sites of the pBluescript II SK(+) vector. The DNA fragment encoding the entire coding region of Amy2A was prepared by digesting the above cloned Amy2A cDNA with XhoI and HindIII, and this fragment was cloned into the XhoI-HindIII sites of the FLAG-tag containing pBluescript II SK(+) vector. From this plasmid, the XhoI-XbaI fragment, which contains the FLAG-tagged entire coding region of Amy- 2A, was prepared. This fragment was subcloned into the XhoI-XbaI sites of the mammalian expression vector pcDNA 3.1/Myc-His A (Invitrogen, Carlsbad, CA), yielding pcDNA-Amy2A. The expression vectors of Amy1 and Amy2B were constructed in the same manner, yielding pcDNA-Amy1 and pcDNA-Amy2B, respectively.
2.7. Site-Directed Mutagenesis
To construct expression vectors for each amylase mutant, site-directed mutagenesis was performed using a KOD-
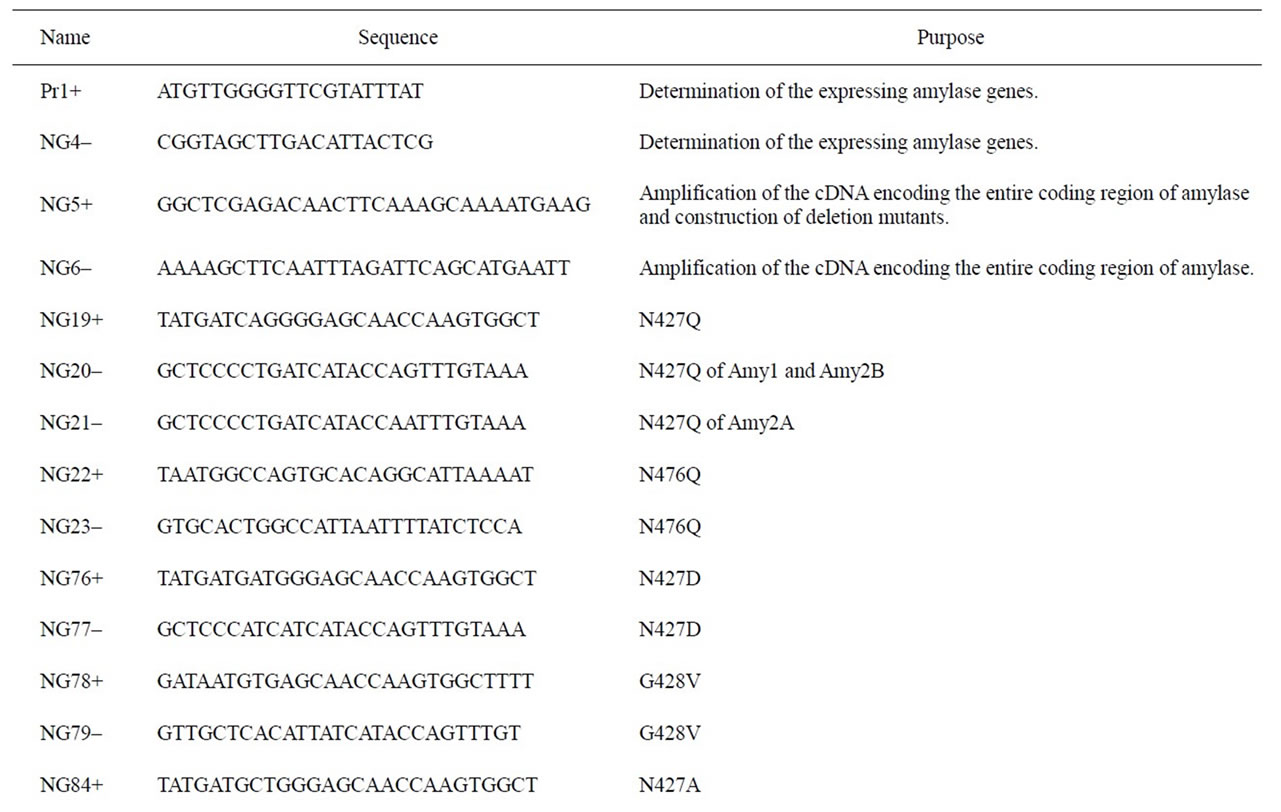
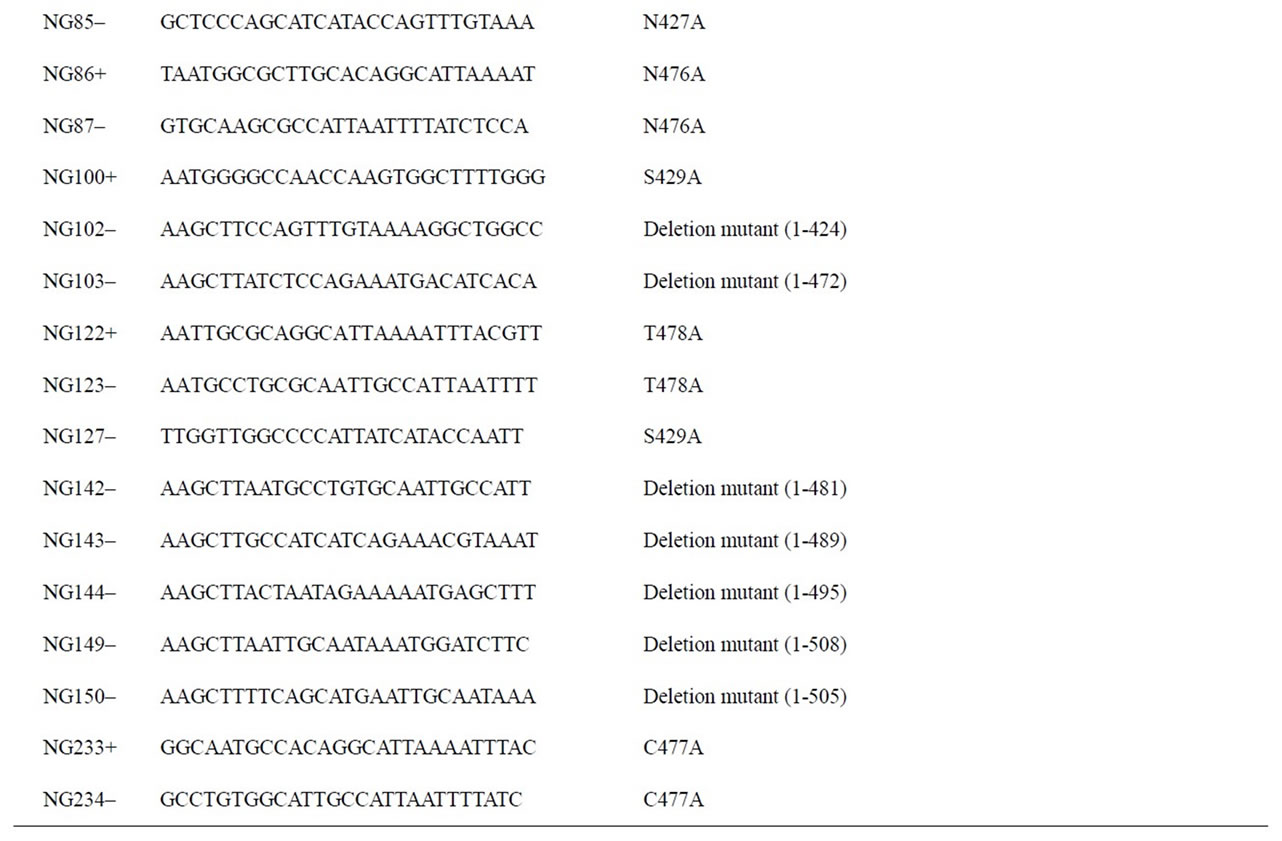
Table 1. Primers used in this study.
Plus-Mutagenesis Kit (Toyobo, Osaka, Japan) and mutagenic primers listed in Table 1.
2.8. Construction of Deletion Mutants
The deletion mutants of the Amy2A C-terminal region were constructed by PCR with the primer sets listed in Table 1 and the cloned Amy2A cDNA as a template. PCRs and construction of expression vectors of the deletion mutants were performed in the same manner as described above.
2.9. Transfection and Preparation of Amylase Samples
Cells were seeded in the 35-mm tissue culture grade dishes 24 h prior to transfection. The expression vectors (0.8 μg each) were transfected into the cells by means of FuGENE 6 Transfection Reagent (Roche, Basel, Switzerland). After 15 h transfection, the cells were washed two times with phosphate-buffered saline and the medium was changed to the same medium but without fetal calf serum. When necessary, 10 μg/ml of