American Journal of Molecular Biology
Vol.05 No.03(2015), Article ID:58160,9 pages
10.4236/ajmb.2015.53007
Identification and Quantification of Corn, Soybean and Cotton Genetically Modified by Real Time PCR
Haiko Enok Sawazaki1, Aildson Pereira Duarte1, Milton Geraldo Fuzatto1, Eduardo Sawazaki1, Silvio Henrique Reginato Grandi2, Jéssica Funari de Ponte2, Larissa Nogueira2
1APTA-Instituto Agronômico de Campinas (IAC), Campinas, Brazil
2Trainee with Scholarship-IAC, Campinas, Brazil
Email: henok@iac.sp.gov.br
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 27 March 2015; accepted 19 July 2015; published 22 July 2015
ABSTRACT
In order to obtain a cheaper method for quantification of transgenic events in corn, soybeans and cotton, primers for real time PCR have been developed and optimized, with fluorescent BRYT Green system. The DNA was extracted from grains, with and without event, by CTAB method. The following events have been studied for corn: MON810, Bt11, MON89034, GA21, TC1507, NK603, MIR162, PRO3; Soybean: GTS-40-3-2, MON87701; MON89788; for cotton: MON1445, MON531, LLCotton25, 281-24-236; 3006-210-23, GHB614, T304-40; GHB119, MON15985, MON88913, besides the respective primers for the endogenous genes of corn, soybean and cotton. The sensitivity was 0.057%, the coefficient of linearity R2 ranged from 0.98 to 0.99 and the efficiency of PCR 0.9 to 1.1. The quantification of events ranged from 92 to 115, with a relative error (RE) from 2 to 18%, and a variance of 0.33 to 3.0. The precision acceptance criterion was observed for all analyses, as well the repeatability and reproducibility. As it was found that the measurement of accuracy and reproducibility were within the international acceptance criterion, it may infer the robustness of the methodology. Therefore, the results from replicates with two different technicians, and validation of results by comparison with those obtained by Eurofins Brazil, showed the possibility of specific and quantitative analysis of transgenic events with a cheaper method with sensitivity, repeatability and robustness.
Keywords:
Transgenic Events, Quantification, Corn, Soybean, Cotton

1. Introduction
The detection and quantification of genetically modified organism (GMO) are required by the countries to which Brazil exports food to. In Brazil, the limit of 1% of GMOs is determined by 4680 Decree of 24 April 2003 [1] being GM labeling mandatory for food with presence above the limit of 1.0% of the final product. Corn, soybean and cotton are genetically modified to express foreign proteins to manage lepidopteran insect pests or to allow application of herbicides (glyphosate and/glufosinate) to control weeds.
The technique of quantitative analysis performed by event-specific real-time PCR, using Taqman is, the official method used in Europe, whose methods are validated by the European Union Reference Laboratory for GM Food and Feed (EU-RL GMFF at http://gmo-crl//.jrc.ec.europa.eu/statusofdoss.htm) and found in the JRC Compendium of Reference Methods for GMO analysis (JRC-ISO/FDIS).
The methodology with fluorescence system Taqman uses probes, in addition to the primers. The fluorescence system BRYT Green has the same principle of detection of PCR products. The advantage of this system is that the fluorescent reagent is cheaper than the Taqman (there are similar dyes as BRYT Green, Evagreen, SYBR Green and the offer of SYBR is high due to be produced by several companies in several countries) and requires no fluorescent probe. The disadvantage is that it can lead to false positive signal when binding to non-specific DNA double strand occurs, requiring the development of specific primer and optimization of reaction to amplify only the desired band.
The objective is to obtain a cheaper and efficient methodology for diagnosis and quantification of transgenic events using BRYT Green (SYBR) in real time PCR, for corn, soybeans and cotton, through the development of specific primers, with efficiency PCR in the range 99% - 101%, in order to facilitate the processes of agribusiness, since the detection and quantification of genetically modified organism (GMO) is required in almost all countries which Brazil exports food to.
For validation, the same DNA samples tested were quantified by Eurofins Brazil (part of the international laboratory which uses certified material and Taqman system). The best reaction conditions were then used in three assays to quantify the event, with the same analyst, and with a different analyst to evaluate the linearity, sensitivity, limit of detection, limit of quantification, accuracy, repeatability, reproducibility and robustness.
2. Materials and Methods
2.1. Material and Events
The events studied are shown in Table 1, the samples are shown in Table 2. After homogenization and grinding the sample, two hundred milligrams were used for DNA extraction by the method of bromide Cetyltrimethyl ammonium bromide (CTAB) as in [2] . The integrity and quantification of extracted DNA were observed using electrophoresis.
