American Journal of Plant Sciences
Vol.4 No.3(2013), Article ID:29007,6 pages DOI:10.4236/ajps.2013.43079
Overexpression of ARAhPR10, a Member of the PR10 Family, Decreases Levels of Aspergillus flavus Infection in Peanut Seeds
![]()
Crops Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China.
Email: 56868600@qq.com, wenshijie1983@163.com, Liu_Haiyan001@126.com, xpchen1011@gmail.com, lihaifen2012@yahoo.cn, hongyanbin1979@yahoo.com.cn, *Liang-804@163.com
Received January 6th, 2013; revised February 8th, 2013; accepted February 20th, 2013
Keywords: Peanut; PR10; Aflatoxin Contamination; Overexpression
ABSTRACT
Peanut (Arachis hypogaea L.) is one of the most susceptible host crops to Aspergillus flavus invasion and subsequent aflatoxin contamination. In this report, a new member of PR10 family putative resistant gene (designated as ARAhPR10, No. EU661964.1) encoding a PR10 protein was isolated and characterized. Analysis of qRT-PCR showed that the expression of ARAhPR10 was induced by pre-harvested A. flavus infection, but no significant difference was observed between resistant genotype “GT-C20” and susceptible genotype “Yueyou 7”. Seven transgenic peanut lines expressing the ARAhPR10 gene under the control of 35S promoter were obtained using the Agrobacterium tumefaciens-mediated method. Real time RT-PCR results showed that the expression level of the ARAhPR10 was significantly higher and the A. flavus infection and aflatoxin content were significantly lower in seeds of transgenic lines than that of the wild type. A significant negative correlation between ARAhPR10 expression at transcript level and seeds aflatoxin production was observed. Combining the previous results, it is suggested that ARAhPR10 expression play an important role in peanut host resistance to A. flavus infection and aflatoxin producing.
1. Introduction
Peanut is one of the most susceptible host crops to Aspergillus flavus invasion and subsequent aflatoxin production. It was suggested that an effective solution to the problem would be the use of peanut varieties that are resistant/tolerant to infection by the aflatoxin-producing fungi, or resistant/tolerant to aflatoxin production if colonized by the fungus [1]. Several peanut genotypes with resistance to A. flavus infection have been identified. However, progress in incorporating resistant trait/genes from these genotypes with desirable agronomical traits has been slow, mainly due to the lack of understanding of resistant mechanism [1-3]. Excitingly, studies on hostresistance mechanisms had make great progress during the past twenty years. With the application of proteomics and genomics technology to high-throughput gene identification, many resistance-associated proteins and genes have been identified in maize [4-6] and peanut [7-9]. These proteins and genes comprise seed storage proteins, antifungal proteins, pathogenesis-related proteins and stress-related proteins [4-9]. Among them, the role of PR10 in host resistant to aflatoxin received more attention.
PR10 family is one of the most important PR families among 17 PR families and a number of cDNAs encoding PR-10 proteins were obtained [10]. So far, there are many reports of cloning, expression and characterization of multiple PR-10 or PR-10-like proteins from plants [10-13]. Based on similarities in their amino acid sequences, the PR-10 family comprises two distinct groups: intracellular pathogenesis related proteins (IPR) with homology to ribonucleases [11], and (S)-norcoclaurine synthases [12]. Although biological function of has not yet been cleared, PR10 proteins in general exhibit allergenic, anti-fungal and ribonuclease activities [10,13,14]. Just like other PR proteins, PR10 is induced by both biotic [10,15] and abiotic stress, such as drought [16], wounding and cold-hardening[17].