tunicamycin, 0.1 mM of Z-VAD-fmk or 0.5 mM of 2-ADN were added to the medium. After a further 24 h of cultivation, the culture medium and cells were collected. The culture medium was centrifuged, and the ANTI-FLAG M2-Agarose (Sigma) was added to the supernatant to adsorb the secreted FLAG-tagged recombinant amylase. After 2 h of rotation of the tube containing the above supernatant at 4˚C, the ANTI-FLAG M2-Agarose was collected by centrifugation, it was washed three times with phosphatebuffered saline, and the sample buffer for SDS-PAGE was added to the M2-Agarose and boiled. For preparation of the cell extract, the lysis buffer containing 10 mM TrisHCl (pH 7.8), 150 mM NaCl, 1 mM EDTA-2Na, 1% Nonidet P-40, 0.1% SDS, and 0.5% sodium cholate was added to the cells and sonicated. The cell lysate was centrifuged and the supernatant was collected. To the appropriate amount of the supernatant, the sample buffer for SDS-PAGE was added and boiled. The boiled samples were then subjected to SDS-PAGE.
2.10. Western Blot Analysis
Western blotting was carried out using a standard protocol. To detect amylase, the mouse anti-human amylase monoclonal antibody (G-10:sc-46657, Santa Cruz, Santa Cruz, CA) was used as a primary antibody (1:5000), and the rabbit anti-mouse IgG HRP-conjugate (Zymed, San Francisco, CA) was used as a secondary antibody (1:5000). The amylase band on the membrane was then visualized with the ChemiDoc XRS System (Bio-Rad, Hercules, CA) using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo, Waltham, MA). Calculation of the rate of protein glycosylation was carried out using a quantification program of the ChemiDoc XRS System. To detect β-actin, the mouse anti-β-actin monoclonal antibody (AC-15:A1978, Sigma) was used as a primary antibody (1:3000), and its detection was carried out as described above.