The purity of DNA was checked with the inhibition test performed with standard curves from a sample called “undiluted” using endogenous primers, i.e., the values of Ct (Threshold cycle: is the cycle in which each amplification curve crosses the threshold line, serving as a basis for comparison between samples; threshold is the detection threshold set by the user to analyze results at the end of a real-time PCR) of the endogenous gene amplification, were compared with the data extrapolated from the calibration curve. The criteria accepted by the Community Reference Laboratory for Genetically Modified Food and Feed (CRL-GMFF) [3] for the absence of PCR inhibitors is when the average difference (ΔCt) between the measured value and the extrapolated Ct value for the “undiluted” sample is <0.5 cycles and the “slope” between −3.6 and −3.1.
2.2. Real Time PCR
Specific primers designed from the region 5' or 3' end of the genome/insert interaction, with the Primer 3 program were optimized. The initiators of endogenous reference genes for corn, soybean and cotton, respectively, adh1 (ADH, maize alcohol dehydrogenase), lec (LEC, lecithin) and adhC (ADH, cotton alcohol dehydrogenase C gene) were used. The reactions and conditions were optimized for the 7500 Fast Real Time Applied Biosystems (APPLIED BIOSYSTEMS) to a volume of 15.0 µl with 7.5 µl of the mix BRYTTM Green (Go Taq qPCR Master Mix of PROMEGA).
The efficiency of PCR standard curve was calculated from the value of “slope” being:
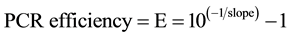
Table 1. GM events for corn, soybean and cotton with their respective proteins.
*TL means lepidopteran tolerance and TG glyphosate tolerance.
2.3. Optimization of Standard Curve
Standard curves were performed for event and endogenous reference. For each sample, the amount of the event was determined from curves standard, and reference.
The standard curve was taken with 20%, 2.86%, 0.41% and 0.057% of DNA event, mixed with none event, for reaction of 100 ng DNA.
The absolute number of copies of the standard curve was determined by dividing the weight of DNA (nano-
Table 2. Samples of corn, soybean and cotton with and without transgenic event.
*Powercore was provided by Down Agroscience, Viptera3 by Syngenta Seeds andIntactaRR2PRO by Nidera Seeds.
grams) by the published average IC value as in [4] of genome DNA, for corn (2725 picograms), soybean (1.13 pg) and cotton (2.33 pg). Table 3 shows the values of copy number of the event in points of standard curve for samples of corn, soybean and cotton
For normalization of quantification of an event in a sample, the event copy number was divided by the copy number of the endogenous gene and multiplied by 100 to yield the percentage value:
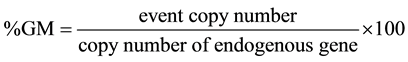
2.4. Validation
The evaluation of linearity, working range, sensitivity, and limit of detection, limit of quantification, repetitiveness, precision, reproducibility, accuracy, and robustness was made according to the parameters defined by CRL- GMFF [3] and DOQ-CGCRE-008-INMETRO [5] . CRL-GMFF [3] gives recommendations to evaluate and validate analytical methods of GMO, according with the Commission regulation (EC) No. 1829/2003 in Europe.
The linear range of work established when the method is linear with an acceptable level of accuracy and precision, is accepted to be 1/10 and at least 5 times the concentration required by legislation (JRRC 56609-Mon810). In Brazil, the GMO limit is 1%, determined by Decree 4680 [1] . Therefore, working range must be from 0.1% to at least 5%.
The parameter used for the sensitivity is the slope, being the acceptance criterion for the standard curve, the average value in the range of −3.1 to −3.6.
By law, the LOQ (limit of quantification) is less than 1/10th and the LOD (limit of detection), at least 1/20th of the threshold value; as in Brazil, the limit of GMOs is 1% the limits correspond to 0.1% and 0.05%.
The precision has been achieved by the repeatability and reproducibility. The repeatability of identification by three replicates for each measurement performed in the same analysis, determined by the coefficient of linearity R2 (correlation coefficient of a standard curve obtained by linear regression analysis) which should be ≥0.98, and the limit of repeatability (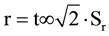 ). To a 95% significance level: r = 2.8∙Sr, where Sr is the standard deviation associated with the Ct readings for the same analysis.
). To a 95% significance level: r = 2.8∙Sr, where Sr is the standard deviation associated with the Ct readings for the same analysis.
