American Journal of Molecular Biology
Vol.04 No.04(2014), Article ID:50749,15 pages
10.4236/ajmb.2014.44020
Leptin Signaling Modulates Expression of Polycomb and Trithorax Complexes in the Brain of Fat Tissue Implanted Polycystic Ovarian Sindrome Mice
Eduardo Henrique da Silva Freitas1,2, Samuel Marcos Ribeiro de Noronha3, Maria Nazareth Gamboa Ritto1,4, Carlos Fernandes Baptista1,4, Ismael Dale Cotrim Guerreiro da Silva1, Silvana Aparecida Alves Correa-Noronha1,3*, Ivaldo da Silva1
1Gynecology Department, Federal University of Sao Paulo, Paulista School of Medicine (UNIFESP-EPM), Sao Paulo, Brazil
2General Medicine Department, Federal University of Rio de Janeiro State (UNIRIO), Rio de Janeiro, Brazil
3Translacional Surgery Division, Surgery Department, Federal University of Sao Paulo, Paulista School of Medicine (UNIFESP-EPM), Sao Paulo, Brazil
4General and Specialized Surgery Department, Federal University of Rio de Janeiro State (UNIRIO), Rio de Janeiro, Brazil
Email: eduardo@eduardohsfreitas.com.br, labgineco@globo.com, nazarethritto@gmail.com, carlosfb@centroin.com.br, ismael.dale@gmail.com, *silaac@globo.com, ivaldo.silva@gmail.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 12 August 2014; revised 10 September 2014; accepted 8 October 2014
The Polycystic Ovary Syndrome (PCOS) is the most common androgenic disorder in women during reproductive life. PCOS may also be accompanied by metabolic syndrome and recent studies point to leptin as playing a role in disrupting infertility and in changing the energy balance in obese mice through its action on the hypothalamus. The aim is to assess the expression of the Polycomb & Trithorax Complexes genes in brain of mice transplanted with fat tissue from normal mice, in order to better understand the neuronal mechanisms underlying the reversion of PCOS. Three B6 V-Lepob/J mouse groups: Normal weight, obese and seven-day-treatment obese had their brain RNA extracted and submitted to an 84 Polycomb & Trithorax Complexes genes PCR Array plate and MetacoreTM pathways localization. Genomic profiles obtained were compared to the ones of the normal-weight-mice group. Differentially expressed genes were 13% and 26% respectively to control and treatment. Major changes were in genes: Snai1/31; Smarca1/−17; Dnmt3b/4.7; Ezh1/ 15. Altered genes were associated to canonical pathways and provided 3 networks related to epigenetics. Underlying neuronal changes caused by leptin in obese mice brain, there is an important role being played by the histone code. Here there is evidence that leptin drives the chromatin packing to a more condensed pattern. Upregulation of methyltransferase genes, like Ezh1, favors this thought. In summary the Polycomb & Trithorax complexes might answer for the silencing of some downregulated genes in the obese mice brain when exposed to leptin.
Keywords:
Polycistic Ovarian Sindrome, Obesity, Polycomb and Trithorax Complexes, Leptin, Fat Mouse

The Polycystic Ovary Syndrome (PCOS) is the most common androgenic disorder in women during reproduc- tive life [1] . PCOS may also be accompanied by metabolic syndrome and recent studies point to leptin as play- ing a role in disrupting infertility and in changing the energy balance in obese mice through its action on the hypothalamus [2] [3] .
It is known that epigenetic mechanisms times the initiation of female puberty in rats and regulates the re- lease of kisspeptins, gonadotropin releasing hormone (GnRH), follicle stimulating hormone (FSH) and luteining hormone (LH) in mouse [4] . DNA methylation is involved in the regulation of many genes expressed in a tissue, playing a crucial role in determining cells fate [5] and it is closely related to chromatin structure [6] [7] . Cellular differentiation involves various genome-wide histone (H) modifications that have been directly linked to active and inactive structures of the chromatin [8] [9] . Acetylation of lysine (K) residues is a prevalent and reversible posttranslational modification and diminishes the interaction between histone and DNA molecules facilitating the binding of non-histone proteins to targeted regions of the DNA [10] . It is also known that nonhistone me- thylation may function as a fingerprint that establishes repressive and active chromatin configuration at target loci causing heritable changes in gene expression [11] . The histone code tightly controls DNA packaging and chromatin remodeling, which leads to repression or activation of gene transcription [12] .
Proteins of the polycomb group exert their function by forming three multi-protein complexes that act as transcriptional repressors, PRC1, PRC2 and PhoRC [11] . The PRC2 complex trimethylates H3K27, a well- known mark for silenced chromatin associated with promoters and regulatory elements of PcG target genes [13] - [15] . Furthermore the PhoRC interacts with H3K27 upstream of PcG target genes and PRC1 complex maintains the chromatin in a silenced state by binding to H3K27me3 [16] .
Trithorax group (TrxG) proteins form multi-subunit complexes to exert their functions as well. Three com- plexes have been purified in mammalian cells so far, SET1-like, BRM and the MLL supercomplex [11] . The SET1-like complex contains HMTase activities and also methylates lysine 4 of histone 3, which is highly asso- ciated with promoter regions of transcription active loci [17] - [19] . The BRM complex contains the SWI/SNF chromatin-remodeling ATPase BRM [20] and the MLL supercomplex has got HMTase and chromatin remode- ling activities [21] .
The objective of this study is to assess the expression of the Polycomb & Trithorax Complexes genes in the brain of mice transplanted with fat tissue from normal mice, in order to better understand the neuronal mechan- isms underlying the reversion of PCOS.
2.1. Experimental Groups
B6.V-Lepob/J mice with 2 and 3 months of age were divided into three groups: normal weight control (lean), obesity control and obese seven days mice transplanted with adipose tissue from lean mice. After treatment, these mice were sacrificed and their brains were used to total RNA extraction.
2.2. RNA Extraction
After using liquid nitrogen for cryogenic soaking, tissues were homogenized in Trizol reagent (Invitrogen) ac- cording to the manufacturer’s protocol. Total RNA was purified with Qiagen RNeasy Mini Kit and treated with DNase A. The quantity and quality of extracted RNA were measured by espectrophotometer (Nanodrop Tech- nologies Inc., Rockland, DE).
2.3. Real Time PCR Array
According to the manufacturer’s (Qiagen) methodology, reverse transcriptase (RT) was carried out for the syn- thesis of cDNA. For each sample we used a PCR array plate containing 84 different pairs of primers as a tem- plate in order to study the expression of genes related to the Polycomb & Trithorax Complexes (RT² Profiler™ PCR Array; SABiosciences).
2.4. Analysis of Relevant Biological Processes and Networks by MetaCore
The MetaCore software (GeneGo, St. Joseph, MI) is a computational resource that uses logic operations for identifying biological processes that are altered because of changes in gene expression. Genes with altered ex- pression were mapped to Gene Ontol’ogy (GO) using MetaCore algorithm. GO annotations were used as indi- cators of the biological functions. GO describes gene products in terms of their associated biological processes, cellular components, and molecular functions. The GO entries are hierarchically linked, thus allowing construc- tion of cluster genes of crossed pathways.
2.5. Statistical Analysis
These results were analyzed by descriptive statistics (means and standard deviation) and inferential statistics through the Student’s t-test, with significance level of 5% (p < 0.05). Real-time PCR array reactions were pro- cessed through the online software RT2 Profiler™ PCR Array Data Analysis (SABiosciences).
3.1. Experimental Results for Treatment with Implant of Gonadal Adipose Tissue during Seven Days
The results presented in this subsection is relate to the treatment for seven days with implantation of gonadal fat in obese ob/ob mice.
