American Journal of Molecular Biology
Vol.4 No.1(2014), Article ID:41626,5 pages DOI:10.4236/ajmb.2014.41002
Unusual metallo-β-lactamases may constitute a new subgroup in this family of enzymes
1Research School of Chemistry, Australian National University, Canberra, Australia
2Department of Chemistry, National University of Ireland-Maynooth, Maynooth, Ireland
3School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Australia
Email: schenk@uq.edu.au, natasa.mitic@nuim.ie
Received 1 October 2013; revised 1 November 2013; accepted 18 November 2013
ABSTRACT
Metallo-β-lactamases (MBLs) are a family of Zn2+- dependent enzymes that have contributed strongly to the emergence and spread of antibiotic resistance. Novel members as well as variants of existing members of this family are discovered continuously, compounding their threat to global health care. MBLs are divided into three subgroups, i.e. B1, B2 and B3. The recent discovery of an unusual MBL from Serratia proteamaculans (SPR-1) suggests the presence of an additional subgroup, i.e. B4. A database search reveals that SPR-1 has only one homologue from Cronobacter sakazakii, CSA-1.These two MBLs have a unique active site and may employ a mechanism distinct from other MBLs, but reminiscent of some organophosphate-degrading hydrolases.
Keywords:Antibiotic Resistance; β-Lactam Antibiotics; Metallo-β-Lactamases; Sequence Homology; Serratia proteamaculans; Cronobacter sakazakii
1. INTRODUCTION
On March 5, 2013, the Centers for Disease Control and Prevention (CDC) published the press release “Action needed now to halt spread of deadly bacteria—Data show more inpatients suffering infections from bacteria resistant to all or nearly all antibiotics” (http://www.cdc.gov/media/releases/2013/p0305_deadly_bacteria.html). According to this article, a family of bacteria has become increasingly resistant to last-resort antibiotics during the past decade, and more hospitalized patients are getting lethal infections that, in some cases, are impossible to cure. The findings published by the CDC are “a call to action for the entire health care community to work urgently—individually, regionally and nationally—to protect patients”. This call is all the more urgent as “the bacteria, Carbapenem-Resistant Enterobacteriaceae (CRE), kill up to half of patients who get bloodstream infections from them”.
One of the most frightening forms of antibiotic resistance occurs through the action of β-lactamases, enzymes capable of breaking down the most widely used antibiotics, and the β-lactams (e.g. carbapenems, penicillins, etc.; Figure 1) [1,2]. Two major groups of β-lactamases are distinguished, the Serine-β-lactamases (SBLs) and the metallo-β-lactamases (MBLs); the latter require Zn2+ to function [1]. Inhibitors for SBLs have been developed (e.g. clavulanic acid) and are frequently co-administered with the antibiotic [1]. Clinically far more dangerous are
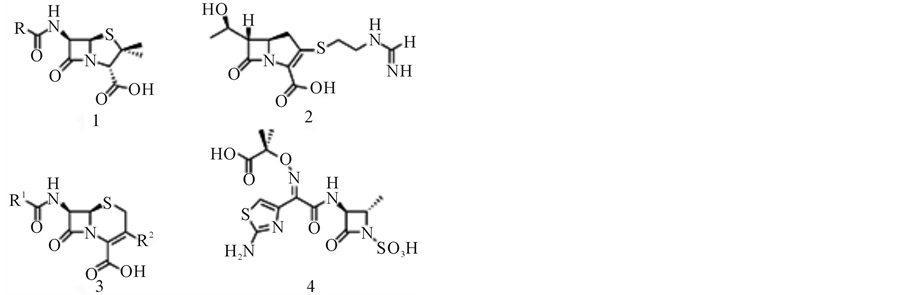
Figure 1. Representatives for the most common β- lactam antibiotic families. Penicillin (1), carbapenem (2), cefalosporin (3) and monobactam (4).
thus the MBLs, for which no clinically useful inhibitors are yet available [1,2]. An example is the MBL NDM-1 that emerged rapidly in 2010; infections by NDM-producing Klebsiella pneumonia have a high risk of being fatal [3]. Thus, a potent and clinically useful inhibitor of MBLs is urgently needed.