The reproducibility was verified by analyses with two different technicians in different days, using the same apparatus under the same conditions of temperature and time previously optimized for each primer; the differences in the percentage of quantitation between the analyses must not be greater than the reproducibility limit R (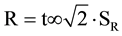 ); or, for a 95% significance level:
); or, for a 95% significance level: 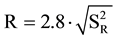 (where
(where 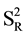 = variance of reproducibility of quantification percentage detected by the two technicians).
= variance of reproducibility of quantification percentage detected by the two technicians).
The accuracy criterion (agreement between the result of the laboratory and the reference value) defined as ±25% as in [3] , requires a reference value. The value used as a reference was the analysis of Eurofins in Brazil (from Eurofins Agroscience Services), although it was not possible to have all events analyzed. The same samples used for quantification studies were analyzed by Eurofins for validation and comparison of studied methodology.
The relative error (RE) was expressed as a percentage by means of the expression:
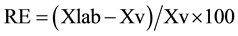
where: Xlab = value obtained experimentally or arithmetical average of obtained values; Xv = value accepted as true.
Table 3. Values and percentage of the number of copies of the events in the standard curve for samples of corn, soybean and cotton.
The robustness by the measures of reproducibility and accuracy was inferred within the limits stipulated by CRL-GMFF [3] that shall not deviate more than ±30%.
3. Results and Discussion
3.1. DNA Extraction Test
The performance of the extraction of DNA, which is essential for the success of PCR analysis, was tested for the presence of inhibitors. By the inhibitor test, no samples of corn, soybean and cotton, showed for the average difference (ΔCt) between the measured value and the extrapolated Ct value, ΔCt > 0.5 cycle, indicating the significant absence of inhibitors.
3.2. Specificity Analysis
The specificity of the primers developed in the region of genome/insert interaction, was tested by using a reaction of 15.0 µl, with 20 ng of DNA on “FAST” method (initial heating at 95˚C/2min followed by 40 cycles of denaturation, annealing and extension at 95˚C/10s and 60˚C/30s). All analysis showed amplification when performed with specific primer of Table 5 and were completely specific in relation to all events of the other samples studied. Also the dissociation peak showed practically only the correspondent peak for the studied event.
The concentration of primers (forward and reverse) used were the same or almost the one used in the quantification analysis (Table 4).
Table 4. PCR conditions of transgenic events in corn, soybeans and cotton and the amount used of primers of event or gene to obtain the standard curve.
Table 5. Specific primers developed by laboratory with original annealing temperature (T) in ˚C, amplification length (A) in base pairs and PCR efficiency (E) for the transgenic events or endogenous genes in corn, soybean, and cotton.
3.3. Quantitative Analysis
3.3.1. Optimization of Real-Time PCR Conditions for Obtaining the Standard Curve
The optimization of reaction conditions in real time pcr to obtain the standard curve was made using as parameter values required for validation by CRL-GMFF [3] . To this end, initially changes were tested in, primer concentration, temperature and time of annealing and extension time. Lower annealing temperature or longer time of annealing and extension can increase the fluorescence signal as it facilitates the annealing or amplification in some cases, however, may increase non-specific amplification. When the dissociation present more than one peak, indicating non-specific annealing, or even curve with the “shoulder” indicating not optimized reaction, the annealing temperature was increased and/or the concentrations of the primers decreased. Later, it was verified that is possible to reduce the time of reaction when the amount of primer was increased. Therefore, it was altered the conditions that requested a long time of reaction by increasing the amount of primer and using the same temperature for annealing and extension.
3.3.2. Real Time PCR
The reactions and conditions optimized for the 7500 Fast Real Time (APPLIED BIOSYSTEMS) to a volume of 15.0 µl with 7.5 µl of the mix BRYTTMGreen (Go Taq qPCR Master Mix of PROMEGA) used 133 - 500 nM of primer, with one step of 95˚C for 2 min followed by 40 cycles of denaturation at 95˚C for 15 sec, annealing and extension at 60˚C for 50 to 90 sec.
Table 4 shows for each transgenic event studied in corn, soybean, and cotton, the amount of primer used in the reactions optimized to obtain the standard curve of each event or endogenous gene and the annealing and extension conditions.
Table 5 shows the best primers developed in the laboratory for the studied events with data used in the standard curve, as the original annealing temperature in ˚C (T), amplification length in base pairs (A) and PCR efficiency (E).
