Journal of Tuberculosis Research
Vol.1 No.2(2013), Article ID:36716,4 pages DOI:10.4236/jtr.2013.12003
Random amplified polymorphic DNA analysis of Mycobacterium tuberculosis isolates resistant to Isoniazid in Indonesia
![]()
1Microbiology Department Faculty of Medicine Hasanuddin University, Makassar, Indonesia; asaad_maidin@yahoo.com
2Biopharmaceutical Laboratory Faculty of Pharmacy Hasanuddin University, Makassar, Indonesia; agnes_lidjaja@yahoo.co.id
3Molecular Biology and Immunology Laboratory, Microbiology Department Faculty of Medicine Hasanuddin University, Makassar, Indonesia; *Corresponding Author: hattaram@indosat.net.id
Copyright © 2013 Asaad Maidin et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 16 April 2013; revised 30 May 2013; accepted 20 June 2013
Keywords: Mycobacterium tuberculosis; RAPD; Isoniazid Resistant
ABSTRACT
Background: M. tuberculosis is the most important etiological factor of tuberculosis. One of the factors that make TB hard to eradicate is the emergence of M. tuberculosis drug resistance. Drug resistance in M. tuberculosis is attributed primarily to the accumulation of mutations in the drug target gene. Objectives: to analyze profile of Random Amplified Polymorphic DNA (RAPD) in M. tuberculosis isolates resistant to Isoniazid and found RAPD marker. Methods: seven Isoniazid resistant isolate of M. tuberculosis from Makassar, Indonesia strain were analyzed by RAPD method using primers OPN 02, OPN 09, OPN 20, BG 65, N 9, that amplification fragment DNA than as molecular marker. Results: The results of the present study showed high degree of polymerphism in the M. tuberculosis strains in the population, and found that specific DNA fragment at Isoniazid resistant isolates using primer N 9 is 1450 bp as a marker. Conclusion: This study gives information about RAPD marker of M. tuberculosis strain to Isoniazid resistant.
1. INTRODUCTION
Tuberculosis (TB) is a bacterial infection disease cause by acid fast bacilli—Mycobacterium tuberculosis (M. tuberculosis). This bacillus currently infect one third of the world’s population. According to the World Health Organization report, 8.8 million new cases of tuberculosis occur each year, resulting in 3 million deaths [1,2]. It is estimated 95% cases of TB and 98% deaths caused by TB occur in developed countries [3].
Indonesia is the fifth country with largest number of TB cases in the world. One of the factors that make TB hard to eradicate is the emergence of M. tuberculosis drug resistance.
The estimated 75% of TB patients are in the economically productive age of 15 to 50 years. Multidrug resistant TB (MDR-TB) is defined as resistance to at least Isoniazid and Rifampicin, the two most important drugs in the treatment of TB [1].
Evidence collected between 1994 and 2002, within the framework of the Global Project on anti TB Drug Resistance Surveillance and from other sources, has confirmed previous assertions that in resource—limited countries, drug resistant TB, including MDR-TB is ubiquitous [4].
China and India, the countries with the highest TB incidence in the world, also have documented MDR-TB among new TB cases in some of their geographical areas [4,5].
Drug resistance in M. tuberculosis is attributed primarily to the accumulation of mutations in the drug target gene; these mutations lead either to an altered target (e.g., RNA polymerase and catalase—peroxidase in Rifampicin and Isoniazid resistance, respectively) [6].
A major development in the diagnosis of TB was the introduction of several nucleic acid amplification (NAA) techniques, such as the polymerase chain reaction (PCR) that has been widely evaluated [2].
Random Amplified Polymorphic DNA (RAPD), also known as arbitrarily primed PCR, allows the detection of polymorphisms without prior knowledge of the nucleotide sequence. The polymorphisms may be used as genetic markers and the construction of genetic maps. This method utilizes short (C10 nucleotides) primers of arbitrary nucleotide sequence that are annealed in the first few cycles of PCR at low stringency. RAPD is also a technique ideally suited to fingerprinting applications because it is fast, requires little material and is technically easy [7].