Figure 1 and Figure 2 show the gene expression of all genes by the PCR array plate of genes related complex Polycomb and Trithorax genes in the form of heat or dispersion points graph, respectively.
After treatment for seven days with the implantation of gonadal fat from lean mices, we performed the analy- sis of gene expression data. The treated mice were compared to untreated controls. And, after this comparison, the values of Fold Regulation (FR), which expresses how many times increase (hyper-regulation) or decreased (hypo-regulating) the expression of a specific gene occurs with which build Tables 1-3 were obtained and the respective Figures 3-5, shown immediately.
The increase in gene expression was determined when the RF is greater than 2.0 or less than −2.0. Only con-
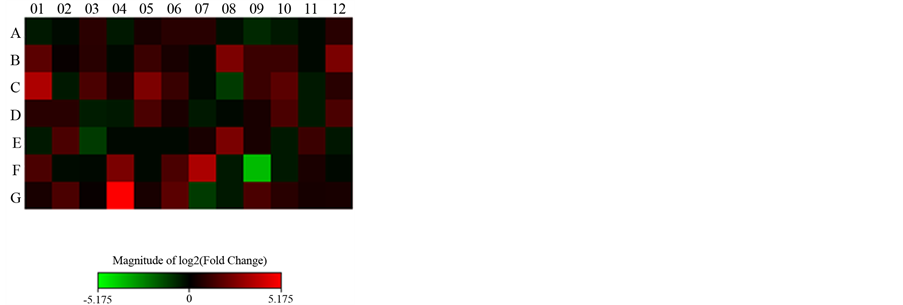
Figure 1. Heat map for treatment after seven days.
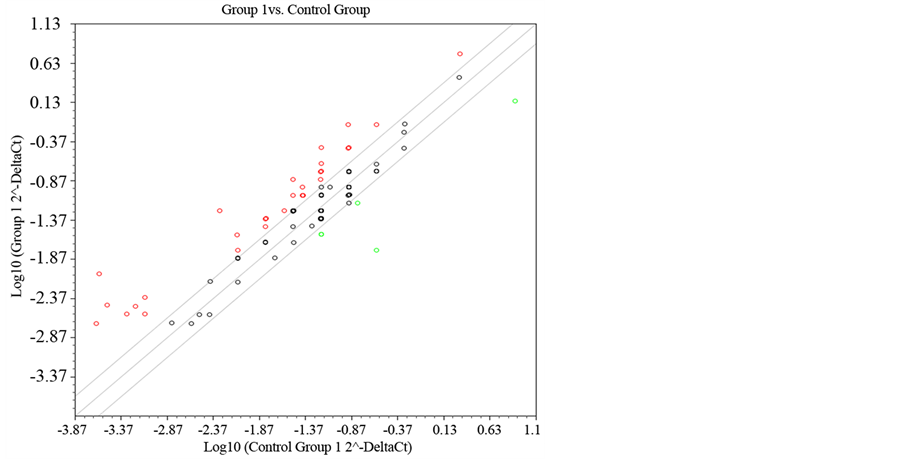
Figure 2. Scatter Plott treatment after seven days.
Table 1. Polycomb complexes: Nuclear Components: Eed and Rbbp4. Alternative Nuclear Components: Ezh1. Gene Complex Interaction: BAP. Additional components of the complex: Pcgf2 (Rnf110). CofatoresPolycomb Complex Assembly: Snai1.
FR = Fold Regulation; *p < 0.05; #p for trend.
Table 2. Polycomb cofactors: Genes Complex Interaction: Bap1, Ctbp2, Htt, Ino80b, Larp7, Mov10, Pcgf5, Trim27, Usp11. Nuclear Complex: Scmh1.
FR = Fold Regulation; *p < 0.05; #p for trend.
Table 3. Trithorax Complexes: Nuclear Components: Rbbp4, Rbbp7, Smarca1, Wbp7. Genes Complex Interaction: Smarcc1.
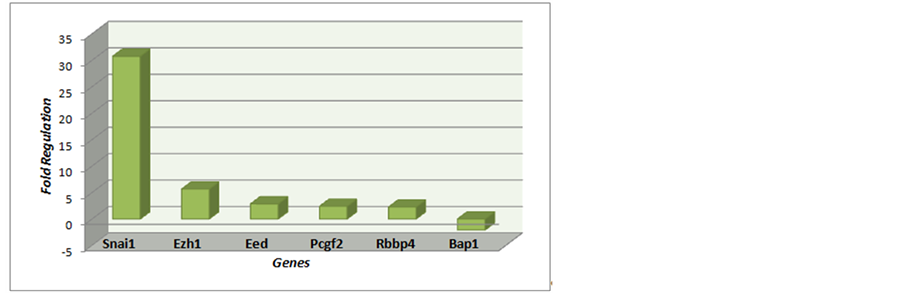
Figure 3. Polycomb complexes: Nuclear Components: Eed and Rbbp4. Alternative Nuclear Components: Ezh1. Gene Complex Interaction: BAP1. Additional components of the complex: Pcgf2 (Rnf110). CofatoresPolycomb Complex Assembly: Snai1.
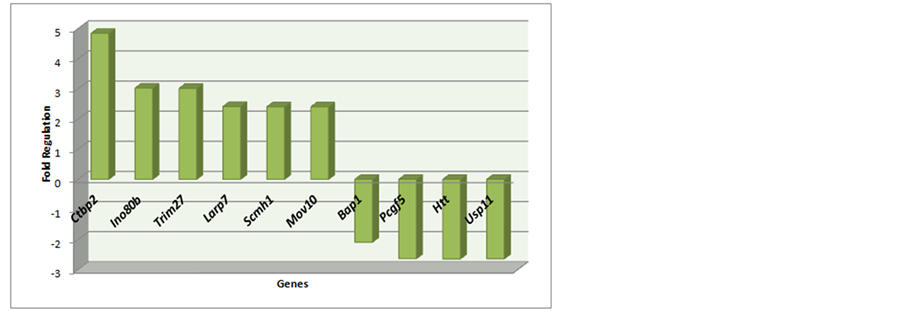
Figure 4. Polycomb cofactors: Genes Complex Interaction: BAP1, Ctbp2, Htt, Ino80b, Larp7, Mov10, Pcgf5, Trim27, and Usp11. Nuclear Complex: Scmh1.
sider the values on which the p value was less than 0.05 and significant.
Table 1, Table 2, Figure 3 and Figure 4 are on the Polycomb genes in Complex and Table 3 and Figure 5 is on the data to Trithorax complex.
Most genes are presented hyper-regulated (71%); furthermore, most belong to the Polycomb complex (71%) than to Trithorax complex (21%).
3.2. Experimental Results for Treatment with Implant of Gonadal Adipose Tissue during 45 Days
The results presented in this subsection relate to the treatment for 45 days in implant gonadal fat in obese mice
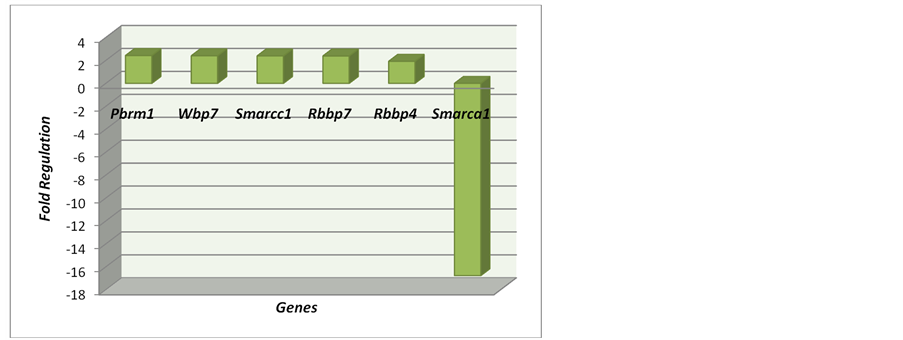
Figure 5. Trithorax Complexes: Nuclear Components: PBRM1, Rbbp4, Rbbp7, Smarca1, Wbp7. Genes Complex Interaction: Smarcc1.
ob/ob mice.