3. RESULTS
3.1. Difference in the Amount of Glycosylated Human Amylase Molecules among Various Cells
Human amylases (Amy1, Amy2A, and Amy2B) commonly have two potential N-glycosylation sites (N427 and N476) in their C-terminal region (Figure 1). In the case of salivary amylase, three molecular species were observed: the unglycosylated form and the glycosylated forms with one N-glycan or two N-glycans (Figure 2). Among these molecules, a small proportion of molecules were fully glycosylated at both the N427 and the N476 sites, about half of the molecules were glycosylated at either of these sites, and the rest of the molecules that showed the same position as PNGase F-treated molecule were unglycosylated. In the case of pancreatic amylase, two molecular species were observed: the unglycosylated form and the glycosylated form with one N-glycan (Figure 2). Unlike salivary amylase, most of the pancreatic amylase was not glycosylated. In the salivary gland, the Amy1 gene was exclusively expressed (Figure 3), indicating that the main component of salivary amylase was Amy1. In pancreas, the Amy2A gene was predominantly expressed and the Amy2B gene was also expressed slightly (Figure 3), indicating that the main component of pancreatic amylase was Amy2A. As Amy1 and Amy2A differ in their amino
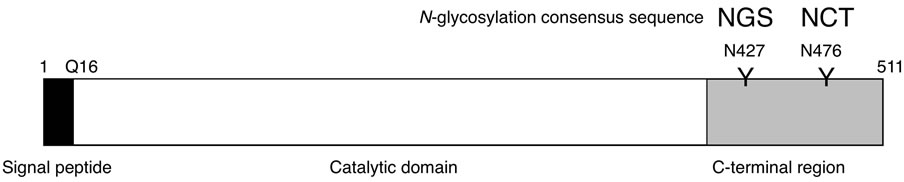
Figure 1. Schematic representation of human amylase. The human amylase family consists of three members, Amy1, Amy2A, and Amy2B. Their amino acid sequences are highly conserved to each other (97% - 99% identity), and all have two potential N-glycosylation sites (N427 and N476) in the C-terminal region.
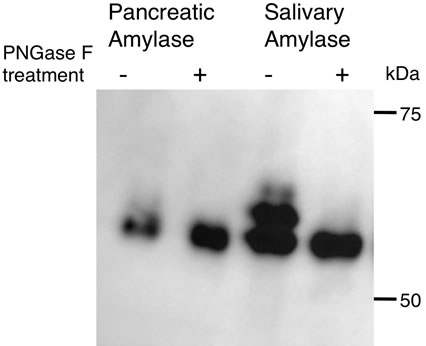
Figure 2. Glycosylation patterns of pancreatic amylase and salivary amylase. Human pancreatic amylase and saliva were treated with or without PNGase F and subjected to SDS-PAGE. The gel was blotted onto a nylon membrane and a Western blot was performed to detect amylase.
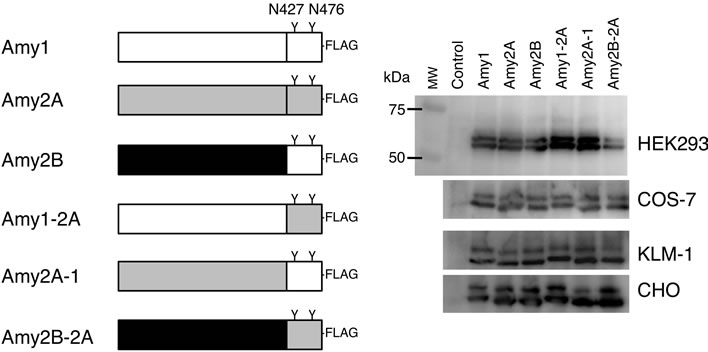
acid sequences at 16 positions (96.9% identity = 495 aa/ 511 aa), we examined whether different glycosylation patterns between salivary and pancreatic amylases were caused by the amino acid sequence discrepancy between Amy1 and Amy2A. As human primary salivary gland cells and pancreatic cells were not available to us, we introduced expression vectors containing coding sequences for Amy1, Amy2A, Amy2B and chimeric enzymes of these amylase isozymes (Figure 4(a)) into HEK293, COS-7, CHO and KLM-1 cells and analyzed glycosylation patterns of secreted recombinant amylases, which were affinity-purified from the medium by the ANTI-FLAG M2-Agarose.
The recombinant amylases produced by these cells consisted of mainly two molecular species, i.e. the unglycosylated form and the glycosylated form with one N-glycan (Figure 4(b)); however, a very small amount of the glycosylated form with two N-glycans was also observed in HEK293 cells. We performed a two-way analysis of variance (ANOVA) to evaluate whether the glycosylation of amylase was affected by differences of the amylase isozymes and the cell types producing them. When amylases produced by the same cell line were compared, the glycosylation patterns of these enzymes were barely affected by differences among the amylase isozymes (F(5,48) = 0.413, p > 0.05), and their glycosylation patterns were similar (Figure 4(c)). The ratio of glycosylated to unglycosylated molecules differed according to the cell types (F(3,48) = 49.735, p < 0.05).