The other study that showed correlation with RAPD method and genetic diversity at M. tuberculosis was studied by: 1) Richner, S. et al., in South Africa (1999); 2) Bardakci, in Turkey (2001); 3) Tazi, L. et al., in Casablanca, Morocco, (2004); 4) Korzekwa, K. et al. in Olsztyn (2006); 5) Singh, J. P. N., Verma, R., Chaudhurt, P., in India (2006); 6) Jain, A. et al. in Luknow, India (2008); 7) Va’zquez-Marrufo, G. et al., in Mexico (2008); 8) Rodriguez, J. C., Royo, G. and Rodriguez-Valera, F. in Spain (2009) [8-15].
2. MATERIALS AND METHODS
The samples were seven resistant isolates from human patients (sputum) diagnosed suffering from pulmonary tuberculosis obtained at the Provincial Health Laboratory of South Sulawesi and Biotechnology laboratory of Research Centre Hasanuddin University, Indonesia. All isolates were cultured on Lowenstein-Jensen medium and identified as Mycobacterium tuberculosis, then continued with susceptibility test on Lowenstein-Jensen medium and anti-tuberculosis drug namely Isoniazid, for analyzing resistance or sensitivity to Isoniazid [16,17]. This study used seven Isoniazid resistant isolates of Mycobacterium tuberculosis.
The DNA was extracted by Wizard Genomic DNA [18]. PCR using RAPD method was performed with 5 primers according to Singh et al. (2006) and Tazi et al. (2004) (Table 1). Amplification of Mycobacterium DNA, uses total volume of 25 µl. The reaction mixture contained 1 unit of Taq DNA polymerase 0.125; MgCl2 3, dNTP 0.5; 10 × buffer 2.5; template DNA 5 µl; ddH2O 12,875 µl and primers 30 pmol. Amplification was carried out in a thermal cycler (AB Applied Biosystem). The cycling condition consisted of 45 cycles of an initial denaturation (pre PCR) step for 5 min at 94˚C, follow by denaturation step for 1 min at 94˚C, an annealing step for 1 min at 36˚C, and an extension step for 1 min at 72˚C, and a final
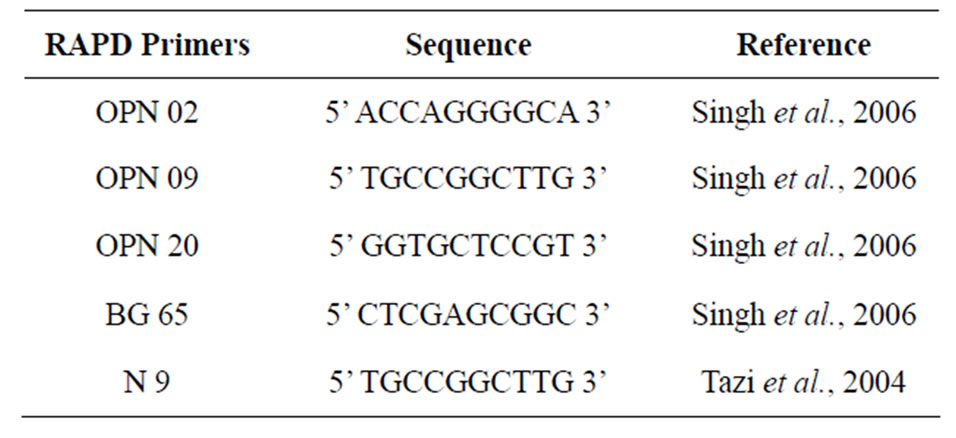
Table 1. Sequence of the 5 RAPD Primers used in this study.
extension (post PCR) for 5 min at 72˚C [12,19,20].
Electrophoresis that was performed after RAPD-PCR, used a 2% agarose gel stained with Etidium bromide and 100 bp marker. Subsequently, the gel was visualized and photographed using a gel documentation and analysis system. The dendrogram program (http://www.expasy.ch/tools//). A data matrix composed of numerals 1 and 0 was built on the basis of presence: 1, or absence: 0.
3. RESULTS
This study used seven Isoniazid resistant isolates, I1, I2, I3, I4, I5, I6, I7, by using five short primers (see Table 1). The primers have been showed in Table 1. The results of RAPD-PCR are shown in Figures 1-5. In this study, all seven isolates of M. tuberculosis showed several fragments. All five primers revealed discriminating patterns.