Figure 6 and Figure 7 show the gene expression of all genes in the PCR array plate of genes related complex Polycomb and Trithorax maps in the form of heat or dispersion points respectively graph.
After treatment for 45 days with the implantation of gonadal fat from lean rats, we performed the analysis of gene expression data. The treated mice were compared to untreated controls. And, after this comparison, the values obtained Fold Regulation (FR), which expresses how many times increase (hyper-regulation) or de- creased (hypo-regulating) the expression of a specific gene occurs, and build Table 4 and corresponding Figure 8, soon displayed. In this table and figure genes related to the Polycomb Complex Complex Trithorax are pre- sented. All hyper-regulated genes were most genes (89%) belong to the Polycomb complex.
Figure 9 shows the comparison between 45 and 7 days treatment of obese mice with gonadal adipose tissue from lean mice.
3.3. Metacore Analysis
In Figures 10-12 we can see the pathways constructed by the genes in the program Metacore® (Thonson Reuters) for the genes related to Polycomb and Thritorax that presented with differential expression.
Polycomb (PcG) proteins were originally identified as part of an epigenetic cellular memory system that controls gene silencing via chromatin structure. However, recent reports suggest that they are also involved in controlling dynamics and plasticity of gene regulation, particularly during differentiation, by interacting with other compo- nents of the transcriptional apparatus [22] .
Enhancer of zeste homolog 1 (Ezh1) is a gene that encodes a PcG protein that initiates repressionin certain chromatin domains and regulates developmental genes expression, which is closely related to cell proliferation [23] . This gene is up-regulated after 7 days of treatment
Others genes related to transcription factors, DNA methyltransferases and chromatin remodeling proteins member were also studied. Snail homolog 1 (SNAl1) proteins are zinc-finger that play important roles in deter- mining cell-fate [24] . This gene isup-regulated after 7 days of treatment.
DNA methyltransferase 3B (Dnmt3b) dynamically regulate chromatin remodeling and gene expression. DNA methyltransferases contribute to the establishment and maturation of cell fate during retinal development [25] . This gene is also upregulated after 7 days of treatment.
SWI/SNF related, matrix associated, actin dependent regulator of chromatin (Smarca1) is a component of the chromatin remodeling complex. This gene is related to neuron differentiation, brain development and chromatin remodeling. In the brain, SMARCA1 is prevalent in proliferating cell populations whereas, it is predominantly expressed in terminally differentiated neurons after birth and in adult animals. These results suggest that SMARCA1 complexes have distinct functions associated with cell maturation or differentiation [26] . This gene is downregulated in this study.
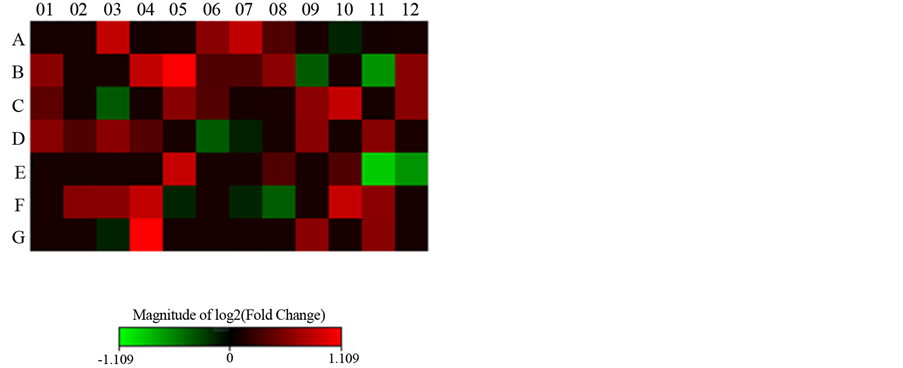
Figure 6. Heat map after treatment for 45 days.
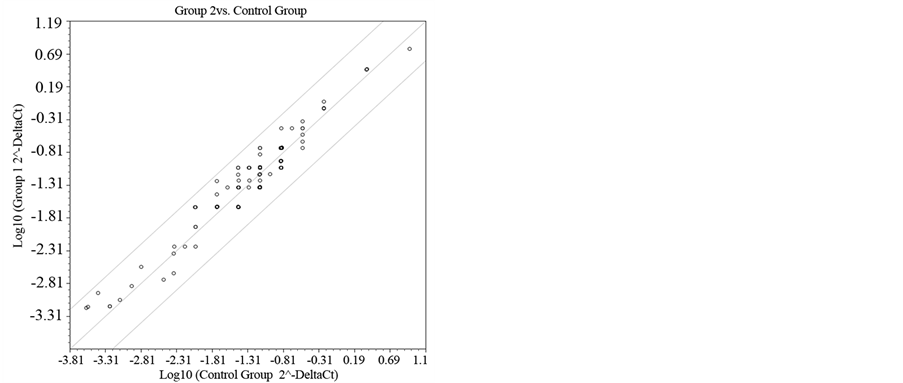
Figure 7. Scatter Plott after treatment for 45 days.
Table 4. Polycomb cofactors: Genes Complex Interaction: Cbx7, Ctbp1, INO80, Ino80b, L3mbtl2. Complex Assembly: Snai1. Genes Complex Interaction: Trim27. Polycomb complex: Alternative Nuclear Components: Ezh1. Trithorax Complexes: Nuclear Components: Wdr5.
FR = Fold Regulation; *p < 0.05.
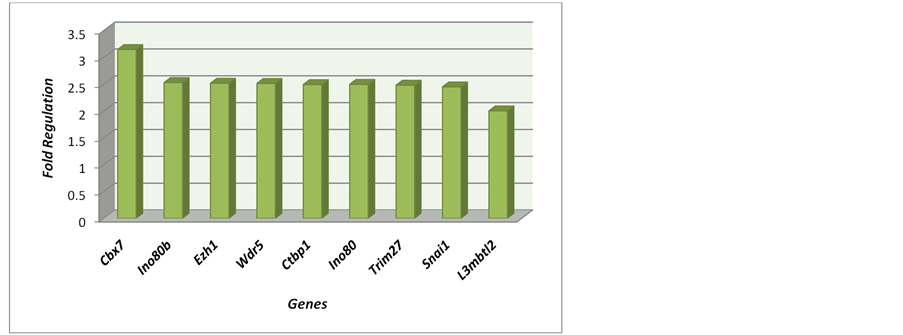
Figure 8. Polycomb cofactors: Genes Complex Interaction: Cbx7, Ctbp1, INO80, Ino80b, L3mbtl2. Complex Assembly: Snai1. Genes Complex Interaction: Trim27. Polycomb complex: Alternative Nuclear Components: Ezh1. Trithorax Complexes: Nuclear Components: Wdr5.
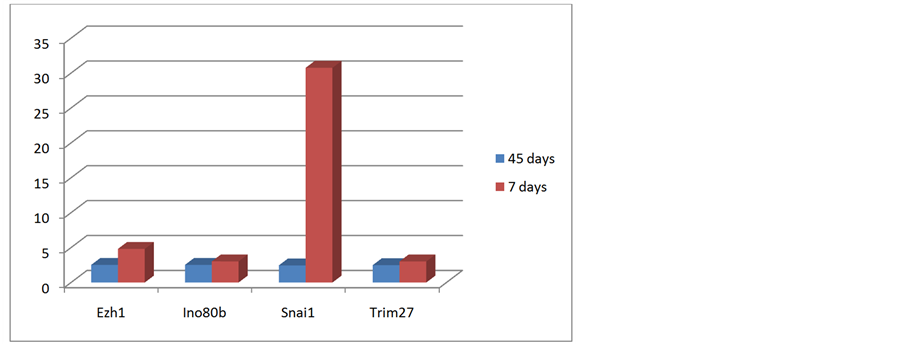
Figure 9. Comparison between 45 and 7 days of treatment with gonadal adipose tissue from lean mice.