The development of such an inhibitor has been rendered difficultly due to the diversity among MBLs with respect to both amino acid sequence and structure. MBLs are divided into three subgroups (B1, B2 and B3) based on overall homology and active site geometry [1,2]. Representatives from the B1 subgroup include NDM-1 [4], IMP-1 [5], BcII [6], Ccr [7] and SPM-1 [8]. The B2 subgroup includes the MBLs CphA [9] and Sfh-1 [10], while enzymes such as AIM-1 [11], L1 [12], SMB-1 [13] and MIM [14] belong to the B3 subgroup. Interestingly, while the MBLs from the B1 and B3 subgroups seem to require two metal ions in their active sites to function optimally, B2-type MBLs are inhibited in the presence of a second metal ion (Figure 2).
These variations do not only indicate mechanistic differences between the MBL subgroups, but they also complicate strategies to develop universally applicable MBL inhibitors. The recent discovery of a novel MBL from Serratia proteamaculans (SPR-1) may complicate matters further [15]. Based on overall sequence homology, SPR-1 was classified as a B3-type MBL, however, in comparison to all known MBLs, SPR-1 appears to have significant variations in its metal binding site. Only three of the commonly observed Zn2+ ligands are conserved; in particular the presence of an arginine instead of a histidine in position 118 is unusual (as will be discussed below). Metal ion measurements indicated that the resting SPR-1 only binds one Zn2+ ion, similar to B2-type MBLs [15]. In contrast, kinetic measurements led to the hypothesis that SPR-1 may bind a second metal ion when a substrate (i.e. antibiotic) is added, indicating that the enzyme may operate like B1- and B3-type MBLs. Insofar, SPR-1 may represent a distinct MBL, mechanistically different from other known members of this family of enzymes. SPR-1 may lie dormant until it is needed. This may prove to be an effective strategy to circumvent inhi-
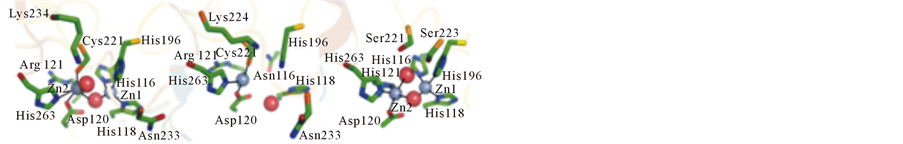
Figure 2. Active site structures of representative MBLs. Left: B1- type BcII from B. cereus; center: B2-type CphA from Aeromonas hydrophila; right: B3-type L1 from Stenotrophomonas maltophilia. Zinc ions are rendered as grey spheres, and water molecules are shown as red spheres. Coordination bonds are shown as solid lines. Copied with permission from reference [14].
bition. Here, we endeavored to identify enzymes that may be SPR-1-like in order to investigate their evolutionary origins.
2. METHODS
2.1. Protein Database Search Using BLAST
SPR-1 from S. proteamaculans (UniProt code: A8GGW7) was used as query sequence for a Conserf protein database search [http://consurf.tau.ac.il /verify.php].
2.2. Multiple Sequence Alignments
Multiple sequence alignments including representatives from the B1, B2 and B3 subgroups of the MBL family were carried out using ClustalW2.1 [http://www.ebi.ac.uk/Tools/msa/clustalw2]. The alignment also includes SPR-1 and its only close homologue, CSA-1, which was discovered in this study. Sequence identities and similarities were also determined from pairwise sequence comparisons using ClustalW software.