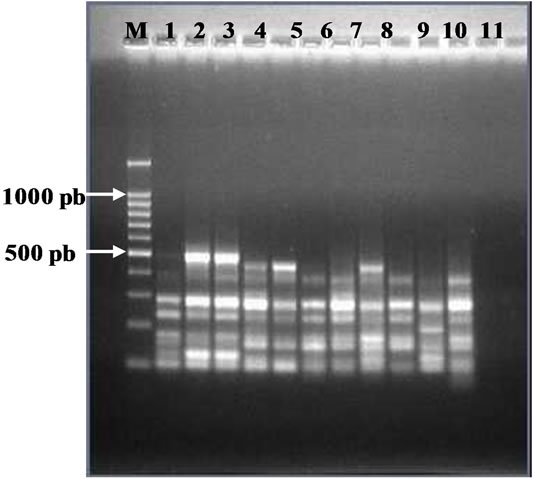
The dendrogram analysis, using primer OPN 02, formed five clusters. The larger cluster consisted of 3 isolates,
 (a)
(a)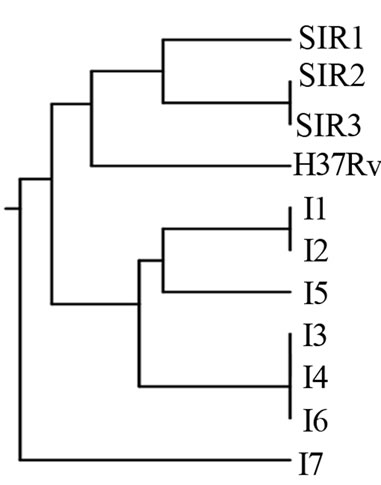 (b)
(b)
Figure 1. (a) RAPD Profiles of M. tuberculosis with primer OPN-02. M = 100 bp DNA marker, 1 - 3 Isoniazid and Rifampicin sensitive, 4 -10: Isoniazid resistant, 11: H37Rv. (b). Dendrogram M. tuberculosis Isoniazid resistant isolates, SIR1 - SIR3: Isoniazid and Rifampicin sensitive, I1 - I7: Isoniazid resistant.
 (a)
(a)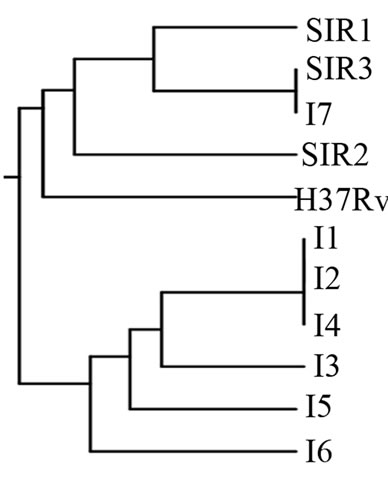 (b)
(b)
Figure 2. (a). RAPD Profiles of M. tuberculosis with primer OPN-09. M = 100 bp DNA marker, 1 - 3: Isoniazid and Rifampicin sensitive, 4 - 10: Isoniazid resistant, 11: H37Rv. (b). Dendrogram M. tuberculosis Isoniazid resistant isolates, SIR1 - SIR3: Isoniazid and Rifampicin sensitive, I1 - I7: Isoniazid resistant.
