American Journal of Molecular Biology
Vol. 3 No. 3 (2013) , Article ID: 34686 , 5 pages DOI:10.4236/ajmb.2013.33019
Quantitative detection of Streptococcus mutans from saliva using FTATM elute cards and real-time polymerase chain reaction
![]()
1Department of Biomedical Laboratory Science, Malmö University, Malmö, Sweden
2Department of Cariology, Faculty of Odontology, Malmö University, Malmö, Sweden
Email: *sepideh.seghat@gmail.com
Copyright © 2013 Sepideh Seghatoleslami et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 29 March 2013; revised 12 May 2013; accepted 15 June 2013
Keywords: Streptococcus mutans; FTATM Elute cards; Real-Time PCR
ABSTRACT
Dental caries is a localized, transmissible, pathological infection process that ends up in the destruction of hard dental tissue. Numerous reports have shown the close relationship between salivary levels of Streptococcus mutans and dental caries. As S. mutans, is considered to be the principle etiological agent of dental caries, the development of a quick and convenient method for detection and quantification of these bacteria from patient saliva samples would simplify diagnosis and treatment. The purpose of this study was to compare a new means of quantifying bacteria using FTATM Elute cards and Real-Time Polymerase Chain Reaction to a conventional culture-based assay using oral S. mutans as a model sample. A total of 60 different saliva samples were investigated. The results show a significant negative correlation between the two methods, with a correlation coefficient of −0.577 (Spearman’s Correlation) and p < 0.01. The method demonstrates a high sensitivity, specificity and reliable quantitative results, covering a large range of bacterial concentrations.
1. INTRODUCTION
Dental caries is one of the most common and costly diseases in the world, and although rarely life threatening it is a major problem for health service providers [1]. Streptococcus mutans is considered a major etiologic agent of dental caries. The differentiation between various strains of oral streptococci using selective culturing media, such as Mitis-salivarius or Mitissalivarius-bacitracin agar, is complicated and time-consuming. Nevertheless, a polymerase chain reaction (PCR) method for simple, rapid and reliable identification of S. mutans was developed [2,3]. Igarash et al. utilized primers formulated from the dextranase (dexA) gene for the detection of S. mutans by PCR. Dextranase is an enzyme that hydrolyses glucans in plaque and is involved in the virulence of S. mutans [3].
Flinders Technology Associates (FTA) matrix card is a membrane intended for rapid collection, purification and storage of genetic material from a wide range of biological sources [4]. The FTATM card is a cellulose membrane which is impregnated with protein denaturants that cause the lysis of cells and microorganism upon contact. Moreover, these chemicals inhibit degradation of the DNA during drying to ensure safe handling of the cards without risk of biohazards and also protect the sample from UV radiation, nucleases, oxidation and microbialor fungal attacks [5]. Nucleic acids are physically entrapped, immobilized and stabilized at room temperature (RT) for several years, using minimal storage area [6].
There are different kinds of FTA cards on the market. FTATM Elute cards are also intended for DNA extraction. As it uses a similar technology as its predecessor, FTATM Classic cards, but instead of keeping DNA attached to the membrane, it allows the release of DNA into water by using heat. DNA extraction was made easier because the only solution needed from beginning to end is ultrapure water [7]. Inhibitory components are retained on the FTATM Elute cards or disposed of during a short washing step [8].
The aim of this study was to compare a new means of quantifying bacteria using FTATM Elute cards and qPCR to a conventional culture-based assay for the determination of oral S. mutans.
2. MATERIALS AND METHODS
2.1. Materials
Dentocult SM strip® was purchased from Orion Diagnostica Oy (Espoo, Finland). FTATM Elute and FTATM Classic cards were purchased from Whatman (BioSciences Ltd., Abington, Cambridge, UK). LightCycler 480 Real-Time PCR System (Roche, Indianapolis, USA) was used. The primers were purchased from Invitrogen (Life Technologies, Carlsbad, USA).
2.2. Sample Collection
Stimulated saliva was obtained from voluntary donors by chewing a piece of sterile paraffin wax (approximately 1 g) and swallowing for 2 min. The saliva was then collected by dwelling for a period of 5 min period into a sterile container. If the saliva samples were not assayed directly they were frozen at −20˚C until further processing. The rough surface of the Dentocult SM strip® was then pressed on the dorsal side of the donor’s tongue 10 times, before being removed between gently closed lips. The strip® was then incubated in a vial of selective culture broth under 37˚C for 48 h. To suppress the growth of other bacteria than the streptococci group, a bacitracin disc was added to the medium broth, according to the manufacturer’s instructions. S. mutans adhere to the rough area of the strip in proportion to their density in saliva sample. After incubation, they are visible as light to dark blue colonies on the rough area of the test strip. The densities of bacterial colony forming units (CFU) were rated into four groups (Class 0-3) according to the standard reference chart provided by the manufacturer [9].