2.3. Phylogenetic Analysis
A phylogenetic tree based on the multiple sequence alignment was generated using the program PhyML 3.0 [16]. Selected bootstrap numbers, which indicate the statistical reliability of the branching, with 1000 bootstrap replicates, are shown. The sequence distances can be estimated from the bar underneath the tree.
3. RESULTS AND DISCUSSION
3.1. Protein Database Search, Nomenclature and Classification of Novel MBLs
Using SPR-1 as query the database search revealed only one close relative, a sequence from the Gram-negative pathogenic bacterium Cronobacter sakazakii [17]. C. sakazakii can cause meningitis and necrotizing enterocolitis in particular in infants with a mortality rate estimated as high as 80% [18]. To our knowledge no cases of antibiotic resistance has yet been reported. This putative MBL from C. sakazakii, CSA-1, shares approximately 60% sequence identity with SPR-1 (Table 1).
Among the MBLs of the well-established subgroups the B3-type ones are the closest relatives with ~30% sequence identity (Table 1). This is not surprising since SPR-1 was initially identified due to its homology to B3 MBLs [15]. In contrast, the homology between CSA-1 and members of the B1 and B2 subfamilies is generally less than 20% sequence identity (Table 1). Also included in the comparison are two more distantly related enzymes with functions that differ greatly from that of MBLs. Methyl-paraoxonhydrolase (MPH) is an organophosphate pesticide-degrading enzyme [19], while glyoxalase II is an enzyme involved in the detoxification
Table 1 . Pairwise sequence comparisons between CSA-1 and selected MBLs representing the different subgroups of this family of enzymes. Also included are MPH and glyoxalase II, two enzymes with an overall MBL fold but different biological function.
process of methylglyoxal and other reactive aldehydes produced in metabolism [20]. Both share less than 15% sequence identity with CSA-1. In Figure 3 a portion of a multiple sequence alignment involving all the enzymes listed in Table 1 is shown. Focus is on amino acid residues that are relevant for binding of the catalytically essential metal ions (i.e. Zn2+ ions). These metal ion-coordinating residues are spread across four short sequence motifs. Within the three subgroups a high degree of conservation is observed. For the B1-type MBLs residues His116, His118 and His196 form one metal binding site (i.e. the M1 site), while Asp120, Cys221 and His263 form the M2 site. In B2-type MBLs His116 in the M1 site is replaced by an asparagine while the M2 site is unaltered. In contrast, B3-type MBLs have an M1 site identical to that of B1 MBLs but Cys221 is replaced by a histidine (i.e. His121) [5]. MPH and glyoxalase II have metal binding sites identical to those in B3 MBLs plus an additional metal ion-bridging ligand (i.e. Asp221; Figure 3). In contrast, SPR-1 was found to differ significantly from all known MBLs [15]. In its M1 site an arginine is proposed to replace His118, while the M2 site is even more diverse. No crystal structure of SPR-1 is yet available, but based on a homology model only Asp120 may be conserved. The remaining ligands may be Gln121 and Asn262, thus replacing Cys221/His121 and His263 observed in B1 and B3 MBLs, respectively. However, Ser221 instead of Cys221/His121 may also be a possible alternative. The unusual composition of the metal ion
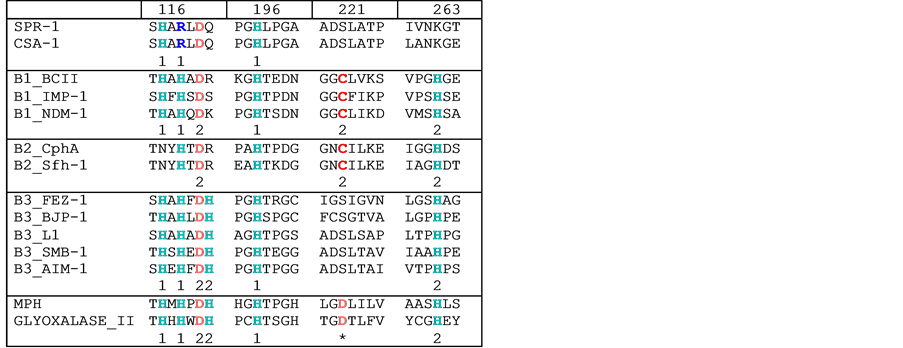
Figure 3. Partial multiple sequence alignment of MBLs representing different subgroups. Metal ion ligands are shown in color. The numbers underneath the residues indicate the metal ion binding site (i.e. M1 or M2). *indicates the metal-bridging ligand Asp221.