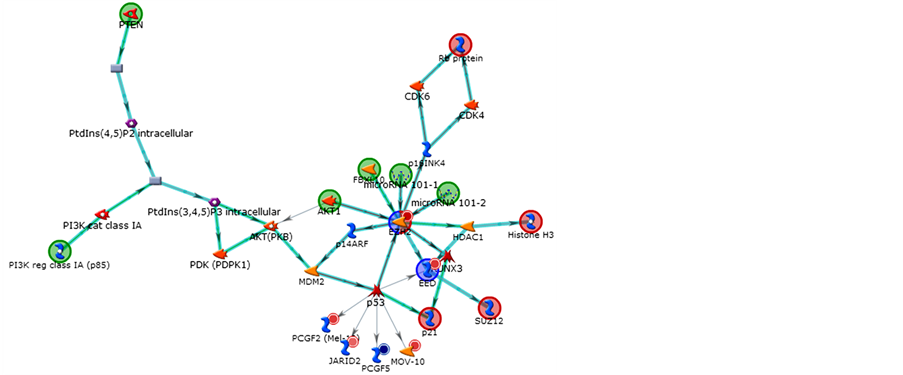
Figure 10. Frst network obtained by Metacore analysis of control vs. 7-day treatment: EZH2, p21, Rb protein, Histone H3, SUZ12. Main GO Process related: regulation of biological process (90.2%). Organic (Hub-centric).
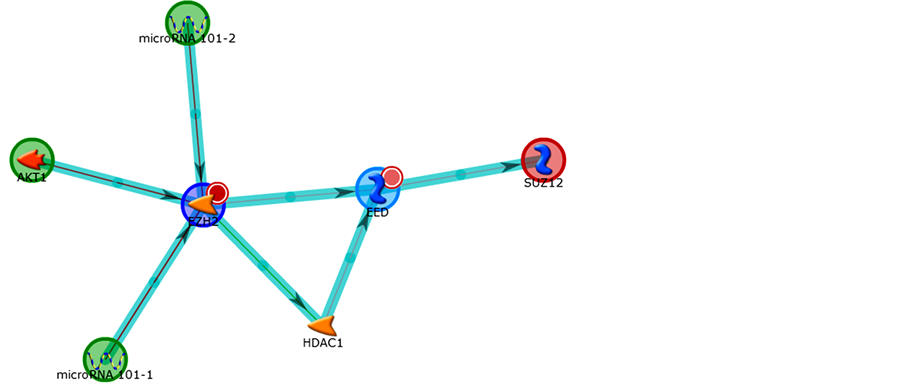
Figure 11. Second network obtained by Metacore analysis of control vs 7- day treatment: SUZ12, EZH2, AKT1, microRNA 101-2, microRNA 101-1. Main GO Process related: covalent chromatin modification (66.7%). Organic (Hub-centric).
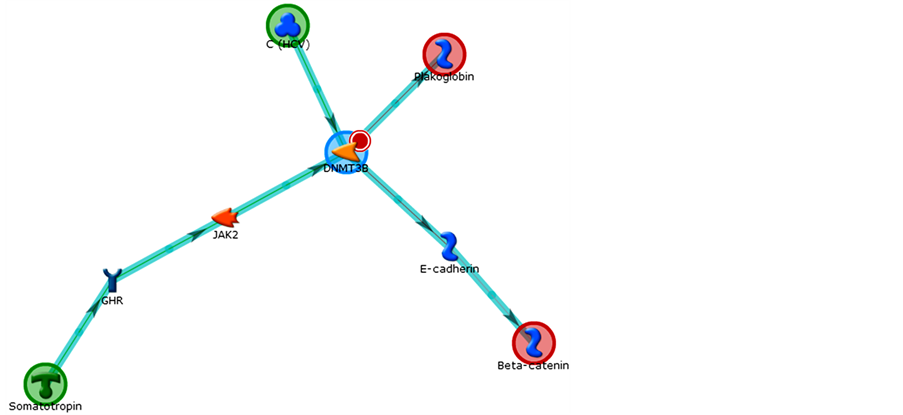
Figure 12. Third network obtained by Metacore analysis of control vs. 7- day treatment: Plakoglobin, Beta-catenin, C (HCV), Somatotropin, DNM- T3B. Main GO Process related: response to oxygen-containing compound (100%). Organic (Hub-centric).
In summary, exogenous leptin drives the phisyological variables related to POS to a normal status (glycemia, ovulation and fertility, body weight and food intake) only after a 45-day treatment period. Exogenous leptin reestablishes a normal pattern of ovarian cycle controlling hormones release after a 7-day treatment period. Un- derlying neuronal changes caused by leptin in obese mice brain, there is an important role being played by the histone code. Here there is evidence that leptin drives the chromatin packing to a more condensed pattern. Upregulation of methyltransferase genes, like Ezh1, favors this thought. In summary the Polycomb & Trithorax complexes might answer for the silencing of some downregulated genes in the obese mice brain when exposed to leptin, like Neu4 and Scg2.
15 genes (68%) of that differentially expressed were related to polycomb and seven genes (32%) were related to trithorax.
Chromatin consists of proteins that serve as the structural organizer of DNA, binding DNA into higher order structures and ultimately forming the chromosome itself. Chromatin restricts the access of DNA to transcription factors. Both Polycomb and trithorax group proteins act to remodel chromatin altering the accessibility of DNA to factors required for gene transcription. Polycomb group genes are involved in chromatin based gene silencing, while trithorax group genes counteract the silencing effects of chromatin to maintain gene activity.
Begun et al. (2013) hypothetised that maternal undernutrition around conception (UN) in sheep would lead to epigenetic changes in hypothalamic neurones regulating energy balance in the offspring, up to five years after the maternal insult. This authors found striking evidence of decreased glucocorticoid receptor (GR) promoter methylation, decreased H3K27 trimethylation and increased H3K9 acetylation in hypothalami from male and female adult offspring of UN mothers. These findings are entirely compatible with the increased GR mRNA and protein observed in the hypothalami. The increased GR predicted the decreased hypothalamic pro-opiomela- nocortin expression and increased obesity we observed in the 5-year-old adult males. Theepigenetic and ex- pression changes in GR were specific to the hypothalamus. Hippocampal GR mRNA and protein were de- creased in UN offspring, whereas pituitary GR was altered in a sex-specific manner. In peripheral polymor- phonuclear leukocytes there were no changes in GR methylation or protein, indicating that this epigenetic analysis did not predict changes in the brain. Overall, these results suggest that moderate changes in maternal nutrition, around the time of conception, signal life-long and tissue-specific epigenetic alterations in a key gene regulating energy balance in the hypothalamus [27] .
De Giorgio et al. (2009) investigated the modifications in the hypothalamic gene expression induced by high-fat (HF) and low-fat (LF) meal ingestion in mice, in order to identify the signals rapidly mediating the hy- pothalamic control on energy intake. The hypothalamus was sampled and the serial analysis of gene expression method was performed. Approximately 254,588 tags, which correspond to 65,548 tag species, were isolated from the 3 groups. The data showed twelve transcripts regulated by food intake. Among these, 2 transcripts have mitochondrial functions (MtCo1, Ppid), 3 are involved in protein transport and regulation (Ube2q2, Mup1, Sec13), 1 in cellular pH control (Slc4a3) and another 1 has a role in the epigenetic control of gene expression (Setd3). In addition, 5 potentially novel transcripts were differentially modulated. So, these authors identified genes that may regulate hypothalamic circuits governing the early response to food intake 3 genes were specifically mod- ulated by high-fat intake [28] .