2.3. Preparation of DNA Samples from FTATM Elute and FTATM Classic Cards
The saliva samples were vigorously mixed using vortex. Then 20 µL of each sample was transferred onto FTATM Elute and Classic cards allowed to dry overnight at RT. Discs of 5 mm in diameter were punched out of the each FTATM cards, using a paper puncher and transferred to individual 1.5 mL tubes. The tip of the puncher was cleaned between each punching with 95% ethanol to prohibit cross contamination. The discs were processed according to the manufacture’s instructions as described below.
2.3.1. FTATM Elute Cards
The discs were washed once by addition of 500 µL of sterile water and pulse vortexed 3 times for 5 s. Then the discs were transferred into clean tubes containing 40 µL of sterile water and incubated at 98˚C for 25 min. At the end of the incubation step, tubes were pulse vortex briefly 60 times for one second and finally the discs were removed from the tubes. The extracts were stored at 4˚C until analyzes.
2.3.2. FTATM Classic Cards
Each disc was washed 3 times with 200 µL FTA purification reagent for 5 min and rinsed twice with TE buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0) for 5 min at RT and then the FTA disc was left to dry overnight. The dry discs were finally transferred into wells of a 96 well PCR plate (LightCycler 480 multiwell plate, Roche, Germany) for qPCR amplification.
To elute the DNA from FTATM Classic discs, 35 µL of a solution containing 0.1 N NaOH, 0.3 mM EDTA, pH 13.0 was added and incubated for 5 min at RT. Subsequently, 65 µL of 0.1 M Tris-HCl solution, pH 7.0 was added and the tube was inverted several times. The sample was incubated for 10 min at RT and then vortexed for 5 min. The FTA card was removed and the supernatant was transferred to a new sterile tube and stored at 4˚C until they were analyzed.
2.4. qPCR Amplification
The primers sequences were selected to amplify specific parts of the S. mutans dexA gene (Table 1). Amplification and quantification was done in a 96 well PCR plate. Each reaction contained 5 µL DNA template from disc extracts, 10 µL of LightCycler 480 SYBR Green I Master (Roche diagnostics Gmbh, Germany), 1 µL of 10 µM forward and reverse primer respectively. Double-distilled water was added to adjust the reaction mix to a final volume of 20 µL. Amplification and detection were performed using the LightCycler 480 Real-Time PCR System with the following cycle profile: Initial denaturation at 95˚C for 10 min, followed by 40 cycles of amplification at 95˚C for 10 s, 65˚C for 5 s, 72˚C for 11 s. Finally, a melting curve analysis was performed using the following cycling parameters: 95˚C for 5 s, 65˚C for 1 min and a slow increase of temperature until the final temperature of 97˚C was reached. Each sample was run in triplicates.

Table 1. Oligonucleotides used for qPCR amplification of S. mutans.
2.5. Generation of qPCR Standards with Bacterial Genomic DNA
To determine the concentration range, a 5-fold dilution series of a bacterial suspension were prepared from 108 CFU/mL to 2.4 × 105 CFU/mL. Then 20 µL of each bacterial solution was loaded onto the FTATM Elute cards. Three different cards were loaded with identical samples. FTATM Elute card discs of 5 mm were punched from each card and bacterial DNA was extracted as described. A standard curve was constructed using the cycle threshold (CT) values from the qPCR and the corresponding CFU/mL. The number of CFU/mL in the saliva samples were then quantified using this curve.
2.6. Statistical Methods
The median number of PCR cycles for 60 samples in two separate runs was analyzed against the Dentocult SM strip® (SM) score using Spearman’s rank correlation. A value of p < 0.01 was considered significant. The data were transformed into nominal variables, SM scores 0 and 1 into a “Low SM” group and SM scores 2 and 3 into “High SM” group. Data from qPCR analysis were transformed in a similar way, samples with no detectable product or a product detectable after more than 32 cycles were assigned to a “Low qPCR” group and samples which a product identified after less than 32 cycles were assigned to a “High qPCR” group. The resulting nominal variables were subject to Chi-Square test. A value of p < 0.01 was considered significant. The tests were carried out using the SPSS statistical package (IBM, New York, USA).
