 Open Journal of Applied Sciences, 2012, 2, 1-10 doi:10.4236/ojapps.2012.21001 Published Online March 2012 (http://www.SciRP.org/journal/ojapps) p54nrb, a PSF Protein Partner, Contributes to Meningitic Escherichia coli K1-Mediated Pathogenicities Lina He1, Feng Chi1, Tao Bo1,2, Lin Wang1,3, Chunhua Wu1, Ambrose Jong1, Shenghe Huang1* 1 Department of Pediatrics, Saban Research Institute of Children’s Hospital Los Angeles, University of Southern California, Los Angeles, USA 2 Department of Pediatrics, Division of Neonatology, The Second Xiangya Hospital, Central South University, Changsha, China 3Department of Histology and Embryology, School of Basic Medical Science, Wuhan University, Wuhan, China Email: *shhuang@hsc.usc.edu Received January 14, 2012; revised February 16, 2012; accepted February 27, 2012 ABSTRACT IbeA is an important invasion determinant contributing to Escherichia coli K1 entry into brain microvascular endothe- lial cells (BMEC) that is a key step in the pathogenesis of E. coli meningitis. Our previous studies have shown that IbeA-induced signaling and E. coli K1 invasion is mediated by two IbeA-binding proteins, vimentin, which is constitu- tively present in the surface of human BMECs (HBMECs), and PSF, which is inducibly expressed in both mesenchymal (endothelium) and non-mesenchymal (epithelium) cells. However, it is unknown whether p54nrb, a PSF partner protein, could contribute to the pathogenesis of E. coli K1 meningitis. Here, we reported that a 54-kDa protein was identified by copurification with PSF through IbeA-affinity chromatography as an IbeA-binding protein, which is identical to p54nrb. Both p54nrb and PSF are RNA-binding proteins and share significant sequence homology. The specific interaction be- tween IbeA and p54nrb was confirmed by Western blot and ligand overlay assays. Recombinant p54nrb blocked E. coli K1 invasion of human BMEC very effectively. Overexpressed p54nrb as a GFP fusion protein in the transfected 293T cells significantly enhanced E. coli K1 invasion. Furthermore, higher levels of surface p54nrb in the transfected 293T cells were detected by flow cytometry. These results suggest that the IbeA invasion protein of E. coli K1 interacts with p54nrb for bacterial invasion of human cells. Keywords: Meningitis; Escherichia coli; BMEC; IbeA; p54nrb; Invasion Protein; Receptor; Protein Interaction 1. Introduction Despite the availability of effective bactericidal antibiot- ics over the last sixty years neonatal bacterial meningitis remains an important cause of high mortality and mor- bidity [1-4]. Escherichia coli is the most common cause of neonatal meningitis [2] and the K1 encapsulated strains are most commonly acquired by neonates via vertical transmission from the maternal genital tract in utero or during passage through the birth canal [2]. Most cases of bacterial meningitis in newborns develop as a result of hematogenous spread. One of the most important issues in the pathogenesis of E. coli meningitis is how circulat- ing pathogens cross the blood-brain barrier (BBB). Strains causing meningitis possess traits that distinguish them from commensal strains of E. coli and other pathogenic strains such as those causing diarrhea and urinary tract infection. Characteristically, meningitic strains of E. coli are composed of a restricted number of O serogroups (O1, O2, O7, O18, O83), produce S fimbriae, express IbeA, and predominately carry K1 capsule (over 84%) [1-3]. The presence of these features implies that meningitic strains possess a defined set of virulence determinants that allow the bacterium to penetrate the BBB and enter the central nervous system. An in vitro brain microvascular endothelial cells (BMEC) culture model with characteris- tics of the BBB has been established for dissecting the molecular and cellular mechanisms of E. coli crossing of the BBB. Using this cell culture model, we and others have shown that efficient penetration of E. coli across the BBB is mediated by multiple factors. Several genetic de- terminants, including ibeA, ibeB, yijP, aslA, traJ, cnfI and ompA, are required for E. coli crossing of the BBB [1-4]. However, the ibeA gene encoding a 50-kDa protein has been found to be unique to pathogenic E. coli K1 strains (e.g., C5 and RS218), while laboratory strains of E. coli K-12 (e.g., DH5 and HB101) as well as noninvasive E. coli K1 (e.g., E412) lack ibeA [1-3,5-8]. In support of the role of IbeA in E. coli K1 invasion, bacterial entry of human BMECs involves a complex in- terplay of IbeA with the host cell receptors and is associ- ated with activation of specific signaling pathways [9-11]. Receptor-mediated mechanisms also play an important *Corresponding autho . Copyright © 2012 SciRes. OJAppS  L. N. HE ET AL. 2 role in biological functions of nicotine [12]. The in- volvement of α7 nicotinic acetylcholine receptors (α7 nAChRs) and IbeA-binding proteins (IBPs) in nicotine- induced stimulation of E. coli invasion has been demon- strated by using α7 antagonists and IbeA. Soluble IbeA protein markedly abolished both the nicotine-induced enhancement and the baseline activity of bacterial inva- sion in human BMECs. These findings suggest that IbeA and its receptors play an important role in both the nico- tine-induced enhancement and the baseline activity of IbeA+ E. coli K1 invasion. These studies suggest that E. coli invasion proteins (i.e., IbeA) and their receptors transduce their signal to a common pathway that can be synergistically enhanced by nicotine-mediated signaling by α7 nAChRs during E. coli invasion of nicotine-stimu- lated human BMEC. Our studies have further shown that signal transductions and cytoskeletal rearrangements me- diated by phosphatidylinositol 3-kinase (PI3K), and cal- cium signaling-mediated by calmodulin kinase II (Cam- KII) are required for IbeA+ E. coli K1 invasion of human BMEC under stimulation with and without nicotine [9-11]. Blocking of PI3K (LY294002) and CamKII (KN93) abolished the entry of IbeA+ E. coli K1 in BMECs in a dose-dependent manner. CamKII-mediated calcium sig- naling is required for both the IbeA receptor vimentin- and α7 nAChR-induced inflammatory responses [10-11]. These data suggest that the IbeA receptors and α7 nAChR could contribute to meningitic inflammation through the common pathway of CamKII-mediated signaling. In order to understand how E. coli invasion proteins interact with BMEC to initiate the signaling required for bacterial entry, a 50-kDa IbeA-binding protein on the surface of human BMECs was identified as vimentin [13]. A similar human BMEC surface protein (IBP55) was pre- viously revealed by immunoblotting with an anti-bovine IBP55 antibody [14]. IbeA, IBP55 and the anti-IBP55 antibody were able to block E. coli invasion of human BMEC [14]. However, the identity of IBP55 is unknown. In our further effort to elucidate the role of IbeA-binding proteins in the pathogenesis of E. coli K1 meningitis, a 100-kDa IbeA-binding protein (IBP100) was purified and identified as polypyrimidine tract-binding protein (PTB)- associated splicing factor (PSF) [15]. Protein sequence comparison shows that PSF has significant sequence ho- mology with another RNA-binding protein p54nrb, which is encoded by a different gene. Bladen et al. demonstrated that p54nrb is a pre-mRNA splicing factor [16]. Besides RNA-binding and DNA-binding properties, p54nrb is able to interact with several proteins including PSF and DNA topoisomerase I [15]. In this study we identify IBP55 as p54nrb and demonstrate the role of human p54nrb protein in IbeA-mediated E. coli K1 invasion of human cells. Like another multi-functional intracellular protein nucleolin which is translocated to the Hep-2 cell surface as a receptor for E. coli O157:H7 [17], p54nrb can serve as a binding protein on the BMEC surface for IbeA. 2. Materials and Methods 2.1. Bacterial Strains, Mammalian Cell Lines and Reagents E. coli E44 is a rifampin-resistant virulent strain derived from RS218 (O18:K1:H7) [1,2]. E. coli DH5α was used for subcloning and BL21 (DE3) for expressing recombinant proteins. The mammalian cells used are human BMEC [2] and human embryonic kidney cell 293T, which were grown at 37˚C in 5% CO2 in RPMI1640 medium (Media- tech) with 10% fetal calf serum. Restriction endonucleases and other enzymes were purchased from New England Biolabs (Beverly, Mass). The anti-IBP55 (Ibe10R) anti- body was prepared by immunizing rabbits with bovine BMEC membrane proteins bound onto IbeA-Ni-Sepharose column [14]. Anti-His6 and GST monoclonal antibodies were generous gifts from Dr. Yu-Zhou Yang. All other chemicals and antibodies were obtained from Sigma (St. Louis, Mo.) unless otherwise stated. 2.2. Bioinformatics Approaches DNA and protein sequence analysis was done with pro- grams developed by Scientific & Education Software (Durham, NC). DNA and deduced protein sequences were used to search the DNA and protein databases at NCBI/NIH by using BLAST and PSI-BLAST [18]. SIM alignment program was used for alignment of two pro- tein sequences [6]. 