3.2. Site-Specificity of Glycosylation
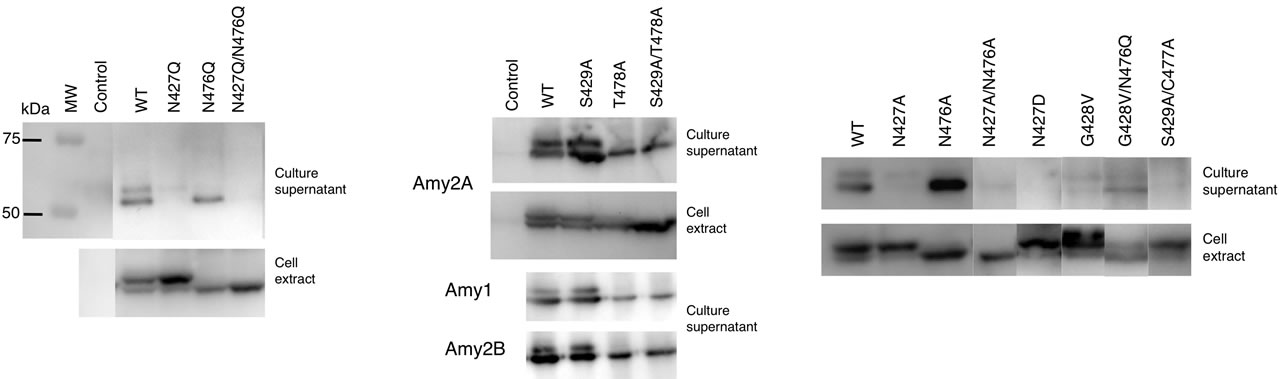
To further investigate the glycosylated form of the amylase containing one N-glycan produced by cultured cells, we examined which potential N-glycosylation site was predominantly glycosylated. The N427Q, N476Q and N427Q/N476Q mutants of Amy2A were produced in order to disrupt each N-glycosylation consensus sequence, and the resulting glycosylation patterns were analyzed in HEK293 cells. As shown in Figure 5(a), the N427Q mutant in the cell extract was more glycosylated than the wild type Amy2A, but its secretion was impaired. On the other hand, the N476Q mutant was barely glycosylated, and it was successfully secreted. The N427Q/N476Q mutant was neither glycosylated nor secreted. These results indicated N476 was the primary glycosylation site and led
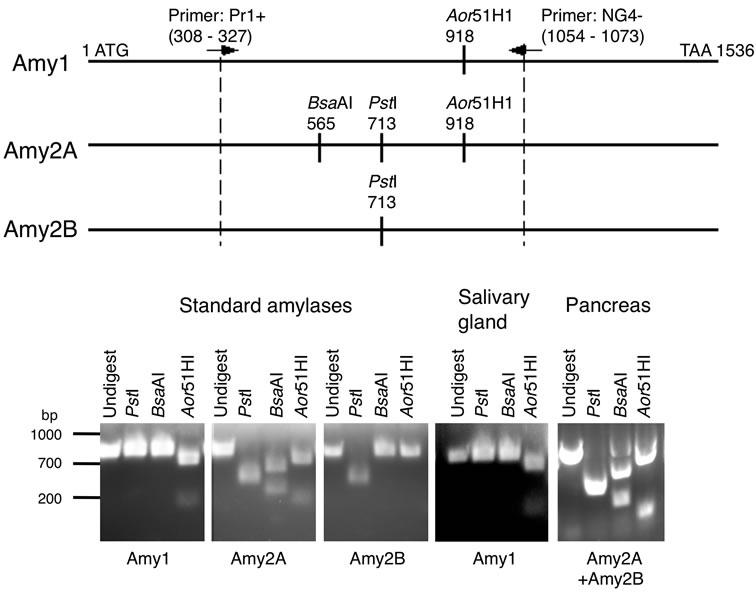
Figure 3. Determination of the expressing amylase genes. The expressed amylase genes were determined by the restriction enzyme digestion patterns of the PCR products.
 (a)
(a)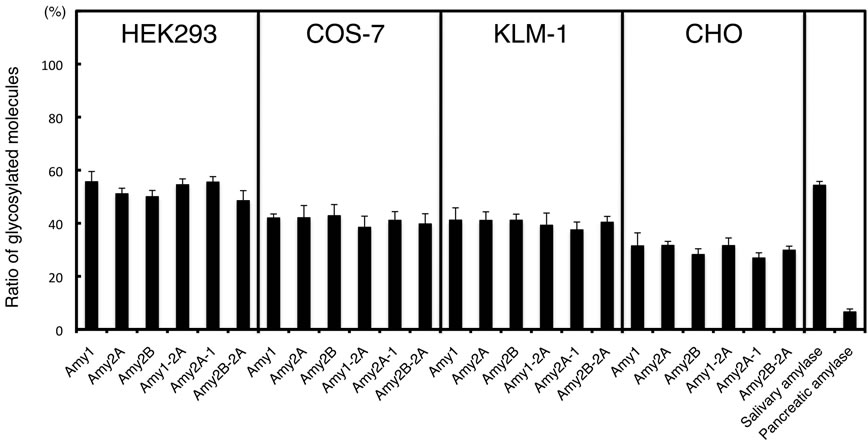 (b)
(b)
Figure 4. Glycosylation of amylase produced by cultured cells. (a) Schematic representation of FLAG-epitope tagged Amy1, Amy2A, Amy2B and their chimeric enzymes. The C-terminal regions of Amy1 and Amy2B have the same amino acid sequence; (b) Western blot analysis of culture supernatants of various cells that were transiently transfected with amylase expression vectors. As a control, an empty pcDNA vector was used for transfection. MW, molecular weight marker; (c) Estimation of the ratio of glycosylated molecules with one and two glycans in Figures 2 and 4(b). Error bars indicate standard error of the mean (n = 3).