The possible relationship of PR10 and host resistant to aflatoxin was first reported by Luo et al. [7]. PR10 gene showed significantly up-regulated in peanut cultivar A13 and responded to A. parasiticus attack. Recently, a PR10 gene (ZmPR10) was found to be associated with maize kernel resistance to A. flavus infection/aflatoxin contamination, and was cloned from maize based on partial peptide sequences of the protein [18]. The ZmPR10 protein over-expressed in E. coli possessed ribonuclease activity and inhibited A. flavus growth [18]. Subsequently, Xie et al. [19] cloned a novel PR10 gene (ZmPR10.1) from maize and found it was induced by A. flavus infection. ZmPR10.1 showed a strong inhibition against A. flavus in vitro. Interestingly, in our earlier research, a PR10 up-regulated unique EST sequences was observed only in resistant genotypes “GT-C20” libraries after comparing the gene expression profiling in developing seeds at different reproduction stages during A. parasiticus infection between “Tifrunner”, susceptible to A. parasiticus and “GT-C20” [8]. Xie et al. [20] was isolated and characterized this up-regulated PR10 (designated as ARAhPR10, No. EU661964.1). Sequence alignments showed that ARAhPR10 showed high identity with the reported AhPR10 (gbAY726607) in peanut by 49.3% and with other PR10 proteins above 40% in other plants. The purified recombinant ARAhPR10 protein showed RNase activity, but could not inhibit A. flavus growth in vitro. This result was consistent with Chadha P and Das RH’s reports [13]. In addition, in another earlier research, PR 10 was not listed in significantly up-regulated proteins in the resistant cultivar challenged by A. flavus under drought stress [9]. A raised question was whether or not the PR10 can play a role in host resistant to aflatoxin contamination in peanut. The objective of this paper was to functionally characterize its role in alleviating aflatoxin contamination in peanut by overexpression of ARAhPR10.
2. Materials and Methods
2.1. Plant Material and Treatment
A resistant cultivar “GT-C20” and susceptible cultivar “Yueyou 7” were collected and treated according to the method described by Wang et al. (2009). Aspergillus flavus isolate As 3.2890, a wild-type strain known to produce high levels of aflatoxin in peanut, was provided by Institute of Microbiology, Chinese Academy of Sciences. Both resistant cultivars “CT-20” and susceptible “Yueyou 7” were subjected to three treatments as Table 1 showed (Table 1). The mature seeds were collected and immediately frozen in liquid nitrogen, and then stored in a freezer at −80˚C.
2.2. Construction of the pCAMBIA1301- ARAhPR10 Overexpression Plasmid and Peanut Transformation
To construct the chimeric genes consisting of the ARAhPR10 coding sequence driven by cauliflower mosaic virus 35S (CaMV35S) promoter, 471 bp ARAhPR10 ORF was amplified with sense prime: 5’-GGG CAT GGG CGT CTT CAC TTT TGA GGA-3’ and antisense prime: 5’-AAG GCC TAT, GTG TTC TAA TAT TGA GAA GGG TT-3’. The PCR product with an NcoI site at the 5’ end and a StuI site at the 3’ end was subcloned into pMD18-T simple vector (TaKaRa). The resulting constructs were digested with NcoI plus StuI, and then cloned into the binary expression vector pCAMBIA1301 digested with NcoI plus PmlI. After verification by PCR amplification and digested with NcoI plus BstⅡ, the recombinant expression vector was transformed into A. tumefaciens strain LBA4404 and transformed into susceptible cultivars “Yueyou 7” by Agrobacterium-mediated transformation method. The transformed plants were screened on hygromycin (50 mg/L) selection medium and identified by PCR amplification with the hygromycin phosphotransferase (hpt) gene primers, sense primer: 5’-ATG CCT GAA CTC ACC GCG AC-3’ and antisense primer: 5’-CTA TTC CTT GCC CTC GCAC. Total 45 positive events were determined and the seeds were harvested for further studies. The plasmid pCAMBIA1301 was also transformed into peanut as control. The structure of recombinant pCAMBIA1301-ARAhPR- 10 overexpression plasmid was showed in Figure 1.