Makedonski et al. (2005) hypothesize that part of the Rett syndrome (RS), a neurodevelopmental disorder, phenotype is due to MeCP2-associated silencing of UBE3A. Indeed, UBE3A mRNA and protein are shown here to be significantly reduced in human and mouse MECP2 deficient brains. This reduced UBE3A level was asso- ciated with biallelic production of the UBE3A antisense RNA. In addition, MeCP2 deficiency resulted in ele- vated histone H3 acetylation and H3 (K4) methylation and reduced H3 (K9) methylation at the PWS/AS im- printing center, with no effect on DNA methylation or SNRPN expression. We conclude, therefore, that MeCP2 deficiency causes epigenetic aberrations at the PWS imprinting center. These changes in histonemodifications result in loss of imprinting of the UBE3A antisense gene in the brain, increase in UBE3A antisense RNA level and, consequently reduction in UBE3A production [29] .
There are no studies in the literature reporting the study of epigenetic modifications in brain of mice PCOS, diabetic or infertile. This present work is the first to study the alterations of genes related to these changes.
The genes of the Polycomb Complexes and cofactors that changed were: CBX2, Ctbp2, DNMT3B, Eed, EzH1, INO80B, JARID2, Larp7, Mov10, Pcgf2, Pcgf5, RNase I, Scmh1, Snai1 and Trim27.All genes in this group were up-regulated, except for Pcgf5. The most over-expressed genes were Snai1, Ctbp2, DNMT3B, and Ezh1 RnaseI.
Cromobox 2 (CBX2) is a component of Polycomb repressive complex 1 (PRC1) [30] .
C-terminal binding protein 2 (CTBP2), in mammals, are among the best characterized transcriptional core- pressors [31] . CtBPs were suggested to regulate neuronal differentiation and they were implied in the control of gene expression during epileptogenesis. However, the expression patterns of CtBP family members in specific brain areas and their subcellular localizations in neurons in situ are largely unknown. It is a component trans- criptional repressor complex.
Embryonic ectoderm development (EED) is related to chromatin modification and histone mthylation.
Preventing the eviction of EED from the Kiss1 promoter disrupted pulsatile gonadotropin-releasing hormone release, delayed puberty and compromised fecundity. This results identify epigenetic silencing as a mechanism underlying the neuroendocrine control of female puberty [4] [33] .
Enhancer of zeste homolog 1 (EZH1) belongs to PcG complex in promoting mRNA transcription [34] .
The genes related to Trithorax Complexes were Pbrm1, Rbbp4, Rbbp7, Smarca 1, Smarcc1 e Wbp7. Also, most of the genes were hyper-regulated, except for the gene Smarca, which was the most drastic change in the expression.
Polybromo 1 (PBRM1) has a function of chromatin binding and is related to process of chromatin modifi- cation and regulation of transcription, DNA-dependent. PBRM1 is envolved in inhibition of cell proliferation [43] .
Smarcc1, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 [Mus musculus, is related to process to chromatin modification and chromatin remodeling. SWI/SNF DNA chromatin remodeling complex family controlling epigenetic modifications, and signaling pathways might indi- cate differences in the therapeutic potential of our examined two cell populations in relation to in cell survival, proliferation, migration, and differentiation following engraftments in different CNS insults [45] .
Wbp7, a lysine (K)-specific methyltransferase 2B, has a function as DNA binding, histone methyltransferase activity (H3-K4 specific) and histone-lysine N-methyltransferase activity. The process related to chromatin mo- difycation, gene silencing, histone H3-K4 methylation, histone H3-K4 trimethylation, histone lysine methylation and methylation. There are a possible link between histone-modifying enzyme to a biosynthetic pathway and in- dicate a specialized biological role for Wbp7 in macrophage function and antimicrobial response [46] .
The Polycomb group (PcG) and Trithorax group (TrxG) of proteins have long been recognized as regulators that maintain the gene expression pattern established during development [47] . Moreover, PcG and TrxG pro- teins perform opposing functions by safeguarding the silenced or active transcriptional states, respectively [47] [48] .
Three families of complexes containing PcG proteins have been identified in Drosophila to date: Polycomb Repressive Complex 1 and 2 (PRC1 and PRC2) and PhoRC. PRC2 is composed of four core components, the mammalian counterparts of which are: Ezh2, Suz12, RbAp46/48 and Eed. Ezh2 is the catalytic subunit and harbors histone lysine methyltransferase activity within its SET domain that gives rise to di- and tri-methylated versions of lysine residue 27 within histone H3 (H3K27me2/3) [47] . The other core components are required for such Ezh2 enzymatic activity. However, whether they play additional roles independent of Ezh2 remains unclear. The core components of Polycomb Repressive Complex 1 (PRC1) include HPC, HPH, Bmi1/Mel18 and Ring1A/ B [49] . PRC1 prevents the ATP dependent remodeling activity of Swi/Snf in vitro [50] and is able to condense chromatin in the absence of histone tails [51] . In addition, PRC1 has a mono-ubiquitylase activity directed to- wards lysine residue 119 of histone H2A (H2AK119) and this is mediated by the E3 ligase activity of its RING1B component [52] . Of note, HPC and its mammalian homologs contain a chromodomain that specifically binds the product of PRC2-catalysis, H3K27me2/3 [53] - [55] . Given this, PRC1 was proposed to act downstream of PRC2 [55] . Yet this scenario does not seem to be universal as Xist RNA can recruit PRC1 in the absence of PRC2 and chromatin regions depleted of H3K27me3 can be bound by PRC1 [56] . Finally, a third polycomb group complex, PhoRC was characterized recently in Drosophila, however its exact function and mammalian counterpart are not yet clear.
PcG proteins bind to Polycomb Response Elements (PRE) that have been identified and characterized in Drosophila. Several DNA binding proteins were shown to be required for PcG recruitment such as GAF, Pips- queak, Zeste or PHO. Surprisingly, no mammalian counterparts were found for these recruiters and, despite ex- tensive searches, PREs have not been identified to date in mammals [47] . Nonetheless, genome-wide analyses identified genes targeted by PRC2 in a variety of cell lines and animal models [57] [58] . As expected, a strong overlap between PRC2, PRC1 and H3K27me2/3 was observed [59] . Moreover, PRC target genes were found to extend far beyond the historically recognized HOX loci. Gene ontology of the target genes revealed a strong enrichment for developmental factors as perhaps expected, although glycoprotein and immunoglobulin related genes were also identified depending on the cell model analyzed [60] .
With few exceptions, invertebrates such as Drosophila or sea urchins have only one copy of PcG genes [61] . However, vertebrates have several paralogs of most PcG genes. Interestingly, among the PRC2 components two genes were not duplicated: Suz12 and Eed. However, different isoforms of Eed do arise from alternative transla- tion start sites [62] and these might play an important role in creating diversity among the PRC2 complexes [14] . Although Drosophila E (z) and its closest mammalian homolog Ezh2 have been well characterized, very little is known about mammalian Ezh1 although it was the first Ez homolog to be cloned. The RNA levels of Ezh1 and Ezh2 appear to be inversely correlated in that Ezh1 is highly expressed in kidney, brain and skeletal muscle tis- sues where Ezh2 RNA is barely detectable [63] . However, two other studies analyzing Ezh1 expression in tis- sues reported slightly divergent results [64] . Ezh1 was also shown to interact with Eed in vitro [65] [66] .