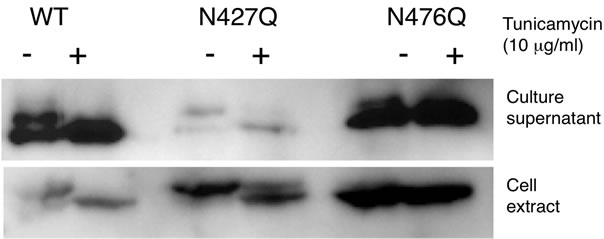
us to speculate that the N-glycan on N427 was necessary for secretion and that it was removed immediately during the course of or after secretion, similar to reports that Nglycans in salivary amylase were removed by the action of endo-β-N-acetylglucosaminidase (ENGase) after secretion [6,7]. We therefore generated another set of mutants, S429A, T478A and S429A/T478A, to examine whether or not this hypothesis is correct. Both the S429A and T478A mutations were designed to impair N-glycosylation at the N427 and N476 sites, respectively, by disrupting the N-glycosylation consensus sequence. As shown in Figure 5(b), the glycosylation patterns of the T478A and S429A/T478A mutants were similar to those of the N476Q and N427Q/ N476Q mutants, respectively, but the S429A mutant in the cell extract was less glycosylated than the N427Q mutant. Unlike the N427Q and N427Q/N476Q mutants, the S429A, T478A and S429A/T478A mutants were properly secreted. This tendency was also observed for Amy1 and Amy2B mutants (Figure 5(b)). These results indicated that the N-glycosylation of N427 site was not necessary for secretion. We further used tunicamycin to inhibit N-glycan biosynthesis and analyzed its effect on secretion of Amy2A mutants, but it did not affect amylase secretion (Figure 6), indicating that N-glycans of amylase were not necessary for secretion.
As secretion of the N427Q mutant was impaired, we further examined the importance of N427 for secretion. We generated N427A and N427D mutants and analyzed their secretion (Figure 5(c)). Similar to the N427Q mutant, secretion of the N427A and N427D mutants was also impaired. The glycosylation efficiencies of N476 site of N427Q, N427A, and N427D mutants in the cell extracts were estimated to be 72%, 70%, and 72%, respectively (Figure 5(d)). On the other hand, the glycosylation efficiency of N476 site of the S429A mutant was estimated to be 52% in the cell and 40% after secretion.
 (a)
(a)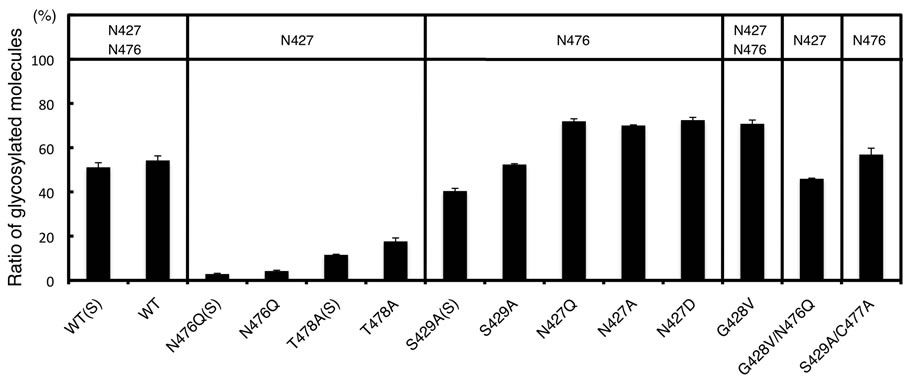 (b)
(b)
Figure 5. Glycosylation and secretion of recombinant amylases. The recombinant amylases in culture supernatants and cell extracts of HEK293 transfectants were detected by Western blot. As a control, an empty pcDNA vector was used for transfection, and a molecular weight marker, MW, was used as before. (a) Secretion of Amy2A mutants containing the N427Q amino acid substitution was impaired; (b) Mutants containing the S429A amino acid substitution were properly secreted; (c) Glycosylation and secretion of Amy2A and its mutants having amino acid substitutions within the N-glycosylation consensus sequences were analyzed by Western blot; (d) Estimation of the ratio of glycosylated molecules. N476Q(S), T478A(S), S429A(S) and WT(S) are indicated as secreted molecules and others are intracellular molecules. The potential N-glycosylation sites are shown in the upper panal. Error bars indicate standard error of the mean (n = 3).