2.3. Evaluation of Transgenic Peanut Seeds for Changes in Resistance to A. flavus Colonization and Aflatoxin Production
To determine the resistance of 7 transgenic peanut lines to A. flavus colonization. The mature seeds of T3 progenies was inoculated with A. flavus isolate As3.2890 and incubated according to Liang et al. [21]. At the 7th day of incubation, A. flavus colonization on the surface of inoculated seeds was rated according to a 0 - 5 scale. After visual rating of A. flavus colonization, the seeds (5 g)
Table 1. plant material and treatment of CT-20 and Yueyou7.
awatered normally; bstarting on the 60th day after sowing; cA. flavus (As3.2890)-contaminated corn powder was sprayed to pots at 60th days after planting and covered with soil, and watered with only 20 ml of water per day. All treatments were conducted simultaneously.

Figure 1. The structure of recombinant pCAMBIA1301- ARAhPR10 plasmid.
were roasted in 115˚C and powdered, defatted with 20 ml of n-hexane, The defatted powders were mixed with 15 ml aflatoxin B1 extract liquid (methanol: H2O = 1:1) and the aflatoxin B1 was extract by the protocol of the B1 ELISA kit (Produced by the institute of microorganism research of Jiangshu Province, China). Seeds of no-transgenic peanut and peanut transformed with vector plasmid pCAMBIA1301 were used as control.
2.4. Quantification of ARAhPR10 Expression Accumulation in Seeds Responded to Pre-Harvested A. flavus Attacks and Transgenic Seeds Using Real Time RT-PCR
Total RNA was isolated from seeds subjected to preharvested A. flavus attacks and drought stress and seeds of 10 transgenic peanut lines using Trizol reagent (Invitrogen, Carlsbad, CA), and genomic DNA was removed by adding RNase-free DNase I (Takara). And then, the RNA samples were purified with the RNeasy Cleanup Kit (Qiagen). Nano drop ND-1000 Spectrophotometer and agarose gel electrophoresis was performed to test RNA quality as described by Aranda, et al. [22]. For all the samples, 4 μg of total RNA was converted to cDNA using PrimeScript II 1st Strand cDNA Synthesis kit (Takara) according to the manufacturer’s protocols. Quantitative real-time RT-PCR was performed with SYBRR Premix Ex Taq™II kit (Takara) and a LightCycler 480 instrument (Roche) equipped with Light-Cycler Software Version 1.5 (Roche) based on the manufacturer’s instructions [23]. Amplifications reactions were carried out in a total volume of 20 μl. PCR cycling was: 95˚C for 10 s, followed by 45 cycles of 95˚C for 10 s, 60˚C for 10 s, and 72˚C for 20 s. Data collection was performed during the annealing phase of the each amplification. Then processing of the melting curve was from 62˚C to 95˚C with reading the intensity of fluorescence every 0.2. The actin gene was used as an internal control for calculating relative expression abundance. The amplicon of this gene is 104 bp and the primers are: forward (5’- GTTCC ACTAT GTTCC CAGGC A-3’) and reverse (5’-CTTCC TCTCT GGTGG TGCTA CA-3’). All real-time PCR reactions were technically repeated three times. The relative quantification of RNA expression was calibrated using formula 2-ΔΔCt method [24].
3. Results
3.1. Expression of ARAhPR10 Following Pre-Harvested Aflatoxin Contamination
To validate whether the expression of the ARAhPR 10 genes had relationship with pre-harvested A. flavus infection, total RNA were extracted from pods of resistant cultivar “CT-20” and the susceptible cultivar “Yueyou7” under well-watered (control), drought stress and A. flavus infection accompanied with drought stress on the 30th days after treatments, and the expression levels of ARAhPR10 in resistant and susceptible genotypes were analyzed through real time RT-PCR (Figure 2). The results demonstrated that the expression levels of ARAhPR10 were not significant changes in both resistant and susceptible cultivar with drought treatment. However, the significantly elevated expression levels of ARAhPR 10 genes were measured in both resistant and susceptible cultivars with A. flavus infection under drought stress, and there was not significant difference in the induced expression levels between resistant cultivar “CT-20” and susceptible cultivar “Yueyou 7” (Figure 2). The results indicated that ARAhPR10 gene in preharvested seeds could induced by A. flavus infection.