The regression curves obtained for all events showed the coefficient of linearity R2 from 0.98 to 0.99 with Cts from 21.04 to 29.62 and 24.59 to 33.67, and variation of PCR efficiencies from 0.9 to 1.1. Regarding the curves of the endogenous gene made for each event, the R2 was almost 0.99 with efficiency of PCR, of 0.91 to 1.1. Those values are within the acceptance criterion of CRL-GMFF [3] .
3.4. Validation
The linearity measured by PCR efficiency was from 0.9 to 1.01 indicating that the PCR product doubled every
cycle when all reagents are also available, demonstrating the linear response.
The criterion working range 0.1% to until at least 5% was observed for the working range of 0.057% to 20% for the standard curve reaction using 100 ng DNA.
The values of LOD and LOQ (0.1% and 0.05%) were practically followed, as the minimum used was 0.057%.
For all three analysis repetitions, the differences in the percentage of quantitation between the analyses were not greater than the repeatability limit and the R2 was ≥0.98.
The variance of reproducibility of quantification for all events (0.33 to 3.0) originated by the variance of the average results of repeating in three days or by different technicians, was lower than the limits of reproducibility and within the parameter set by CRL-GMFF [3] which is ≤30% showing the reproducibility.
The precision was observed for all analysis, as well the repeatability and reproducibility.
Table 6 shows data for validation by comparison the quantification average results (some analyses could not be made by Eurofins) obtained by Eurofins and our laboratory, with the average variance, the coefficient of linearity R2 and the relative error (RE). The variance is related to comparison of at least three laboratory analyses on different days.
The average quantification of laboratory tests carried at least three times on different days (at least two repetition tests with the same technician, and one with different technician) ranged from 92 to 115. By CRL-GMFF [3] , the accuracy criterion is ±25%. The results of the relative errors (RE) methodologies between our laboratory and Eurofins (except for PRO, NK603 and GA21, which clearly have had problems because all samples were pure event), were from 2 to 18, below the stipulated by the CRL-GMFF [3] , showing the accuracy of the laboratory
Table 6. Validation by comparison quantification percentage averages of laboratory and Eurofins.
method. As it was found according to the measurement of accuracy and reproducibility that the method was within the limits set by the CRL-GMFF [3] , it may infer the robustness of the methodology.
4. Conclusion
For twenty-one transgenic events tested in corn, soybean and cotton have been observed for all developed primers, the overall specificity for each event, the limit of quantification (LOQ) of 0.057%, PCR efficiency in the range 0.9 to 1.1. The R2 ranged from 0.98 to 0.99. The relative error (ER) for quantification samples with events ranged from 2% to 18%. The precision was observed for all analyses, as well the repeatability and reproducibility. As it was found according to the measurement of accuracy and reproducibility that the method was within the international acceptance criterion, it might infer the robustness of the methodology. Therefore, the results from replicates with two different technicians, and validation of results by comparison with those obtained by Eurofins Brazil, showed the possibility of specific and quantitative analysis of transgenic events with a cheaper method with sensitivity, repeatability and robustness.
Acknowledgments
To Fundação de Amparo a Pesquisa no Estado de São Paulo-FAPESP by financial support and TT-2 scholarship, and CNPq for the PIBIC scholarship.
Cite this paper
Haiko EnokSawazaki,Aildson PereiraDuarte,Milton GeraldoFuzatto,EduardoSawazaki,Silvio HenriqueReginato Grandi,Jéssica Funaride Ponte,LarissaNogueira, (2015) Identification and Quantification of Corn, Soybean and Cotton Genetically Modified by Real Time PCR. American Journal of Molecular Biology,05,84-93. doi: 10.4236/ajmb.2015.53007
References
- 1. DOU (2003) Decree n° 4680 on April 2003. Section I, Diário Oficial da União, 2.
- 2. Cardarelli, P., Branquinho, M.R., Ferreira, R.T.B., Cruz, F.P. and Gemal, A.L. (2005) Detection of GMO in Food products in Brazil: The INCQS Experience. Food Control, 16, 859-866.
http://dx.doi.org/10.1016/j.foodcont.2004.07.010 - 3. CRL-GMFF (2009) Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing.
http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requirements_Analytical_methods.pdf - 4. Arumuganathan, K. and Earle, E.D. (1991) Nuclear DNA Content of Some Important Plant Species. Plant Molecular Biology Reporter, 9, 208-218.
http://dx.doi.org/10.1007/BF02672069 - 5. DOQ-CGCRE-008-INMETRO (2010) Orientação sobre validação de métodos de ensaios químicos. Revisão 03. 20 p.
http://www.inmetro.gov.br/Sidoq/Arquivos/CGCRE/DOQ/DOQ-CGCRE-8_03.pdf