 (a)
(a)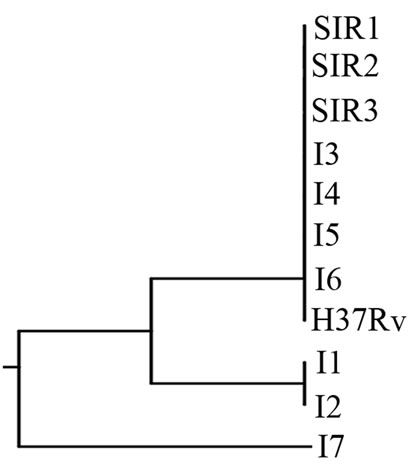 (b)
(b)
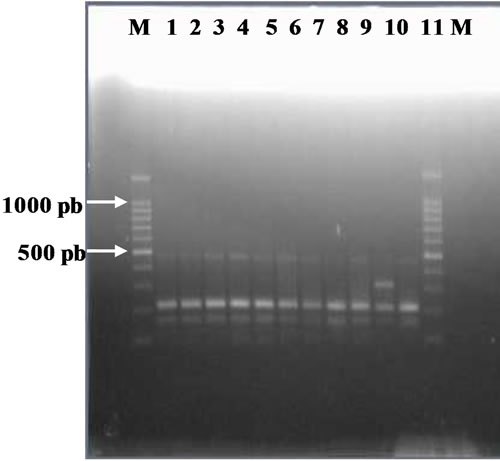
Figure 3. (a). RAPD Profiles of M. tuberculosis with primer OPN-20. M = 100 bp DNA marker, 1 - 3: Isoniazid and Rifampicin sensitive, 4 - 10: Isoniazid resistant, 11: H37Rv. (b). Dendrogram M. tuberculosis Isoniazid resistant isolates, SIR1 - SIR3: Isoniazid and Rifampicin sensitive, I1 - I7: Isoniazid resistant.
the smaller cluster consisted of one isolate. The genetic relatedness was closest among isolates within clusters (Figure 1).
Figure 2(b) using primer OPN 09 were divided into two populations that were sensitive (comparison) and H37Rv (reference) was with Isoniazid resistant, whereas each population were divided into three subpopulations.
Figure 3(b) using primer OPN 20, formed two clusters. The larger cluster consisted of 8 isolates (3 sensitive isolates, 1 H37Rv, 4 Isoniazid resistant isolates), and the smaller cluster consisted of one isolate (resistant to Isoniazid).
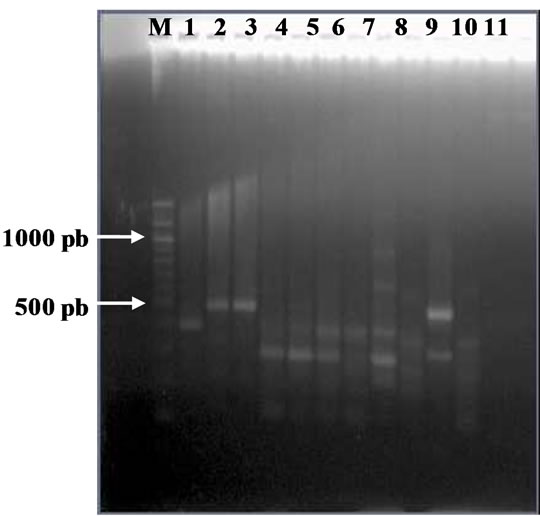
Figure 4(b) using primer BG 65 showed four clusters, whereas the larger cluster consisted of 3 isolates (resistant to Isoniazid), and the smaller cluster consisted of one isolate (resistant to Isoniazid).
The dendrogram analysis with primer N 9, formed six clusters with two populations that are sensitive, H37Rv
 (a)
(a)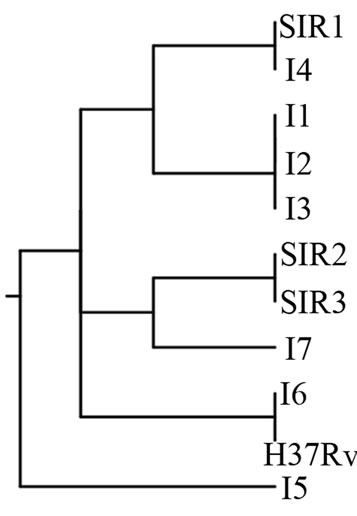 (b)
(b)
Figure 4. (a). RAPD Profiles of M. tuberculosis with primer BG-65. M = 100 bp DNA marker, 1 - 3: Isoniazid and Rifampicin sensitive, 4 - 10: Isoniazid resistant, 11: H37Rv. (b). Dendrogram M. tuberculosis Isoniazid resistant isolates, SIR1 - SIR3: Isoniazid and Rifampicin sensitive, I1 - I7: Isoniazid resistant.