3.2. Generation, Verification and Expression Analysis of Transgenic Plants
Overexpression approach was employed to investigate
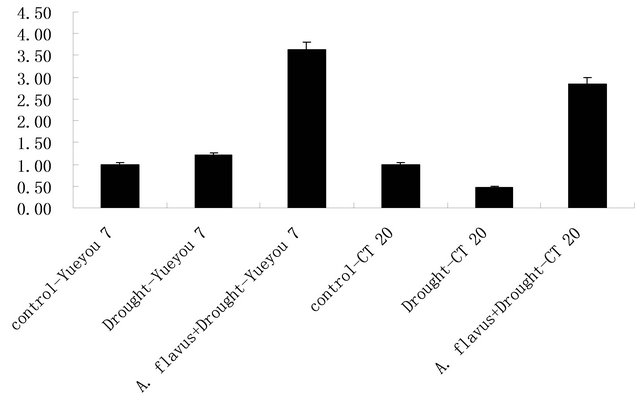
Figure 2. ARAhPR10 gene expression in young pods of resistant and susceptible cultivar in response to drought stress and pre harvested A. flavus inoculation by Real time RTPCR.
the role of ARAhPR10 in host resistance to A. flavus infection and aflatoxin production in cultivar peanut. A total of 45 transgenic peanut plants carrying ARAhPR10 gene were regenerated after the screening with hygromycin (50 mg/L). Seven of T3 transgenic lines were selected to investigate further, compared with non-transgenic plants and transgenic plants with control plasmid pCAMBIA1301, hereinafter refer to control plants. Genomic DNA and total RNA were isolated from the mature pods to confirm the transformation and to determine the level of ARAhPR10 expression. All of 7 transgenic lines were confirmed to be positive for the presence of ARAhPR10 gene and hpt gene by PCR (Table 2). The ARAhPR10 transcript level varied greatly among the transgenic lines, ranging from 54.86244 -fold to 1317.026- fold. The lowest expression is in transgenic line PR-957 and the highest expression is in transgenic line PR-975. The average transcript level of the transgenic lines is 776.6068, significantly higher than that in control plants (Table 2).
3.3. Evaluation of Fungal Growth and Aflatoxin Production in Overexpression Transgenic Seeds
To evaluate the peanut seeds tolerance to A. flavus colonization and aflatoxin production in seven overexpression transgenic peanut lines, the mature seeds of 7 putative transgenic plants and non-transgenic and transgenic plant with pCAMBIA1301 vector were evaluated (Figure 3). Seeds generated by control plants showed higher A. flavus colonization at the end of 7 d artificial incubation, whereas seeds from transgenic lines showed significantly lower level of A. flavus colonization (Table 2). Aflatoxin content analysis of infected seeds showed control plants had higher aflatoxin production level, however, 6 transgenic lines (PR225, PR962, PR975, PR1083, PR1211, PR1138 had significantly low level of aflatoxin production. A line (PR957) had fairly higher level of aflatoxin production compared to other transgenic lines (Table 2). Analysis of correlation coefficient between the transcript level of ARAhPR10 expression and the changes in fungal colonization or aflatoxin production showed the significant negative correlations (correlation coefficient r = −0.745 to −0.777). These data strongly demonstrated that overexpression of ARAhPR10 can alleviate A. flavus colonization and aflatoxin production in peanut.