3. RESULTS AND DISCUSSION
Saliva contains a large number of bacterial cells, approximately 1.7 × 107/mL. Thus, DNA extracted from saliva may contain a large proportion of microbial DNA [10]. Therefore using a rapid, reliable and convenient technique to extract DNA simplifies the quantification of bacteria present in a sample. In this study, the FTATM Elute cards in combination with qPCR, were used for isolation and quantification of oral S. mutans and the results were compared with the strip assay.
There was a significant correlation between the median number of PCR cycles and the SM, with a correlation coefficient of −0.577 (Spearman’s Correlation) as shown in Table 2.
Despite the advantages and usefulness of the strip assay it possesses some limitations while analyzing low concentrations of bacteria, according to Baca et al. It was demonstrated that some colonies did not remain adhered onto the plastic spatula, which could lead to an underestimation of the counts despite a good interand intraobserver agreement [11]. In order to confirm that the
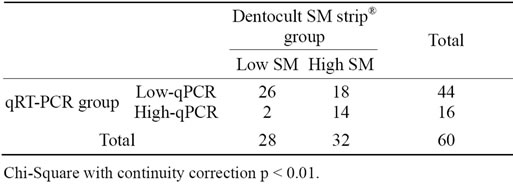
Table 2. Results of Dentocult SM strip® assay and qPCR method.
correct PCR-products are amplified during the reaction, the products from this study were sequenced by GATC (GATC Biotech AG, Germany), and electrophoresed on a 1% agarose gel with GelStar® nucleic acid gel stain (Cambrex Bio Science Rockland, Inc, UK). The results from sequencing and electrophoresis (Figure 1) in combination with the melting curve analysis clearly show that the correct PCR-product was amplified during the reactions.
Various methods, such as biochemical and immunological tests, species-specific DNA probes, 16S rRNA sequencing comparison methods and polymerase chain reaction (PCR) have been used both to detect and identify oral microbes. Among them, PCR provides a rapid, sensitive and specific technique [12,13]. So, in this study, qPCR technique was used for the rapid identification and quantification of S. mutans in salivary samples utilizing the LightCycler 480 Real-Time PCR System. No conclusive results could be obtained using FTATM classic cards neither having the discs directly in the reaction mixtures, nor eluting DNA using high pH elute buffers, according to protocols obtained from the manufacturer. Nozawa et al. found that only instruments equipped with a photomultiplier-tube scanning system (e.g. Stratagene MX 3500P) could be used for real-time PCR assays with FTA filter discs in the reaction mixture, due to the fact that instruments using a charge-coupled device (CCD) camera (e.g. ABI7700) were adversely affected by nonspecific signals from the discs [14]. Since CCD camera is the optic unit of the LightCycler 480 Real-Time PCR System, maybe discs from FTA Classic cards present during the reactions, interfered with instrument signal detection. As it is difficult to determine the exact amount of DNA bound to the membrane, the cell number was determined prior to addition of the cell suspension on to the membrane. This will provide a good estimation of the sensitivity of the method (Figure 2). The FTA card circle zone of 11 mm in diameter was loaded with a 20 μL sample of a 2.4 × 105 CFU/mL bacterial suspension (4800 CFU) whereas 5 mm discs from this area were removed for PCR detection. Approximately 20% of the area containing DNA from the FTA card (20% of 4800 CFU) was used for PCR detection. Subsequent PCR amplification showed a detection limit of around 103 CFU/
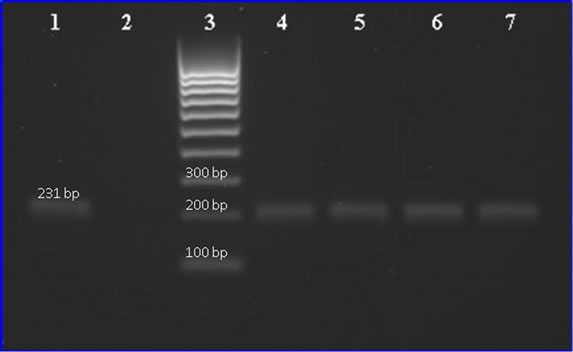
Figure 1. Agarose gel electrophoresis of 231 bp PCR fragments of S. mutans dexA gene from saliva samples archived on FTATM Elute cards. Lane 1: positive control of ative control; lane 3: Molecular weight marker (100 bp ladder); and lanes 4, 5, 6, 7: show the PCR product of saliva samples.