We investigated the cellular role of Ezh1 relative to that of Ezh2. Here, we show that Ezh1 is ubiquitously expressed whereas Ezh2 expression is associated with proliferating tissues. Ezh1 is part of a PRC2 complex quite similar to the one containing Ezh2 and they share an overlapping set of target genes that they appear to co-occupy. Yet surprisingly and in contrast to PRC2-Ezh2, PRC2-Ezh1 exhibits low levels of histone methyl- transferase (HKMT) activity. On the other hand, PRC2-Ezh1 efficiently represses transcription and compact chromatin, in contrast to PRC2-Ezh2. These distinct functional roles for PRC2-Ezh1 and PRC2-Ezh2 in repres- sion might pertain to their differential expression and to sub-functionalization of Ez during evolution.
Altogheter, our results show that the Leptin can change molecularly the expression of genes related to Polycomb group (PcG) and Trithorax group (TrxG) in the brain of mice transplanted with fat tissue from normal mice. This work can help us to better understand the neuronal mechanisms underlying the reversion of PCOS.
To Sao Paulo Research Foundation (FAPESP) for Research Grants number 2008/54383-0.
- Pasquali, R. and Gambineri, A. (2013) Insulin Sensitizers in Polycystic Ovary Syndrome. Frontiers of Hormone Re- search, 40, 83-102. http://dx.doi.org/10.1159/000341837
- Hahn, S., Bering van Halteren, W., Roesler, S., Schmidt, M., Kimmig, R., Tan, S., Mann, K. and Janssen, O.E. (2006) The Combination of Increased Ovarian Volume and Follicle Number Is Associated with More Severe Hyperandr Ogenism in German Women with Polycystic Ovary Syndrome. Experimental and Clinical Endocrinology & Diabetes, 114, 175-181. http://dx.doi.org/10.1055/s-2006-924063
- Premoli, A.C., Santana, L.F., Ferriani, R.A., Moura, M.D., DeSa, M.F. and Reis, R.M. (2005) Growth Hormone Secre- tion and Insulin-Like Growth Factor-1 Are Related to Hyperandrogenism in Nonobese Patients with Polycystic Ovary Syndrome. Fertility and Sterility, 83, 1852-1855. http://dx.doi.org/10.1016/j.fertnstert.2004.10.057
- Lomniczi, A., Loche, A., Castellano, J.M., Ronnekleiv, O.K., Bosch, M., Kaidar, G., Knoll, J.G., Wright, H., Pfeifer, G.P. and Ojeda, S.R. (2003) Epigenetic Control of Female Puberty. Nature Neuroscience, 16, 281-289.
- Yagi, S., Hirabayashi, K., Sato, S., Li, W., Takahashi, Y., Hirakawa, T., Wu, G., Hattori, N., Hattori, N., Ohgane, J., Tanaka, S., Liu, X.S. and Shiota, K. (2008) DNA Methylation Profile of Tissue-Dependent and Differentially Me- thylated Regions (T-DMRs) in Mouse Promoter Regions Demonstrating Tissue-Specific Gene Expression. Genome Research, 18, 1969-1978. http://dx.doi.org/10.1101/gr.074070.107
- Hattori, N., Nishino, K., Ko, Y.G., Hattori, N., Ohgane, J., Tanaka, S. and Shiota, K. (2004). Epigenetic Control of Mouse Oct-4 Gene Expression in Embryonic Stem Cells and Trophoblast Stem Cells. Journal of Biological Chemistry, 279, 17063-17069. http://dx.doi.org/10.1074/jbc.M309002200
- Ikegami, K., Ohgane, J., Tanaka, S., Yagi, S. and Shiota, K. (2009) Interplay between DNA Methylation, Histone Mo- dification and Chromatin Remodeling in Stem Cells and during Development. International Journal of Developmental Biology, 53, 203-214. http://dx.doi.org/10.1387/ijdb.082741ki
- Ohgane, J., Hattori, N., Oda, M., Tanaka, S. and Shiota, K. (2002) Differentiation of Trophoblast Lineage Is Asso- ciated with DNA Methylation and Demethylation. Biochemical and Biophysical Research Communications, 290, 701- 706. http://dx.doi.org/10.1006/bbrc.2001.6258
- Shiota, C., Larsson, O., Shelton, K.D., et al. (2002) Sulfonylurea Receptor Type 1 Knock-Out Mice Have Intact Feed- ing-Stimulated Insulin Secretion Despite Marked Impairment in Their Response to GlucoseJournal of Biological Chemistry, 277, 37176-37183.
- Shogren-Knaak, M., Ishii, H., Sun, J.M., Pazin, M.J., Davie, J.R. and Peterson, C.L. (2006) Histone H4-K16 Acetyla- tion Controls Chromatin Structure and Protein Interactions. Science, 311, 844-847. http://dx.doi.org/10.1126/science.1124000
- Baskind, H.A., Na, L., Ma, Q., Patel, M.P., Geenen, D.L. and Wang, Q.T. (2009) Functional Conservation of Asxl2, a Murine Homolog for the Drosophila Enhancer of Trithorax and Polycomb Gene Asx. PLoS ONE, 4, e4750. http://dx.doi.org/10.1371/journal.pone.0004750
- Escargueil, A.E., Soares, D.G., Salvador, M., Larsen, A.K. and Henriques, J.A. (2008) What Histone Code for DNA Repair? Mutation Research/Reviews in Mutation Research, 658, 259-270. http://dx.doi.org/10.1016/j.mrrev.2008.01.004
- Czermin, B., Melfi, R., McCabe, D., Seitz, V., Imhof, A. and Pirrotta, V. (2002) Drosophila Enhancer of Zeste/ESC Complexes Have a Histone H3 Methyltransferase Activity That Marks Chromosomal Polycomb Sites. Cell, 111, 185- 196. http://dx.doi.org/10.1016/S0092-8674(02)00975-3
- Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P. and Reinberg, D. (2002) Histone Methyltransferase Activity Associated with a Human Multiprotein Complex Containing the Enhancer of Zeste Protein. Genes & Deve- lopment, 16, 2893-2905. http://dx.doi.org/10.1101/gad.1035902
- Ikegami, K., Ohgane, J., Tanaka, S., Yagi, S. and Shiota, K. (2009) Interplay between DNA Methylation, Histone Mo- dification and Chromatin Remodeling in Stem Cells and during Development. International Journal of Developmental Biology, 53, 203-214. http://dx.doi.org/10.1387/ijdb.082741ki
- Schuettengruber, B., Chourrout, D., Vervoort, M., Leblanc, B. and Cavalli, G. (2007) Genome Regulation by Polycomb and Trithorax Proteins. Cell, 128, 735-745. http://dx.doi.org/10.1016/j.cell.2007.02.009
- Yokoyama, A., Wang, Z., Wysocka, J., Sanyal, M., Aufiero, D.J., Kitabayashi, I., et al. (2004) Leukemia Proto-Onco- protein MLL Forms a SET1-Like Histone Methyltransferase Complex with Menin to Regulate Hox Gene Expression. Molecular and Cellular Biology, 24, 5639-5649. http://dx.doi.org/10.1128/MCB.24.13.5639-5649.2004
- Bernstein, B.E., Kamal, M., Lindblad-Toh, K., Bekiranov, S., Bailey, D.K., Huebert, D.J., et al. (2005) Genomic Maps and Comparative Analysis of Histone Modifications in Human and Mouse. Cell, 120, 169-181. http://dx.doi.org/10.1016/j.cell.2005.01.001
- Schneider, R., Bannister, A.J., Myers, F.A., Thorne, A.W., Crane-Robinson, C. and Kouzarides, T. (2004) Histone H3 Lysine 4 Methylation Patterns in Higher Eukaryotic Genes. Nature Cell Biology, 6, 73-77. http://dx.doi.org/10.1038/ncb1076
- Yoo, A.S. and Crabtree, G.R. (2009) ATP-Dependent Chromatin Remodeling in Neural Development. Current Opi- nion in Neurobiology, 19, 120-126. http://dx.doi.org/10.1016/j.conb.2009.04.006
- Nakamura, T., Mori, T., Tada, S., Krajewski, W., Rozovskaia, T., Wassell, R., et al. (2002) ALL-1 Is a Histone Me- thyltransferase That Assembles a Supercomplex of Proteins Involved in Transcriptional Regulation. Molecular Cell, 10, 1119-1128. http://dx.doi.org/10.1016/S1097-2765(02)00740-2
- Prezioso, C. and Orlando, V. (2011) Polycomb Proteins in Mammalian Cell Differentiation and Plasticity. FEBS Let- ters, 585, 2067-2077.