4. Discussion
Earlier functional analysis of PR 10 showed that overexpressing PR-10 genes in transgenic plants are not consistent. Over-expression of STH-2, a member of the Ypr10 family, has not resulted in enhanced resistance of potato to pathogens [25]. Similar negative results have also been observed in the studies of pea PR-10.1 [26]. However, expression of the pea PR-10.1 gene in potato confers resistance to early dying disease [27]. In the recent studies, Chen et al. [28] found RNAi-silenced kernels had a significant reduction in PR10 production and lower in aflatoxin production compared to no-RNAisilenced kernels in maize. In this paper, overexpression of 7 ARAhPR10 transgenic lines resulted in significantly high level of ARAhPR10 expression and low level of A. flavus infection rate and aflatoxin production in peanut. As the expression of ARAhPR10 was induced by preharvested A. flavus infection, but no significant difference was observed between resistant genotype “Yueyou 20” and susceptible genotype “Yueyou 7”, and the purified recombinant ARAhPR10 protein could not inhibit A. flavus growth in vitro.
Table 2. Evaluation of ARAhPR10 expression and fungal colonization and aflatoxin production in 7 putative transgenic peanut lines.
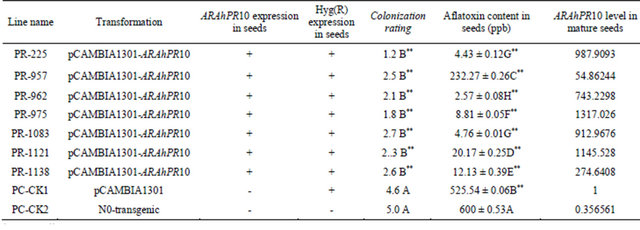
*: t < 0.05; **: t < 0.01.

Figure 3. Antifungal effect of un-transgenic and transgenic peanut seeds (line 2). A. un-transgenic seeds; B. transgenic seeds overexpressed pCAMBIA1301; C & D. transgenic seeds overexpressed pCAMBIA1301-ARAhPR10.
The evidence suggested that the ARAhPR10 proteins might be involved in signal transduction networks and stress responses pathways and play an important role in host resistant to aflatoxin contamination.
5. Conclusion
ARAhPR10 expression play an important role in peanut host resistance to A. flavus infection and aflatoxin producing, overexpression of ARAhPR10 can alleviate A. flavus colonization and aflatoxin production in peanut.
6. Acknowledgements
This research was funded by grants from National Natural Science Foundation of China (No. 31200155 and 31271767), Science and Technology Planning Project of Guangdong Province (2011B010500019), supported by the earmarked fund for Modern Agro-industry Technology Research System (CARS-14) and foundation of CRI/ GDAAS (2012-2). The founders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We declare no conflict of interests.
REFERENCES
- X. Q. Liang, M. Luo and B. Z. Guo, “Resistance Mechanisms to Aspergillus flavus Infection and Aflatoxin Contamination in Peanut (Arachis hypogaea),” Journal of Plant Pathology, Vol. 5, No. 1, 2006, pp. 115-124.
- B. Guo, Z. Y. Chen, R. D. Lee and B. T. Scully, “Drought Stress and Preharvest Aflatoxin Contamination in Agricultural Commodity: Genetics, Genomics and Proteomics,” Journal of Integrative Plant Biology Vol. 50, No. 10, 2008, pp. 1281-1291. doi:10.1111/j.1744-7909.2008.00739.x
- M. Luo, R. L. Brown, Z. Y. Chen and T. E. Cleveland, “Host Genes Involved in the Interaction between Aspergillus flavus and Maize,” Toxin Reviews, Vol. 28, No. 2, 2009, pp. 118-128.
- R. Y. Kelley, W. W. Paul, J. E. Mylroie, D. L. Boykin, L. K. Hawkins, G. L. Windham, T. D. Brooks, S. M. Bridges, B. E. Scheffler and J. R. Wilkinson, “Genomic Profile of Maize Response to Aspergillus flavus Infection,” Toxin Reviews, Vol. 28, No. 2-3, 2009, pp. 129-140.