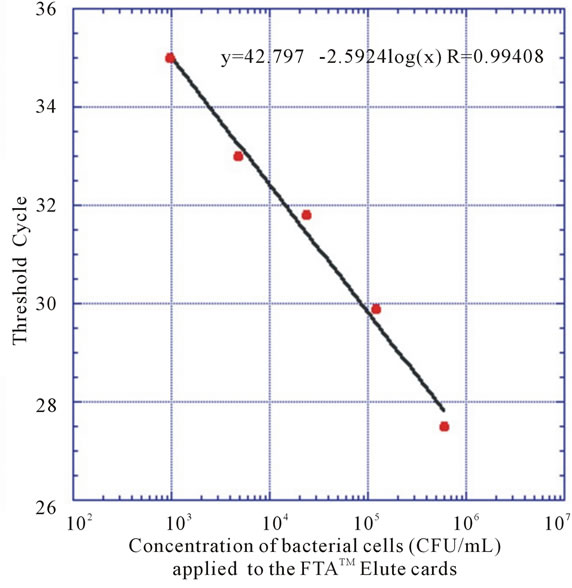
Figure 2. Standard curves of the qPCR assays obtained by the amplification of S. mutans dexA gene using 5-fold serial dilutions of bacteria archived on FTA membranes.
mL indicating good sensitivity of the assay.
In an effort to compare and translate results of bacterial quantification using the conventional strip assay to the qPCR method, a standard curve (Figure 2) was produced, allowing the number of CFU/mL in a sample to be calculated from the CT-values obtained. It is generally accepted that CT-values above 35 are regarded as uncertain, sometimes showing false positive results; the results were treated as previously stated.
These results clearly demonstrate the usefulness of the FTATM Elute cards for rapid archiving and recovery of bacterial DNA aimed for PCR-based applications. This is the first time, known to us, to detect and quantify oral bacteria, including S. mutans in saliva samples archived on FTATM Elute cards, although these cards have been used for a wide range of biological sources such as Schistosoma mansoni [15], Malaria [16,17], human papilomavirus [18-20], cancer [21,22], buccal samples [7], bronchitis virus [23] and microorganisms [4]. It is evident that the FTATM Elute cards technology is a valuable tool for molecular biology considering the ease of use, space saving, long term stability and storage at RT.
In conclusion, quantification of S. mutans in saliva samples archived on FTATM Elute cards by qPCR provides greater efficiency compared to the traditional Dentocult SM strip® assay.
4. ACKNOWLEDGEMENTS
We thank Isaac Kuraishe for valuable technical assistance and Malmö University for providing the grant supporting inter-faculty cooperation, making this study possible.
![]()
![]()
REFERENCES
- Forssten, S., Björklund, M. and Ouwehand, A. (2010) Streptococcus mutans, caries and simulation models. Journal of Nutrients, 2, 290-298.
- E Franco, T.C., Amoroso, P., Marin, J.M. and de Ávilla, F.A. (2007) Detection of Streptococcus mutans and Streptococcus sobrinus in Dental Plaque Samples from Brazilian Preschool Children by Polymerase Chain Reaction. Brazilian Dental Journal, 18, 329-333. doi:10.1590/S0103-64402007000400011
- Igarashi, T., Yamamoto, A. and Goto, N. (1996) Direct detection of Streptococcus mutans in human dental plaque by polymerase chain reaction. Oral Microbiology and Immunology, 11, 294-298. doi:10.1111/j.1399-302X.1996.tb00184.x
- Rajendram, D., Ayenza, R., Holder, F.M, Moran, B., Long, T. and Shah, H.N. (2006) Long-term storage and safe retrieval of DNA from microorganisms for molecular analysis using FTA matrix cards. Journal of Microbiological Methods, 67, 582-592. doi:10.1016/j.mimet.2006.05.010
- Narayanan, M.S., Parthiban, M., Sathiya, P. and Kumanan, K. (2010) Molecular detection of Newcastle disease virus using Flinders Tehnology Associates-PCR. Veterinarski arhiv, 80, 51-60.
- Mullen, M., Howard, D., Powell, R. and Hanrahan, J. (2009) A note on the use of FTATM technology for storage of blood samples for DNA analysis and removal of PCR inhibitors. Irish Journal of Agricultural and Food Research, 48, 109-113.
- Wolfgramm Ede, V., de Carvalho, FM., Aguiar, V.R., Sartori, M.P., Hirschfeld-Campolongo, G., Tsutsumida, W.M., et al. (2009) Simplified buccal DNA extraction with FTA Elute Cards. Journal of Forensic Science International Genetics, 3, 125-127.