This suggests that N427 is an important residue for proper secretion of human amylases and amino acid substitution around N427 site may cause structural changes of human amylases during protein folding process and promote N-glycosylation on the N476 site. The glycosylation efficiencies of N427 site of the N476Q and T478A mutants were estimated to be 4% and 18% in the cell and 3% and 12% after secretion, respectively (Figure 5(d)). We also examined mutants that have an amino acid substitution within the consensus sequences for N-glycosylation, and found that secretion of mutants such as G428V, G428V/N476Q, and S429A/C477A was also impaired, although they were properly glycosylated (Figures 5(c) and 5(d)). The G428V mutation promoted N-glycosylation on the N427 site.
We examined whether N-glycans on Amy2A were removed by endogenous PNGase or ENGase produced by HEK293 cells as the glycosylation patterns of intracellular and extracellular Amy2A differed and there were reports that N-glycans in salivary amylase were removed by the action of ENGase after secretion [6,7]. We used ZVAD-fmk as a PNGase inhibitor [8,9] and 2-ADN as an ENGase inhibitor [10] and analyzed their effects on Nglycosylation of Amy2A. These inhibitors, however, did not show any effect on N-glycosylation of Amy2A and its mutants in HEK293 cells (Figure 7), indicating that direct removal of N-glycans on Amy2A by endogenous PNGase or ENGase did not occur in HEK293 cells.
3.3. Analysis of Deletion Mutants of the Amy2A C-Terminal Region
The mature human amylase consists of the catalytic domain and the C-terminal region. It has been shown that deletion of 65 amino acids of the C-terminal region of rat pancreatic amylase impaired its secretion by COS-7 cells [1]. As the amino acid substitution around the N-glycol-
 (a)
(a)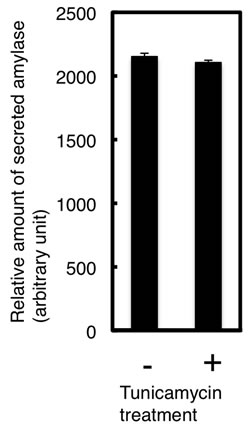 (b)
(b)
Figure 6. Tunicamycin treatment did not affect the secretion of recombinant amylase. (a) Effect of tunicamycin on the secretion of Amy2A and its mutants was analyzed by Western blot; (b) Estimation of the relative amount of secreted Amy2A with and without tunicamycin treatment. Error bars indicate standard error of the mean (n = 3).
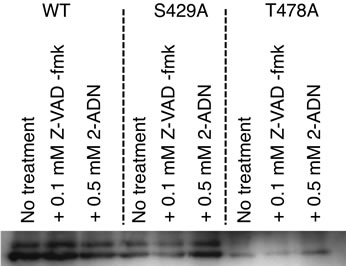
Figure 7. Removal of N-glycans on recombinant amylase did not occur in HEK293 cells. HEK293 cells were transiently transfected with amylase expression vectors. After 15 h of transfection, the culture medium was changed to the medium with or without the inhibitor, and secreted amylase in the medium was analyzed by Western blot.
sylation consensus sequences located in the C-terminal region of human amylase also caused impaired secretion in some cases, we further constructed a series of deletion mutants of the Amy2A C-terminal region to evaluate its importance in secretion. As shown in Figure 8, these mutants were successfully produced in HEK293 cells. Secretion of these deletion mutants, even the mutant that lacks the last 3 residues of the C-terminal amino acids, was almost completely depressed. On the other hand, Nglycosylation of these mutants was promoted.
4. DISCUSSION
The glycosylation patterns of salivary amylase and pancreatic amylase are different. In this study, we have shown that different glycosylation patterns of these amylases were related to the types of amylase-producing cells rather than the types of amylase isozymes. In addition, we have shown that each of the two N-glycosylation sites of human amylase has a different glycosylation efficiency. It seemed that the combination of these different glycosylation efficiencies and the glycosylation ability of amylase-producing cells resulted in the heterogeneous N-glycosylation pattern of human amylase. The environment that the secreted amylase encounters may also contributes to the different glycosylation pattern of human amylase, as parts of the N-glycans of salivary amylase was shown to be removed by ENGase in saliva [6,7]. As shown in Figure 7, N-glycans of recombinant Amy2A produced by HEK293 cells were not removed by ENGase. These remaining N-glycans may be related to the small amount of secreted ENGase in culture medium of HEK293 cells.
Although we have shown some mechanistic aspects of heterogeneous N-glycosylation of human amylases, it is still uncertain why salivary amylase is more glycosylated than pancreatic amylase. There are many glycoproteins in pancreatic juice, indicating that the glycosylation machinery of pancreatic cells works well. There may be amylase-specific glycosylation mechanisms in pancreatic cells and salivary gland cells. The detailed mechanism underlying the tissue specific glycosylation of amylase is under investigation.