- Z. Chen, R. L. Brown, K. Rajasekaran, K. E. Damann and T. E. Cleveland, “Evidence for Involvement of a Pathogenesis-Related Protein in Maize Resistance to Aspergillus flavus Infection/Aflatoxin Production,” Phytopathology, Vol. 96, No. 1, 2006, pp. 87-95.
- Z. Y. Chen, R. L. Brown, K. E. Damann and T. E. Cleveland, “Identification of Maize Kernel Endosperm Proteins Associated with Resistance to Aflatoxin Contamination by Aspergillus flavus,” Phytopathology, Vol. 97, No. 9, 2007, pp. 1094-1103. doi:10.1094/PHYTO-97-9-1094
- M. Luo, X. Q. Liang, P. Dang, C. C. Holbrook, M. G. Bausher, R. D. Lee and B. Z. Guo, “Microarray-Based Screening of Differentially Expressed Genes in Peanut in Response to Aspergillus parasiticus Infection and Drought Stress,” Plant Science, Vol. 169, No. 4, 2005, pp. 695-703.
- B. Z. Guo, X. P. Cheng, P. Dang, B. T. Scully, X. Q Liang, Hol C. C. brook, J. Yu and A. K. Culbreath, “Peanut Gene Expression Profiling in Developing Seeds at Different Reproduction Stages during Aspergillus parasiticus Infection,” BMC Developmental Biology, Vol. 8, 2008, pp. 12-28.
- T. Wang, E. Zhang, X. Chen, L. Li and X. Liang, “Identification of Seed Proteins Associated with Resistance to Pre-Harvested Aflatoxin Contamination in Peanut (Arachis hypogaea L.),” BMC Developmental Biology, Vol. 10, 2010, p. 267. doi:10.1186/1471-2229-10-267
- J. J. Liu and A. K. M. Ekramoddoullah, “The Family 10 of Plant Pathogenesis-Related Proteins: Their Structure, Regulation, and Function in Response to Biotic and Abiotic Stresses,” Physiological and Molecular Plant Pathology, Vol. 68, No. 1-3, 2006, pp. 3-13.
- L. C. Van Loon, W. S. Pierpoint, T. Boller and V. Conejero, “Recommendation for Naming Plant PathogenesisRelated Proteins,” Plant Molecular Biology Reporter, Vol. 12, No. 3,1994, pp. 245-264.
- N. Samanani, D. K. Liscombe and P. J. Facchini, “Molecular Cloning and Characterization of Norcoclaurine Synthase, an Enzyme Catalyzing the First Committed Step in Benzylisoquinoline Alkaloid Biosynthesis,” The Plant Journal, Vol. 40, No. 2, 2004, pp. 302-313. doi:10.1111/j.1365-313X.2004.02210.x
- P. Chadha and R. H. Das, “A Pathogenesis Related Protein, AhPR10 from Peanut: An Insight of Its Mode of Antifungal Activity,” Planta, Vol. 225, No. 1, 2006, pp. 213- 222. doi:10.1007/s00425-006-0344-7
- S. Srivastava, B. Fristensky and N. N. Kav, “Constitutive Expression of a PR10 Protein Enhances the Germination of Brassica napus under Saline Conditions,” Plant and Cell Physiology, Vol. 45, No. 9, 2004, pp. 1320-1324. doi:10.1093/pcp/pch137
- K. M. Koistinen, H. I. Kokko, V. H. Hassinen, A. I. Tervahauta, S. Auriola, S. O. Karenlampi, “Stress-Related RNase PR-10c Is Post-Translationally Modified by Glutathione in Birch,” Plant, Cell and Environment, Vol. 25, No. 6, 2002, pp. 707-715.
- X. Yu, A. K. Ekramoddoullah and S. Misra, “Characterization of Pin m III cDNA in Western White Pine,” Tree Physiology, Vol. 20, No. 10, 2000, pp. 663-671.