2.3. Protein Expression and Purification The expression and purification of recombinant proteins in E. coli were carried out according to protocols of manu- facturer (Novagen). The GST-IbeA expression construct pGEX17A [15] and the control vector pGEX-KG [19] were transformed into E. coli DH5α and the constructs for expression of His tag-containing proteins (pET17A [5], pET28A-p54nrb [20] and pET15b-PSF [21]) were trans- formed into E. coli BL21 (DE3). Purification of GST-IbeA and GST was carried out with glutathione sepharose col- umns (Pharmacia Biotech). His-tagged proteins purifica- tion was performed under denatured condition (8 M urea) using nickel-nitrilotriacetic acid (Ni2+-NTA) agarose col- umns (Qiagen). The eluted proteins were dialyzed against decreasing concentrations of urea in the same buffer as described previously [5]. 2.4. Isolation of IbeA-Binding Protein with FPLC Around 1 ml of Ni2+-NTA agarose (Qiagen) was packed in a HR 5/5 column (Amersham), and washed with 10 Copyright © 2012 SciRes. OJAppS  L. N. HE ET AL. 3 mM phosphate buffer containing 150 mM NaCl and 0.2% Tween 20 at 0.5 ml/min on the Biologic FPLC sys- tem (Bio-Rad). The purified His-IbeA in either active or denatured form was diluted and injected into the column at 0.5 ml/min. After washing for 10 min with the phos- phate buffer, 5 ml of HBMEC crude membrane extract was injected into the IbeA-immobilized column at 0.5 ml/min. The column was washed extensively, and the binding complex of IbeA was eluted with 250 mM imi- dazol at 0.5 ml/min and 1.5 ml of peak was manually collected. The 500 μl of the collected sample was pre- cipitated by 10% TCA and analyzed with SDS-PAGE. 2.5. SDS-PAGE and Western Blot Protein samples were resolved on SDS-12.5% poly- acryamide Phastgels (Amersham) at 16˚C. For Western blot the resolved proteins were transferred onto polyvi- nylidene fluoride (PVDF) membranes (Millipore). The PVDF membrane was blocked with 5% non-fat milk powder in PBS plus 0.05% Tween 20 (PBS-T) at room temperature for 1 h. Then the membrane was incubated in the primary antibody (1:3000 dilution in PBS-T) for 1.5 h at room temperature, and washed in PBS-T. The horseradish peroxidase (HRP) conjugated secondary an- tibody (1:5000 dilution in PBS-T) was incubated with the membrane for 1.5 h at room temperature. After washing the proteins were detected using Lumi-light western blot- ting substrate kit (Roche). 2.6. Ligand Overlay Assays The ligand overlay assay was carried out as previously described [22]. Briefly HBMECs were lysed in Laemmli sample buffer by boiling for 5 min. The HBMEC lysate was resolved by SDS-PAGE and transferred onto a PVDF membrane. After blocking with 5% non-fat milk powder the membrane was incubated with 10 μg/ml of GST-IbeA or GST in PBS overnight at 4˚C. Following extensive washing, the GST-IbeA or GST binding spots on the membrane were detected with anti-GST mono- clonal antibody as described above. 2.7. Transfection and Invasion Assays The 293T of approximately 70% confluency was trans- fected with GFP-p54nrb construct pEGFP-C1-p54nrb or GFP construct pEGFP-C1 using the calcium phosphate transfection protocol [23]. The cells were examined un- der fluorescence microscope after 24 h incubation. The transfected cells were used for E. coli K1 invasion assays and Western blot. Invasion assays were performed in the cultured HBMEC and 293T cells by the method de- scribed previously [1,2]. 2.8. Flow Cytometry for the Detection of Cell Surface p54nrb Transfected cells were grown to approximately 80% con- fluency and harvested with trypsin. Cells were then washed thrice in PBS containing 0.5% bovine serum albumin (BSA) and incubated in PBS containing 5% BSA for 30 min. Approximately 3 × 105 cells per sample were incu- bated with anti-p54nrb monoclonal antibody for 1 hour at room temperature. Following a wash with PBS/BSA solu- tion as described above, cells were incubated with PE-con- jugated goat anti-mouse antibody for 1 hour at room tem- perature in the dark. Again, cells were washed in PBS/FBS and resuspended in PBS/FBS for flow cytometry analysis using a FACSCalibur (BD Biosciences, San Jose, CA). Cells treated with secondary antibody alone served as a negative control for each cell line. A minimum of 10,000 events were scored for each cell line. 3. Results 3.1. p54nrb Shows Significant Sequence Homology with IBP100 Protein (PSF) DNA and protein sequences of IBP100 were used to search the DNA and protein databases at the National Center for Biotechnology Information (National Insti- tutes of Health, National Library of medicine, Washing- ton, D.C.) by using the BLAST algorithm. The results indicate that the major portion of PSF protein (241 to 707 residues) shows 59% identity with almost the entire pro- tein sequence of p54nrb (16 to 471 residues) (Figure 1). Human p54nrb shows a high degree homology with the homologous proteins of rats (96%) and mouse (97%) (data not shown), suggesting that p54nrb is a highly con- served