The glycosylation efficiencies of the N427 and N476 sites of Amy2A produced by HEK293 cells were estimated to be 3% - 18% and 40% - 52%, respectively. These values were estimated by using N476Q and T478A mutants for the N427 site and S429A mutant for the N476 site, respectively. In this estimation, the effects of structural changes caused by the amino acid substitution and the existence of N-glycan at the other site, which might be observed in the wild type amylase, were not considered, so the actual glycosylation efficiencies of these sites in the wild type Amy2A may vary. In fact, the glycosylation efficiency of each site varied among mutants (Figure 5(d)). Regardless, the glycosylation efficiency of the N476 site in the wild type Amy2A seemed to be higher than that of the N427 site. We could detect glycopeptides containing N476 site derived from human pancreatic amylase by mass spectrometry analysis, but we could not
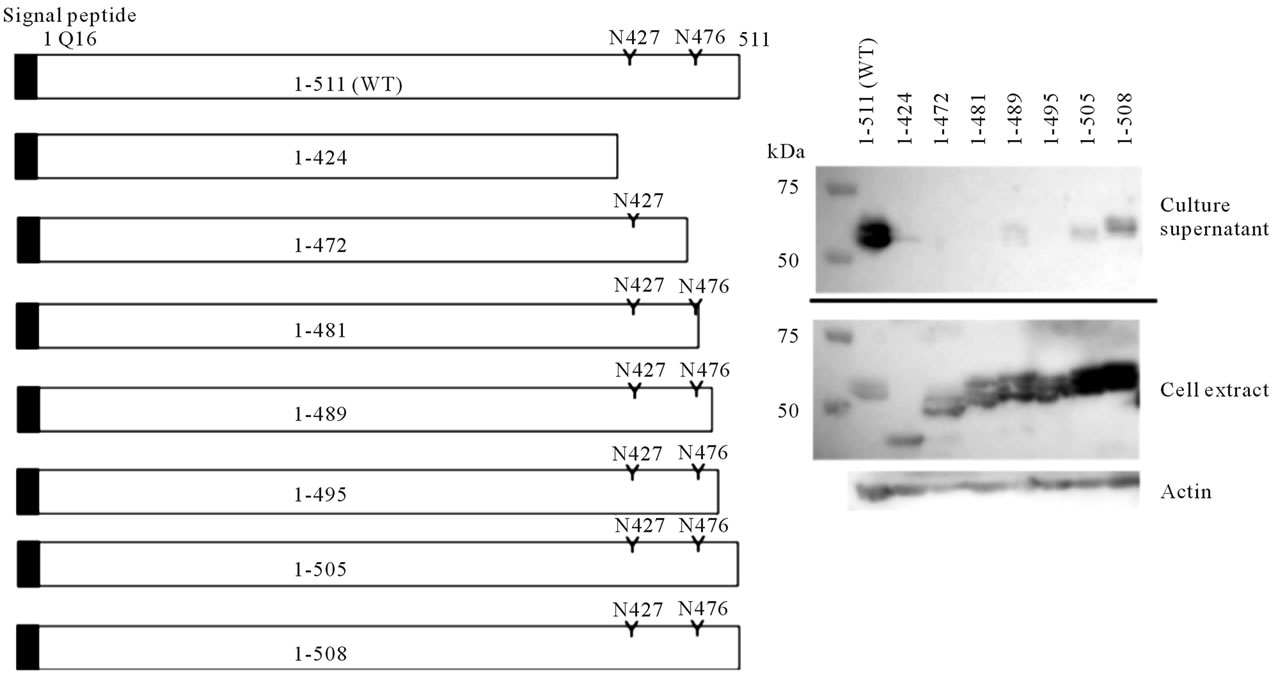
Figure 8. Secretion and glycosylation of deletion mutants at the Amy2A C-terminal region. Schematic representation of deletion mutants of the Amy2A C-terminal region is shown in the left panel. Western blot of these deletion mutants produced by HEK293 cells is shown in the right panel.
detect glycopeptides containing N427 site (Oyama et al., submitted). The low glycosylation efficiency of the N427 site of Amy2A indicates that this site is a poorer substrate for the oligosaccharyltransferase complex than the N476 site. This tendency was also observed for Amy1 and Amy2B (Figure 5(b)). In addition, the corresponding possible N-glycosylation site (N410) of rat pancreatic amylase, whose overall amino acid sequence shows 84.5% identity with human Amy2A, was not glycosylated [1]. The microenvironment around the N427 site is probably structurally more unfavorable for the oligosaccharide transfer by the oligosaccharyltransferase complex than the N476 site. We also speculate that folding of amylase occurred simultaneously during or immediately after the nascent amylase polypeptide passed through the oligosaccharyltransferase complex because amino acid substitution of the N427 site affected the N-glycosylation on the N476 site.
It should be noted that the ratio of N-glycosylated amylase molecules inside the cell was higher than outside the cell. As N-glycans of amylase were not removed by endogenous PNGase or ENGase of HEK293 cells (Figure 7), it seemed that significant amount of N-glycosylated amylase molecules was misfolded and degraded by the quality control system in the ER, where the N-glycan acts as a signal for misfolded proteins [11].
We observed that the introduction of the amino acid substitution around the N-glycosylation sites of Amy2A impaired its secretion by HEK293 cells in some cases. In addition, deletion mutants of the C-terminal region of Amy2A, even the mutant that lacks the last 3 residues of the C-terminal amino acids only, had depressed secretion. The N-glycosylation of these mutants was promoted, suggesting that the structural changes in the C-terminal region favorable for N-glycosylation occurred, but their enhanced N-glycosylation did not affect secretion. These results indicate that the C-terminal region of Amy2A itself, but not the N-glycans, plays an important role in secretion in HEK293 cells. It seemed that the proper conformation of the C-terminal region was necessary for amylase secretion and that increased N-glycosylation observed in Amy2A mutants was highly related to the unfavorable structural changes in the C-terminal region for secretion. As pancreatic enzymes are secreted via secretion granules from the exocrine pancreatic acinar cells, the secretion mechanisms of Amy2A in pancreatic cells and HEK293 cells, which are derived from the human embryonic kidney, may differ. It is, however, possible that the C-terminal region of Amy2A is also necessary for secretion in pancreatic cells.