- J. J. Liu, A. K. M. Ekramoddoullah and X. Yu, “Differential Expression of Multiple PR10 Proteins in Western White Pine Following Wounding, Fungal Infection and Cold-Hardening,” Plant Physiology, Vol. 119, No. 4, 2003, pp. 544-553.
- Z. Chen, R. Brown, K. Rajasekaran and K. Damann, “Evi- dence for Involvement of a Pathogenesis-Related Protein in Maize Resistance to Aspergillus flavus Infection/Aflatoxin Production,” Phytopathology, Vol. 96, No. 1, 2006, pp. 87-95.
- Y. R. Xie, Z. Y. Chen, R. L. Brown and D. Bhatnagar, “Expression and Functional Characterization of Two Pathogenesis-Related Protein 10 Genes from Zea Mays,” Journal of Plant Physiology, Vol. 167, No. 2, 2010, pp. 121- 130. doi:10.1016/j.jplph.2009.07.004
- C. Z. Xie, X. Q. Liang, L. Li and H. Y. Liu, “Cloning and Prokaryotic Expression of ARAhPR10 Gene with Resistance to Aspergillus Flavus in Peanut,” Genomics and Applied Biology in Chinese, Vol. 28, No. 2, 2009, pp. 237- 244.
- X. Q. Liang, C. C. Holbrook, R. E. Lynch and B. Z. Guo, “β-1,3-Glucanase Activity in Peanut Seed (Arachis hypogaea) Is Induced by Inoculation with Aspergillus flavus and Copurifies with a Conglutin-Like Protein,” Phytopathology, Vol. 95, No. 5, 2005, pp. 506-511.
- W. F. Anderson, C. C. Holbrook and D. M. Wilson, “Development of Greenhouse Screening for Resistance to Aspergillus parasiticus Infection and Preharvest Aflatoxin Contamination in Peanut,” Mycopathologia, Vol. 135, No. 2, 1996, pp. 115-118.
- E. Alos, M. Roca, D. J. Iglesias, M. I. Minguez-Mosquera, C. M. Damasceno, T. W. Thannhauser, et al., “An Evaluation of the Basis and Consequences of a Stay-Green Mutation in the Navel Negra Citrus Mutant Using Transcriptomic and Proteomic Profiling and Metabolite Analysis,” Plant Physiology, Vol. 147, No. 3, 2008, pp. 1300- 1315. doi:10.1104/pp.108.119917
- K. J. Livak and T. D. Schmittgen, “Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method,” Methods, Vol. 25, No. 4, 2001, pp. 402-408. doi:10.1006/meth.2001.1262
- C. P. Constabel, C. Bertrand and N. Brisson, “Transgenic Potato Plants Overexpressing the Pathogenesis-Related STH-2 Gene Show Unaltered Susceptibility to Phytophthora infestans and Potato Virus X,” Plant Molecular Biology, Vol. 22, No. 5, 1993, pp. 775-782.
- Y. Wang, G. Nowak, D. Culley, L. A. Hadwiger and B. Fristensky, “Constitutive Expression of Pea Defense Gene DRR206 Confers Resistance to Blackleg (Leptosphaeria maculans) Disease in Transgenic Canola (Brassica napus),” Molecular Plant-Microbe Interactions, Vol. 12, No. 5, 1999, pp. 410-418.
- M. M. Chang, C. C. Chiang, W. M. Martin and L. A. Had- wiger, “Expression of a Pea Disease Resistance Response Gene in the Potato Cultivar Shepody,” American Journal of Potato Research, Vol. 70, No. 9, 1993, pp. 635-647.
- Z. Y. Chen, R. L. Brown, K. E. Damann and T. E. Cleveland, “PR10 Expression in Maize and Its Effect on Host Resistance against Aspergillus flavus Infection and aflatoxin Production,” Molecular Plant Pathology, Vol. 11, No. 1, 2010, pp. 69-81. doi:10.1111/j.1364-3703.2009.00574.x
NOTES
*Corresponding author.