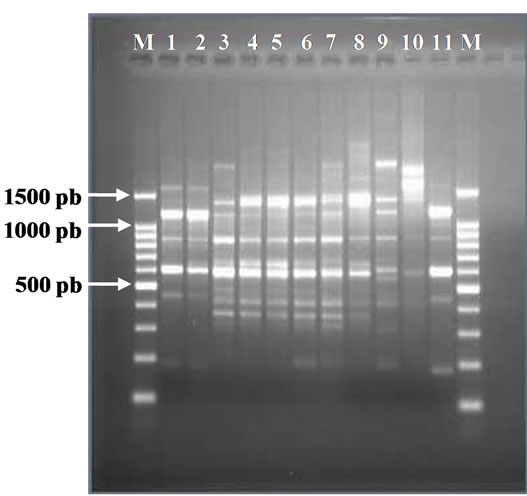 (a)
(a)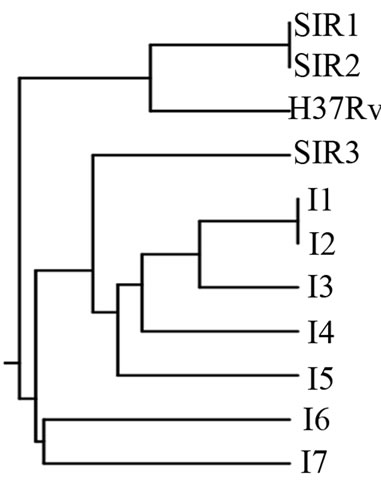 (b)
(b)
Figure 5. (a). RAPD Profiles of M. tuberculosis with primer N-9. M = 100 bp DNA marker, 1 - 3: Isoniazid and Rifampicin sensitive, 4 - 10: Isoniazid resistant, 11: H37Rv. (b). Dendrogram M. tuberculosis Isoniazid resistant isolates, SIR1 - SIR3: Isoniazid and Rifampicin sensitive, I1 - I7: Isoniazid resistant.
and resistant to Isoniazid. The larger cluster consisted of two isolates namely I1, I2, and SIR1, SIR2, and the smaller cluster consisted of one isolate (Figure 5(b)). It was found spesific DNA fragment at Isoniazid resistant isolate using primer N 9 is 1450 bp, there are 5 from the 7 isolates resistant to Isoniazid (Figure 5(a)) that are sample I1, I2, I3, I4, and I5, on location 4 - 8.
4. DISCUSSION
RAPD is faster and technically less demanding than most other molecular typing methods and furthermore, no DNA sequence information is necessary. Also, much smaller amounts of purified DNA are required than for methods such as Restricted Fragment Length Polymorphism (RFLP). Although RAPD is relatively simple and useful for epidemiological analysis, standardization of the PCR mixture and conditions are very important for reproducibility [12]. Finally, even with this standardization, reproducible profiles were difficult to obtain, and it was necessary to perform duplicate analysis for the true profile differences to be differentiated from experimental variation.
Figures 1-5 show band of fragment DNA with different thickness. The thicker fragment represents higher DNA concentration as compared to the thinner bands. Variation of DNA profile is present which can be seen by the difference of fragment number and difference of fragment size. The relatedness of isolates was recovered from different patients, and multi varieting isolates were recovered from the patients from similar location.
The dendrogram of M. tuberculosis Isoniazid resistant isolates showed a high degree of polymorphism with RAPD analysis (Figures 1(b)-5(b)). The dendrogram analysis M. tuberculosis Isoniazid resistant isolates with primer OPN 02, OPN 09, OPN 20, BG 65, and N 9, that have genetic diversity. The highest is primer N 9; and the lowest is primer OPN 20.
I found specific DNA fragment at Isoniazid resistant isolates using primer N 9 is 1450 bp as a marker. This study gives information about RAPD marker of M. tuberculosis strain to Isoniazid resistant.
5. ETHICAL CLEARANCE AND INFORM CONSENT
This study has been ethical clearance before being carried out, with number 1464/H4.8.4.5.31/PP36-KOMETIK/ 2012 under the approval of the head of Ethical clearance Hasanuddin University, Makassar, Indonesia. Notifications were concerned not be done because those samples used are isolate and taken from Provincial Health Laboratory of South Sulawesi, Indonesia and they have got the permission from the head of Provincial Health Laboratory of South Sulawesi, Indonesia with number PL00.03.3.2.749.