binding site in the active center of SPR-1 is also reflected in its metal ion requirement. In the resting form only one Zn2+ may be present; the second metal ion may bind only when substrate is added [15]. It was thus speculated that SPR-1 activity is tightly controlled [15], possibly as a means to minimize undesired side reactions. This hypothesis also suggests that the in vivo function of SPR-1 may be diverse or even different from that of common MBLs. Insofar, the discovery of an SPR-1-like protein in only one other microorganism is significant; the C. sakazakii enzyme has active site residues identical to those of SPR-1 (Figure 3), indicating that these unusual MBLs may have evolved from a common ancestor separate from that of other MBLs.
3.2. Phylogenetic Analysis
In order to assess the evolutionary origin of SPR-1 and CSA-1 a phylogenetic analysis was carried out (Figure 4). MPH and in particular Glyoxalase II, the two enzymes that have a MBL overall fold but have biological functions different from MBLs are the most diverged sequences. Especially in the case of Glyoxalase II the fold similarity may be a reflection of convergent evolution rather than sequence divergence from a common ancestral gene. It is also evident that B1 and B2 MBLs are closely related (indicated by the 96% statistical (i.e. bootstrap) reliability). In contrast, B3 MBLs form a separate cluster. As expected from the pairwise sequence comparisons (Table 1) SPR-1 and CSA-1 cluster with B3 MBLs but form a separate group with high statistical reliability (bootstrap score of 90%). In fact, the separation of these unusual MBLs from the B3 MBLs is of a similar statistical reliability as that of the B2 MBLs from the B1 group (91%; Figure 4). It should also be noted

Figure 4. Maximum-likelihood tree including representatives from each of the MBL subgroups plus MPH and Glyoxalase II. The tree has been constructed using PhyML, with 1000 bootstrap replicates [16].
that the sequence distance between SPR-1/CSA-1 and the B3 subgroup is similar to that between the B1 and B2 subgroups. Hence, it is very likely that SPR-1 and CSA-1 originated from the same ancestral sequence as the well characterized MBLs from the B1, B2 and B3 subgroups, but based on the variations in both the overall sequence homology and the active site (i.e. metal binding site) residues (Figure 4) it is plausible to propose that these two unusual MBLs constitute a separate subgroup. We thus suggest the introduction of a new subgroup, B4, to encompass MBLs such as SPR-1 and CSA-1.
4. CONCLUSION
In summary, our sequence analysis has revealed that an additional subgroup of MBLs, here labeled as subgroup B4, exists. B4 MBLs such as SPR-1 and CSA-1 cluster with the B3 subgroup, are similar to the cluster formed by the B1 and B2 subgroups. It thus appears that an initial evolutionary separation led to the divergence between the B1/B2 and B3/B4 MBLs, before a second, more recent separation led to the formation of four separate subgroups. It is interesting to point out that within the B1/B2 cluster the B1 enzymes are essentially binuclear enzymes (although the MBL BcII from B. cereus can operate in the presence of only one metal ion in the active site [21]) whereas the B2 MBLs are only active in mononuclear form [2,9,22]. Similarly, the B3 MBLs are known to require two Zn2+ ions in the active site to be catalytically active [2,9,12]. For the B4 MBLs, the precise metal ion requirement has not yet been established but it seems likely that these enzymes are mononuclear at least in their resting states [15]. The reason for this observed variation in metal ion requirement between different subgroups of MBLs is poorly understood but may provide essential insight into strategies needed to develop universal MBL inhibitors, compounds that may be employed to combat the spread of antibiotic resistance. Insofar, it is now essential to investigate the molecular properties of SPR-1 and CSA-1. Efforts towards this goal are in progress.