Differences in glycosylation and secretion of amylase are also found when comparing orthologs in rats and humans. The N410Q mutant of rat pancreatic amylase, which corresponds to the N427Q mutant of human Amy2A, was successfully secreted [1], whereas the N410 of rat pancreatic amylase was completely unglycosylated [1]. On the other hand, the N427Q mutant of human Amy2A was not secreted, whereas the N427 of human Amy2A was slightly glycosylated. These suggest that the microenvironment around the first possible N-glycosylation site of amylase is different among animal species.
It should be noted that although both N427Q and S429A mutants have a disrupted consensus sequence for N-glycosylation at the N427 site, only the S429A mutant was secreted. The effect of amino acid substitution and concomitant structural changes, but not the loss of an Nglycan at the N427 site, may have caused impaired secretion of Amy2A mutants containing the N427Q amino acid substitution. Had the N427Q mutant alone been observed, the obvious, yet incorrect, interpretation would have been that an N-glycan on the N427 site is necessary for secretion. To analyze the functions of the N-glycans, amino acid substitution of an asparagine at the N-glycolsylation site has been often adopted. Our result suggests that analysis of only this type of mutant may be insufficient in order to deduce whether glycosylation is necessary or not.
Serum amylase has been used as a biomarker for acute pancreatitis [12]. The glycosylation state and/or the structure of N-glycans of amylase also might be used as a biomarker of some diseases, because the structural changes of amylase N-glycans have been reported in some cancers [3]. To efficiently use amylase as a biomarker, we hope to elucidate the mechanism underlying the tissue and disease specific glycosylation of amylase.
5. CONCLUSION
Different glycosylation pattern of human amylases was related to the types of cells producing the enzymes rather than the types of amylase isozymes and different glycosylation efficiencies of two N-glycosylation sites in human amylases. The C-terminal region of human amylase, but not the N-glycans, plays an important role in secretion.
REFERENCES
- Doyon, Y., Home, W., Daull, P. and LeBel, D. (2002) Effect of C-domain N-glycosylation and deletion on rat pancreatic α-amylase secretion and activity. Biochemical Journal, 362, 259-264.
- Yamashita, K., Tachibana, Y., Nakayama, T., Kitamura, M., Endo, Y. and Kobata, A. (1980) Structural studies of the sugar chains of human parotid α-amylase. The Journal of Biological Chemistry, 255, 5635-5642.
- Yamashita, K., Tachibana, Y., Takeuchi, T. and Kobata, A. (1981) Structural study of the sugar chains of α-amylases produced ectopically in tumors. The Journal of Biochemistry, 90, 1281-1289.
- Moriyama, T. (2008) Sialyl salivary-type amylase associated with ovarian cancer. Clinica Chimica Acta, 391, 106- 111. doi:10.1016/j.cca.2008.01.025
- Stiefel, D.J. and Keller, P.J. (1973) Preparation and some properties of human pancreatic amylase including a comparison with human parotid amylase. Biochimica et Biophysica Acta, 302, 345-361.
- Ito, K. (2006) Formation of glycoisoforms of human salivary α-amylase and endo-β-N-acetylglucosaminidase HS. Trends in Glyososcience and Glycotechnology, 18, 73-84.
- Ito, K., Okada, Y., Ishida, K. and Minamiura, N. (1993) Human salivary endo-β-N-acetylglucosaminidase HS specific for complex type sugar chains of glycoproteins. The Journal of Biological Chemistry, 268, 16074-16081.
- Misaghi, S., Pacold, M.E., Blom, D., Ploegh, H.L. and Korbel, G.A. (2004) Using a small molecule inhibitor of peptide: N-glycanase to probe its role in glycoprotein turn-over. Chemistry & Biology, 11, 1677-1687. doi:10.1016/j.chembiol.2004.11.010
- Migashi, S., Korbel, G.A., Kessler, B., Spooner, E. and Ploegh, H.L. (2006) z-VAD-fmk inhibits peptide: N-glycanase and may result in ER stress. Cell Death & Differentiation, 13, 163-165. doi:10.1038/sj.cdd.4401716
- Kim, Y.K., Kim, K.R., Kang, D.G., Jang, S.Y., Kim, Y.H. and Cha, H.J. (2009) Suppression of β-N-acetylglucosaminidase in the N-glycosylation pathway for complex glycoprotein formation in Drosophila S2 cells. Glycobiology, 19, 301-308. doi:10.1093/glycob/cwn138
- Aebi, M., Bernasconi, R., Clerc, S. and Molinari, M. (2010) N-glycan structures: recognition and processing in the ER. Trends in Biochemical Sciences, 35, 74-82. doi:10.1016/j.tibs.2009.10.001
- Panteghini, M. and Pagani, F. (1989) Time course of changes in serum activity of the P3 isoform of pancreatic amylase isoenzyme in patients with acute pancreatitis. Clinical Biochemistry, 22, 479-482.