- Margueron, R., Li, G., Sarma, K., Blais, A., Zavadil, J., Woodcock, C.L., Dynlacht, B.D. and Reinberg, D. (2008) Ezh1 and Ezh2 Maintain Repressive Chromatin through Different Mechanisms. Molecular Cell, 32, 503-518.
- Zhuge, X., Kataoka, H., Tanaka, M., Murayama, T., Kawamoto, T., Sano, H., Togi, K., Yamauchi, R., Ueda, Y., Xu, Y., Nishikawa, S., Kita, T. and Yokode, M. (2005) Expression of the Novel Snai-Related Zinc-Finger Transcription Factor Gene Smuc during Mouse Development. International Journal of Molecular Medicine, 15, 945-948.
- Nasonkin, I.O., Lazo, K., Hambright, D., Brooks, M., Fariss, R. and Swaroop, A. (2011) Distinct Nuclear Localization Patterns of DNA Methyltransferases in Developing and Mature Mammalian Retina. Journal of Comparative Neurology, 519, 1914-1930.
- Lazzaro, M.A. and Picketts, D.J. (2001) Cloning and Characterization of the Murine Imitation Switch (ISWI) Genes: Differential Expression Patterns Suggest Distinct Developmental Roles for Snf2h and Snf2l. Journal of Neurochemis- try, 77, 1145-1156.
- Begum, G., Davies, A., Stevens, A., Oliver, M., Jaquiery, A., Challis, J., Harding, J., Bloomfield, F. and White, A. (2013) Maternal Undernutrition Programs Tissue Specific Epigenetic Changes in the Glucocorticoid Receptor in Adult Offspring. Endocrinology, 154, 4560-4569.
- De Giorgio, M.R., Yoshioka, M. and St-Amand, J. (2009) Feeding Induced Changes in the Hypothalamic Transcrip- tome. Clinica Chimica Acta, 406, 103-107.
- Makedonski, K., Abuhatzira, L., Kaufman, Y., Razin, A. and Shemer, R. (2005) MeCP2 Deficiency in Rett Syndrome Causes Epigenetic Aberrations at the PWS/AS Imprinting Center That Affects UBE3A Expression. Human Molecular Genetics, 14, 1049-1058.
- Vincenz, C. and Kerppola, T.K. (2008) Different Polycomb Group CBX Family Proteins Associate with Distinct Re- gions of Chromatin Using Nonhomologous Protein Sequences. Proceedings of the National Academy of Sciences of the United States of America, 105, 16572-16577.
- Zhao, L.J., Subramanian, T. and Chinnadurai, G. (2006) Changes in C-Terminal Binding Protein 2 (CtBP2) Corepre- ssor Complex Induced by E1A and Modulation of E1A Transcriptional Activity by CtBP2. The Journal of Biological Chemistry, 281, 36613-36623.
- Murphy, T.M., Mullins, N., Ryan, M., Foster, T., Kelly, C., McClelland, R., O’Grady, J., Corcoran, E., Brady, J., Reilly, M., Jeffers, A., Brown, K., Maher, A., Bannan, N., Casement, A., Lynch, D., Bolger, S., Buckley, A., Quinlivan, L., Daly, L., Kelleher, C. and Malone, K.M. (2013) Genetic Variation in DNMT3B and Increased Global DNA Me- thylation Is Associated with Suicide Attempts in Psychiatric Patients. Genes, Brain and Behavior, 12, 125-132.
- Lomniczi, A., Wright, H., Castellano, J.M., Sonmez, K. and Ojeda, S.R. (2013) A System Biology Approach to Iden- tify Regulatory Pathways Underlying the Neuroendocrine Control of Femalepuberty in Rats and Nonhuman Primates. Hormones and Behavior, 64, 175-186. http://dx.doi.org/10.1016/j.yhbeh.2012.09.013
- Mousavi, K., Zare, H., Wang, A.H. and Sartorelli, V. (2012) Polycomb Protein Ezh1 Promotes RNA Polymerase II Elongation. Molecular Cell, 45, 255-262.
- Seo, J., Kim, K., Chang, D.Y., Kang, H.B., Shin, E.C., Kwon, J. and Choi, J.K. (2014) Genome Wide Reorganization of Histone H2AX toward Particular Fragile Sites on Cell Activation. Nucleic Acids Research, 42, 1016-1025.
- Takeuchi, T., Watanabe, Y., Takano-Shimizu, T. and Kondo, S. (2006) Roles of Jumonji and Jumonji Family Genes in Chromatin Regulation and Development. Developmental Dynamics, 235, 2449-2459.
- Peterlin, B.M., Brogie, J.E. and Price, D.H. (2012) 7SK snRNA: A Noncoding RNA That Plays a Major Role in Regu- lating Eukaryotic Transcription. Wiley Interdisciplinary Reviews: RNA, 3, 92-103.
- LaFave, M.C., Varshney, G.K., Gildea, D.E., Wolfsberg, T.G., Baxevanis, A.D. and Burgess, S.M. (2014) MLV Inte- gration Site Selection Is Driven by Strong Enhancers and Active Promoters. Nucleic Acids Research, 42, 4257-4269. http://dx.doi.org/10.1093/nar/gkt1399
- Junco, S.E., Wang, R., Gaipa, J.C., Taylor, A.B., Schirf, V., Gearhart, M.D., Bardwell, V.J., Demeler, B., Hart, P.J. and Kim, C.A. (2013) Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs. Structure, 21, 665-671.
- Stapels, M., Piper, C., Yang, T., Li, M., Stowell, C., Xiong, Z.G., Saugstad, J., Simon, R.P., Geromanos, S., Langridge, J., Lan, J.Q. and Zhou, A. (2010) Polycomb Group Proteins as Epigenetic Mediators of Neuroprotection in Ischemic Tolerance. Science Signaling, 3, ra15.
- Lee, Y.H., Kim, S.H., Lee, Y.J., Kang, E.S., Lee, B.W., Cha, B.S., Kim, J.W., Song, D.H. and Lee, H.C. (2013) Trans- cription Factor Snail Is a Novel Regulator of Adipocyte Differentiation via Inhibiting the Expression of Peroxisome Proliferator-Activated Receptor γ. Cellular and Molecular Life Sciences, 70, 3959-3971.
- Zaman, M.M., Nomura, T., Takagi, T., Okamura, T., Jin, W., Shinagawa, T., Tanaka, Y. and Ishii, S. (2013) Ubiquiti- nation-Deubiquitination by the TRIM27-USP7 Complex Regulates Tumor Necrosis Factor α-Induced Apoptosis. Mo- lecular and Cellular Biology, 33, 4971-4984.