ACKNOWLEDGEMENTS
N. M. thanks the Science Foundation Ireland (SFI) for financial support in the form of a President of Ireland Young Researcher Award (PIYRA) and G. S. acknowledges the award of a Future Fellowship from the Australian Research Council (FT120100694). D. O. and G. S. are grateful to the National Health and Medical Research Council of Australia for funding.
REFERENCES
- Fisher, J.F., Meroueh, S.O. and Mobashery, S. (2005) Bacterial resistance to beta-lactam antibiotics: Compelling opportunism, compelling opportunity. Chemical Reviews, 105, 395-424. http://dx.doi.org/10.1021/cr030102i
- Walsh, T.R., Toleman, M.A., Poirel, L. and Nordmann, P. (2005) Metallo-beta-lactamases: The quiet before the storm? Clinical Microbiology Reviews, 182, 306-325. http://dx.doi.org/10.1128/CMR.18.2.306-325.2005
- Escobar Perez, J.A., Olarte Escobar, N.M., Castro-Cardozo, B., Valderrama Marquez, I.A., Garzon Aguilar, M.I., Martinez de la Barrera, L., Barrero Barreto, E.R., Marqeuz-Ortiz, R.A., Moncada Guayazan, M.V. and Vanegas Gomez, N. (2013) Outbreak of NDM-1 producing Klebsiella pneumoniae in a neonatal unit in Colombia. Antimicrobial Agents and Chemotherapy, 57, 1957-1960.
- Thomas, P.W., Zheng, M., Wu, S.S., Guo, H., Liu, D.L., Xu, D.G. and Fast, W. (2011) Characterization of Purified New Delhi Metallo-beta-lactamase-1. Biochemistry, 5046, 10102-10113. http://dx.doi.org/10.1021/bi201449r
- Laraki, N., Franceschini, N., Rossolini, G.M., Santucci, P., Meunier, C., de Pauw, E., Amicosante, G., Frere, J.M. and Galleni, M. (1999) Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-beta-lactamase IMP-1 produced by Escherichia coli. Antimicrobial Agents and Chemotherapy, 434, 902-906.
- Carfi, A., Pares, S., Duee, E., Galleni, M., Duez, C., Frere, J. and Dideberg, O. (1995) The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new-type of protein fold. The EMBO Journal, 1420, 4914- 4921.
- Crowder, M.W., Wang, Z.G., Franklin, S.L., Zovinka, E. P. and Benkovic, S.J. (1996) Characterization of the metal-binding sites of the beta-lactamase from Bacteroides fragilis. Biochemistry, 3537, 12126-12132. http://dx.doi.org/10.1021/bi960976h
- Murphy, T.A., Catto, L.E., Halford, S.E., Hadfield, A.T., Minor, W., Walsh, T.R. and Specer, J. (2006) Crystal structure of Pseudomonas aeruginosa SPM-1 provides insights into variable zinc affinity of metallo-beta-lactamases. Journal of Molecular Biology, 3573, 890-903. http://dx.doi.org/10.1016/j.jmb.2006.01.003
- Bebrone, C., Delbruck, H., Kupper, M.B., Scholmer, P., Wilmann, C., Frere, J.M., Fischer, R., Galleni, M. and Hoffmann, K.M. (2009) The structure of the dizinc subclass B2 metallo-beta-lactamase CphA reveals that the second inhibitory zinc ion binds in the histidine site. Antimicrobial Agents and Chemotherapy, 53, 4464-4471. http://dx.doi.org/10.1128/AAC.00288-09
- Fonseca, F., Bromley, E.H., Saavedra, M.J., Correia, A. and Spencer, J. (2011) Crystal structure of Serratia fonticolaSfh-I: Activation of the nucleophile in mono-zinc metallo-beta-lactamases. Journal of Molecular Biology, 411, 951-959. http://dx.doi.org/10.1016/j.jmb.2011.06.043
- Leiros, H.K., Borra, P.S., Brandsdal, B.O., Edvardsen, K.S.W., Spencer, J., Walsh, T.R. and Samuelsen, O. (2012) Crystal structure of the mobile metallo-beta-lactamase AIM-1 from Pseudomonas aeruginosa: Insights into antibiotic binding and the role of Gln157. Antimicrobial Agents and Chemotherapy, 568, 4341-4353. http://dx.doi.org/10.1128/AAC.00448-12
- Crowder, M.W., Walsh, T.R., Banovic, L., Pettit, M. and Spencer, J. (1998) Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. Antimicrobial Agents and Chemotherapy, 424, 921-926.