6. ACKNOWLEDGEMENTS
The authors are grateful to Provincial Health Laboratory of South Sulawesi, and Biotechnology laboratory of Research Centre Hasanuddin University, Indonesia; the authors thank Debbie Soefie Retnoningrum, PhD and Dr. Fatmawaty Badaruddin for their critical review in this manuscript. This study was supported by Hibah Pascasarjana of Hasanuddin University, Makassar, Indonesia.
REFERENCES
- Mendez, J.C. (2001) Multi drug resistance in tuberculosis and the use of PCR for defining molecular markers of resistance, Florida.
- Palomino, J.C. (2006) Newer diagnostic for tuberculosis and multi-drug resistant tuberculosis, mycobacteriology unit. Institute of Tropical Medicine, Antwerp.
- Health Department Republic of Indonesia (2011) pedoman nasional pengendalian tuberkulosis. jakarta.
- Cole, S.T., et al. (2005) Tuberculosis and the tubercle bacillus. ASM Press, Washington DC.
- World Health Organization (2010) Multidrug and extensively drug-resistant TB (M/XDR-TB). Global Report on Surveillance and Response, Geneva.
- Rattan, A., et al. (2006) Multidrug-resistant mycobacterium tuberculosis: Molecular perspectives. All India Institute of Medical Sciences, Ansari Nagar, New Delhi.
- Newton, C.R. and Graham, A. (1994) Introduction to biotechniques PCR. Bios Scientific Publishers, Oxford.
- Richner, S., Meining, J. and Kirby, R. (1999) DNA profiling of mycobacterium tuberculosis from the Eastern Cape Province of South Africa and the detection of a high level of genetic diversity, department of biochemistry and microbiology. Rhodesh University, Grahamstown.
- Bardakci, F. (2001) Random Amplified Polymorphic DNA (RAPD) markers. Cumhuriyet University, Faculty of Arts and Science, Department of Biology, Tubitak.
- Tazi, L., et al. (2004) Genetic diversity and population structure of mycobacterium tuberculosis in casablanca, a moroccan city with high incidence of tuberculosis. Journal of Clinical Microbiology, 42, 461-466.
- Korzekwa, K., Polok, K. and Zielinski, R. (2006) Application of DNA markers to estimate genetic diversity of mycobacterium tuberculosis strains. Poland Journal of Microbiology, 55, 19-24.
- Singh, J.P.N., Verma, R. and Chaudhurt, P. (2006) Random Amplified Polymorphic DNA (RAPD) analysis of mycobacterium tuberculosis strains in India. Journal of Veterinary Science, 7, 181-187.
- Jain, A., et al. (2008) Prevalence of multidrug resistant mycobacterium tuberculosis in Lucknow, Uttar Pradesh. Department of Microbiology & Clinical Epidemiology Unit, CSM Medical University, Lucknow.
- Va’zquez-Marrufo, G., et al. (2008) Genetic diversity among mycobacterium tuberculosis isolates from Mexican patients. Canadian Journal of Microbiology, 54, Ingenta Connect.
- Rodriguez, J.C., Royo, G. and Rodriguez-Valera, F. (2009) Application of four molecular techniques for typing outbreak-associated mycobacterium tuberculosis strains. Acta Pathologica Microbiologica Et Immunologica Scandinavica.
- Baron, E.J., Peterson, L.R. and Finegold, S.M. (1994) Baily & scotts diagnostic microbiology. 9th Edition, Mosby—Year Book, Toronto.
- Mahon, C.R. and Manuselis, G. (1995) Textbook of diagnostic microbiology. W.B. Saunders Company, Tokyo.
- Promega Corporation (2012) Wizard genomic DNA purification Kit, isolating genomic DNA from gram positive and gram negative bacteria. Madison, 16-17.
- Harn, H.-J., et al. (1997) Evidence of transmission of mycobacterium tuberculosis by random amplified polymorphic DNA (RAPD) fingerprinting in Taipei City, Taiwan. Department of Pathology, General Hospital, Ting-Chow Rd, Taipei.
- Linton, C.J., et al. (1995) Comparison of random amplified polymorphic DNA with restriction fragment length polymorphism as epidemiological typing methods for mycobacterium tuberculosis, Bristol.