- Xiao, X., Tang, C., Xiao, S., Fu, C. and Yu, P. (2013) Enhancement of Proliferation and Invasion by MicroRNA-590- 5p via Targeting PBRM1 in Clear Cell Renal Carcinoma Cells. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics, 20, 537-544. http://dx.doi.org/10.3727/096504013X13775486749335
- Shore, A.N., Kabotyanski, E.B., Roarty, K., Smith, M.A., Zhang, Y., Creighton, C.J., Dinger, M.E. and Rosen, J.M. (2012) Pregnancy-Induced Noncoding RNA (PINC) Associates with Polycomb Repressive Complex 2 and Regulates Mammary Epithelial Differentiation. Plos Genetics, 8, e1002840. http://dx.doi.org/10.1371/journal.pgen.1002840
- Marei, H.E,. Ahmed, A.E., Michetti, F., Pescatori, M., Pallini, R., Casalbore, P., Cenciarelli, C. and Elhadidy, M. (2012) Gene Expression Profile of Adult Human Olfactory Bulb and Embryonic Neural Stem Cell Suggests Distinct Signaling Pathways and Epigenetic Control. PLoS ONE, 7, e33542. http://dx.doi.org/10.1371/journal.pone.0033542
- Austenaa, L., Barozzi, I., Chronowska, A., Termanini, A., Ostuni, R., Prosperini, E., Stewart, A.F., Testa, G. and Nato- li, G. (2012) The Histone Methyltransferase Wbp7 Controls Macrophage Function through GPI Glycolipid Anchor Synthesis. Immunity, 36, 572-585.
- Kingston, R.E. and Tamkun, J.W. (2014) Transcriptional Regulation by Trithorax-Group Proteins. Cold Spring Harbor Perspectives in Biology, 6, pii: a019349. http://dx.doi.org/10.1101/cshperspect.a019349
- Ringrose, L. and Paro, R. (2007) Polycomb/Trithorax Response Elements and Epigenetic Memory of Cell Identity. De- Velopment, 134, 223-232. http://dx.doi.org/10.1242/dev.02723
- Levine, S.S., Weiss, A., Erdjument-Bromage, H., Shao, Z., Tempst, P. and Kingston, R.E. (2002) The Core of the Po- lycomb Repressive Complex Is Compositionally and Functionally Conserved in Flies and Humans. Molecular and Ce- llular Biology, 22, 6070-6078. http://dx.doi.org/10.1128/MCB.22.17.6070-6078.2002
- Kia, S.K., Gorski, M.M., Giannakopoulos, S. and Verrijzer, C.P. (2008) SWI/SNF Mediates Polycomb Eviction and Epigenetic Reprogramming of the INK4b-ARF-INK4a Locus. Molecular and Cellular Biology, 28, 3457-3464. http://dx.doi.org/10.1128/MCB.02019-07
- Francis, N.J., Kingston, R.E. and Woodcock, C.L. (2004) Chromatin Compaction by a Polycomb Group Protein Com- plex. Science, 306, 1574-1577.
- Wang, H., Wang, L., Erdjument-Bromage, H., Vidal, M., Tempst, P., Jones, R.S. and Zhang, Y. (2004) Role of Histone H2A Ubiquitination in Polycomb Silencing. Nature, 431, 873-878. http://dx.doi.org/10.1038/nature02985
- Bernstein, E., Duncan, E.M., Masui, O., Gil, J., Heard, E. and Allis, C.D. (2006) Mouse Polycomb Proteins Bind Di- fferentially to Methylated Histone H3 and RNA and Are Enriched in Facultative Heterochromatin. Molecular and Cel- lular Biology, 26, 2560-2569. http://dx.doi.org/10.1128/MCB.26.7.2560-2569.2006
- Fiskus, W., Pranpat, M., Balasis, M., Herger, B., Rao, R., Chinnaiyan, A., Atadja, P. and Bhalla, K. (2006) Histone Deacetylase Inhibitors Deplete Enhancer of Zeste 2 and Associated Polycomb Repressive Complex 2 Proteins in Human Acute Leukemia Cells. Molecular Cancer Therapeutics, 5, 3096-3104.
- Wang, L., Brown, J.L., Cao, R., Zhang, Y., Kassis, J.A. and Jones, R.S. (2004) Hierarchical Recruitment of Polycomb Group Silencing Complexes. Molecular Cell, 14, 637-646. http://dx.doi.org/10.1016/j.molcel.2004.05.009
- Schoeftner, S., Sengupta, A.K., Kubicek, S., Mechtler, K., Spahn, L., Koseki, H., Jenuwein, T. and Wutz, A. (2006) Recruitment of PRC1 Function at the Initiation of X Inactivation Independent of PRC2 and Silencing. The EMBO Journal, 25, 3110-3122. http://dx.doi.org/10.1038/sj.emboj.7601187
- Boyer, L.A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L.A., Lee, T.I., Levine, S.S., Wernig, M., Tajonar, A., Ray, M.K., et al. (2006) Polycomb Complexes Repress Developmental Regulators in Murine Embryonic Stem Cells. Nature, 441, 349-353. http://dx.doi.org/10.1038/nature04733
- Bracken, A.P., Dietrich, N., Pasini, D., Hansen, K.H. and Helin, K. (2006) Genome-Wide Mapping of Polycomb Tar- get Genes Unravels Their Roles in Cell Fate Transitions. Genes & Development, 20, 1123-1136. http://dx.doi.org/10.1101/gad.381706
- Lee, T.I., Jenner, R.G., Boyer, L.A., Guenther, M.G., Levine, S.S., Kumar, R.M., Chevalier, B., Johnstone, S.E., Cole, M.F., Isono, K., et al. (2006) Control of Developmental Regulators by Polycomb in Human Embryonic Stem Cells. Cell, 125, 301-313. http://dx.doi.org/10.1016/j.cell.2006.02.043
- Squazzo, S.L., O’Geen, H., Komashko, V.M., Krig, S.R., Jin, V.X., Jang, S.W., Margueron, R., Reinberg, D., Green, R. and Farnham, P.J. (2006) Suz12 Binds to Silenced Regions of the Genome in a Cell-Type-Specific Manner. Genome Research, 16, 890-900. http://dx.doi.org/10.1101/gr.5306606
- Whitcomb, S.J., Basu, A., Allis, C.D. and Bernstein, E. (2007) Polycomb Group Proteins: An Evolutionary Perspective. Trends in Genetics, 23, 494-502. http://dx.doi.org/10.1016/j.tig.2007.08.006
- Cao, Q., Wang, X., Zhao, M., Yang, R., Malik, R., Qiao, Y., Poliakov, A., Yocum, A.K., Li, Y., Chen, W., Cao, X., Jiang, X., Dahiya, A., Harris, C., Feng, F.Y., Kalantry, S., Qin, Z.S., Dhanasekaran, S.M. and Chinnaiyan, A.M. (2014) The Central Role of EED in the Orchestration of Polycomb Group Complexes. Nature Communications, 5, Article No. 3127. http://dx.doi.org/10.1038/ncomms4127
- Ho, L. and Crabtree, G.R. (2008) An EZ Mark to Miss. Cell Stem Cell, 3, 577-578.
- Blumenberg, M., Gao, S., Dickman, K., Grollman, A.P., Bottinger, E.P. and Zavadil, J. (2007) Chromatin Structure Regulation in Transforming Growth Factor-β-Directed Epithelial-Mesenchymal Transition. Cells Tissues Organs, 185, 162-174. http://dx.doi.org/10.1159/000101317
- Han, Z., Xing, X., Hu, M., Zhang, Y., Liu, P. and Chai, J. (2007) Structural Basis of EZH2 Recognition by EED. Struc- ture, 15, 1306-1315. http://dx.doi.org/10.1016/j.str.2007.08.007
- MuhChyi, C., Juliandi, B., Matsuda, T. and Nakashima, K. (2013) Epigenetic Regulation of Neural Stem Cell Fate du- ring Corticogenesis. International Journal of Developmental Neuroscience, 31, 424-433. http://dx.doi.org/10.1016/j.ijdevneu.2013.02.006
*Corresponding author.