- Tongeren, S., Degener, J. and Harmsen, H. (2011) Comparison of three rapid and easy bacterial DNA extraction methods for use with quantitative real-time PCR. European Journal of Clinical Microbiology & Infectious Diseases, 30, 1053-1061. doi:10.1007/s10096-011-1191-4
- Jensen, B. and Bratithhall, D. (1989) A New method for the estimation of mutans Streptococci in human saliva. Journal of Dental Research, 68, 468-471. doi:10.1177/00220345890680030601
- Quinque, D., Kittler, R., Kayser, M., Stoneking, M. and Nasidze, I. (2006) Evaluation of saliva as a source of human DNA for population and association studies. Analytical Biochemistry, 353, 272-277. doi:10.1016/j.ab.2006.03.021
- Baca, P., Parejo, E., Bravo, M., Castillo, A. and Liébana, J. (2011) Discriminant ability for caries risk of modified colorimetric tests. Medicina Oral Patologia Oral y Cirugia Bucal, 16, 978-983. doi:10.4317/medoral.17358
- Kang, M.S., Oh, J.S., Kim, O., Kim, H., Lee, K., Choi, H., et al. (2009) Prevalence of oral microbes in the saliva of oncological patients. Journal of Bacteriology and Virology, 39, 277-285.
- da Silva, A.C., Cruz, J., Sampaio, F. and de Araújo, D. (2008) Detection of oral streptococci in dental biofilm from caries-active and caries-free children. Brazilian Journal of Microbiology, 39, 648-651. doi:10.1590/S1517-83822008000400009
- Nozawa, N., Koyano, S., Yamamoto, Y., Inami, Y., Kurane, I. and Inoue, N. (2007) Real-Time PCR assay using specimens on filter disks as a template for detection of cytomegalovirus in urine. Journal of Clinical Microbiolgy, 45, 1305-1307. doi:10.1128/JCM.02502-06
- Van den Broeck, F., Geldof, S., Polman, K., Volckaert, F. and Huyse, T. (2011) Optimal sample storage and extraction procotols for reliable multilocus genotyping of the human parasite Schistosoma mansoni. Infection Genetics and Evolution, 11, 1413-1418. doi:10.1016/j.meegid.2011.05.006
- Sultan, D.M., Khalil, M.M., Abdouh, A., Doleh, W.F. and Al Muthanna, A. (2009) Imported malaria in United Arab Emirates: Evaluation of a new DNA extraction technique using nested PCR. The Korean Journal of Parasitology, 47, 227-233. doi:10.3347/kjp.2009.47.3.227
- Zhong, K.J., Salas, C.J., Shafer, R., Gubanov, A., Gasser, J., Magill, A.J., et al. (2001) Comparison of IsoCode STIX and FTA Gene Guard collection matrices as wholeblood storage and processing devices for diagnosis of malaria by PCR. Journal of Clinical Microbiolgy, 39, 1195- 1196. doi:10.1128/JCM.39.3.1195-1196.2001
- Gustavsson, I., Lindell, M., Wilander, E., Strand, A. and Gyllensten, U. (2009) Use of FTA card for dry collection, transportation and storage of cervical cell specimen to detect high-risk HPV. Journal of Clinical Virology, 46, 112-116. doi:10.1016/j.jcv.2009.06.021
- Gyllensten, U., Gustavsson, I., Lindell, M. and Wilander, E. (2012) Primary high-risk HPV screening for cervical cancer in post-menopausal women. Gynecologic Oncology, 125, 343-345. doi:10.1016/j.ygyno.2012.01.036
- Lenselink, C.H., de Bie, R.P., Van Hamont, D., Bakkers, J.M., Quint, W.J., Massuger, L.F., et al. (2009) Detection and genotyping of human papillomavirus in self-obtained cervicovaginal samples by using the FTA cartridge: New possibilities for cervical cancer Screening. Journal of Clinical Microbiolgy, 47, 2564-2570.
- Saieg, M.A., Geddie, W.R., Boerner, S.L., Liu, N., Tsao, M., Zhang, T., et al. (2012) The use of FTA cards for preserving unfixed cytological material for high-throughput molecular analysis. Cancer Cytopathology Journal, 120, 206-214. doi:10.1002/cncy.20205
- Dobbs, L.J., Madigan, M.N., Carter, A.B. and Earls, L. (2002) Use of FTA gene guard filter paper for the storage and transportation of tumor cells for molecular testing. Archives of Pathology & Laboratory Medicine, 126, 56- 63.
- Moscoso, H., Raybon, E., Thayer, S.G. and Hofacre, C.L. (2005) Molecular detection and serotyping of infectious bronchitis virus from FTA filter paper. Avian Diseases, 49, 24-29. doi:10.1637/7220
NOTES
*Corresponding author.