- Wachino, J., Yoshida, H., Yamane, K., Suzuki, S., Matsui, M., Yamagishi, T., Tsutsui, A., Konda, T., Shibayama, K. and Arakawa, Y. (2011) SMB-1, a novel subclass B3 metallo-beta-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrobial Agents and Chemotherapy, 55, 5143-5149. http://dx.doi.org/10.1128/AAC.05045-11
- Miraula, M., Brunton, C., Schenk, G. and Mitic, N. (2013) Identification and preliminary characterization of novel B3-type metallo-β-lactamases. American Journal of Molecular Biology, 3, 198-203.
- Vella, P., Miraula, M., Phelan, E., Leung, E.W., Ely, F., Ollis, D.L., McGeary, R.P., Schenk, G. and Mitic, N. (2013) Identification and characterization of an unusual metallo-beta-lactamase from Serratia proteamaculans. Journal of Biological Inorganic Chemistry, 18, 855-863. http://dx.doi.org/10.1007/s00775-013-1035-z
- Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. and Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59, 307-321. http://dx.doi.org/10.1093/sysbio/syq010
- Ivy, R.A., Farber, J.M., Pagotto, F. and Wiedmann, M. (2013) International life science institute north America Cronobacter (formerly Enterobacter sakazakii) isolate set. Journal of Food Protection, 76, 40-51. http://dx.doi.org/10.4315/0362-028X.JFP-11-546
- Friedemann, M. (2009) Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. European Journal of Clinical Microbiology & Infectious Diseases, 28, 1297-1304. http://dx.doi.org/10.1007/s10096-009-0779-4
- Dong, Y.J., Bartlam, M., Sun, L., Zhou, Y.F., Zhang, Z. P., Zhang, C.G., Rao, Z. and Zhang, X.E. (2005) Crystal structure of methyl parathion hydrolase from Pseudomonas sp. WBC-3. Journal of Molecular Biology, 353, 655- 663. http://dx.doi.org/10.1016/j.jmb.2005.08.057
- Cameron, A.D., Ridderstrom, M., Olin, B., and Mannervik, B. (1999) Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. Structure, 7, 1067-1078. http://dx.doi.org/10.1016/S0969-2126(99)80174-9
- Rasia, R.M. and Vila, A.J. (2002) Exploring the role and the binding affinity of a second zinc equivalent in B. cereus metallo-beta-lactamase. Biochemistry, 41, 1853-1860. http://dx.doi.org/10.1021/bi010933n
- Hernandez Valladares, M., Felici, A., Weber, G., Adolph, H.W., Zeppezauer, M., Rossolini, G.M., Amicosante, G., Frere, J.M. and Galleni, M. (1997) Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-beta-lactamase activity and stability. Biochemistry, 36, 11534-11541. http://dx.doi.org/10.1021/bi971056h