protein. However, no homology was found with any other recognized invasion protein receptors, sug- gesting that PSF and p54nrb represents novel IbeA-bind- ing proteins contributing to E. coli K1 invasion of BMEC. 3.2. Interaction between p54nrb and IbeA PSF has been demonstrated to interact with IbeA and contribute to meningitic E. coli K1 invasion in vitro [15]. As p54nrb shows significant homology to PSF and also forms a complex with PSF, it might be involved in E. coli K1 invasion during the development of meningitis. In our previous studies, a polyclonal antibody against bovine IBP55 was generated [14]. To examine whether the anti- IBP55 antibody can recognize both PSF and p54nrb, the purified His-PSF and His-p54nrb were blotted with anti- IBP55 antibody (Figure 2a). The anti-IBP55 antibody was unable to interact with His-PSF. However, His-p54nrb was clearly recognized by the anti-IBP55 antibody. The recognition of p54nrb was further confirmed by blotting GFP-p54nrb with anti-IBP55 antibody (data not shown). To investigate whether IbeA is able to bind both p54nrb Copyright © 2012 SciRes. OJAppS  L. N. HE ET AL. Copyright © 2012 SciRes. OJAppS 4 Figure 1. Protein sequence comparison of human IBP100 with IBP55. Amino acid sequences are denoted in single-letter code. The symbol * indicates the identical amino acids. and PSF, we employed an overlay of Human BMEC lysate with purified GST-IbeA (Figure 2b). Two major bands (55-kDa and 100-kDa) were observed in the GST-IbeA overlay. In contrast, no distinguish bands appeared in the GST overlay control. The positions of 55-kDa and 100-kDa protein bands correspond to the IbeA-binding proteins p54nrb and PSF, respectively. This suggests that IbeA spe- cifically interacts with both p54nrb and PSF. To further examine the interaction between IbeA and p54nrb, we employed ligand affinity purification. The HBMEC lysate was incubated with Ni-NTA-agarose attached active IbeA or Ni-NTA-agarose alone in ep- pendorf tubes. After extensive washing, the IbeA-binding proteins were eluted, and p54nrb in the eluates was de- tected with anti-p54 monoclonal antibody on a western blot (Figure 2c). In contrast to the Ni-NTA-agarose, the eluate from IbeA attached Ni-NTA-agarose contains much stronger signal of p54nrb. To improve the results of affinity purification, we performed the large-scale purification processes on the Biologic FPLC workstation. The HBMEC membrane crude extract was applied onto active IbeA or denatured IbeA saturated Ni-NTA agarose columns. After completion of washing, the IbeA-binding proteins were eluted and analyzed on western blots (Fig- ure 2d). The active IbeA was able to capture p54nrb from crude HBMEC membrane extract. However, no binding of p54nrb from the crude extract to the column bound with denatured His-tagged IbeA was observed. 3.3. Soluble p54nrb Competitively Inhibits E. coli K1 Invasion of Human BMEC To gain further insight into the relevance of p54nrb in the IbeA-mediated entry process, we examined the ability of soluble p54nrb to compete with the human BMEC surface receptor for binding of IbeA+ E. coli K1 during the proc- ess of bacterial entry. As shown in Figure 3, p54nrb blocked E. coli invasion in a dose-dependent manner with a 50% inhibition at 2.5 µg/well (P < 0.01) and an 80% inhibition at 10 µg/well (P < 0.001). In contrast, BSA did not show significant effect on E. coli K1 invasion. We further verified whether p54nrb exhibited any bactericidal effect by incubating the E. coli with p54nrb, followed by colony plating. No bactericidal activity was observed (data not shown). These results suggest that the soluble p54nrb is able to prevent bacterial binding to the cell sur- face of human BMEC, thus blocking E. coli K1 entry. 3.4. Overexpressed p54nrb in Mammalian Cells Enhances E. coli K1 Invasion To investigate whether overexpression of human p54nrb in 293T cells would affect binding and entry of IbeA+ E. coli K1, we transfected 293T cells with a plasmid expressing a green fluorescent protein (GFP)-tagged human p54nrb. The GFP intrinsic fluorescence allows direct visualization of transfected cells (Figures 4a and 4b). Transfected cells were tested for their ability to promote binding and entry of IbeA+ E. coli K1. As shown in Figure 4c, IbeA+ E. coli K1 invasion was 2 times more efficient in transfected cells than in cells transfected with the vector alone. Western blot was used to verify the GFP-p54nrb expression. The native p54nrb of 54 kDa was detected in both GFP-p54nrb and GFP transfected cells, and the GFP-p54nrb of approximate 80 kDa was only detected in the transfected cells with the GFP-p54nrb plasmid (Figure 4d). As a result of these 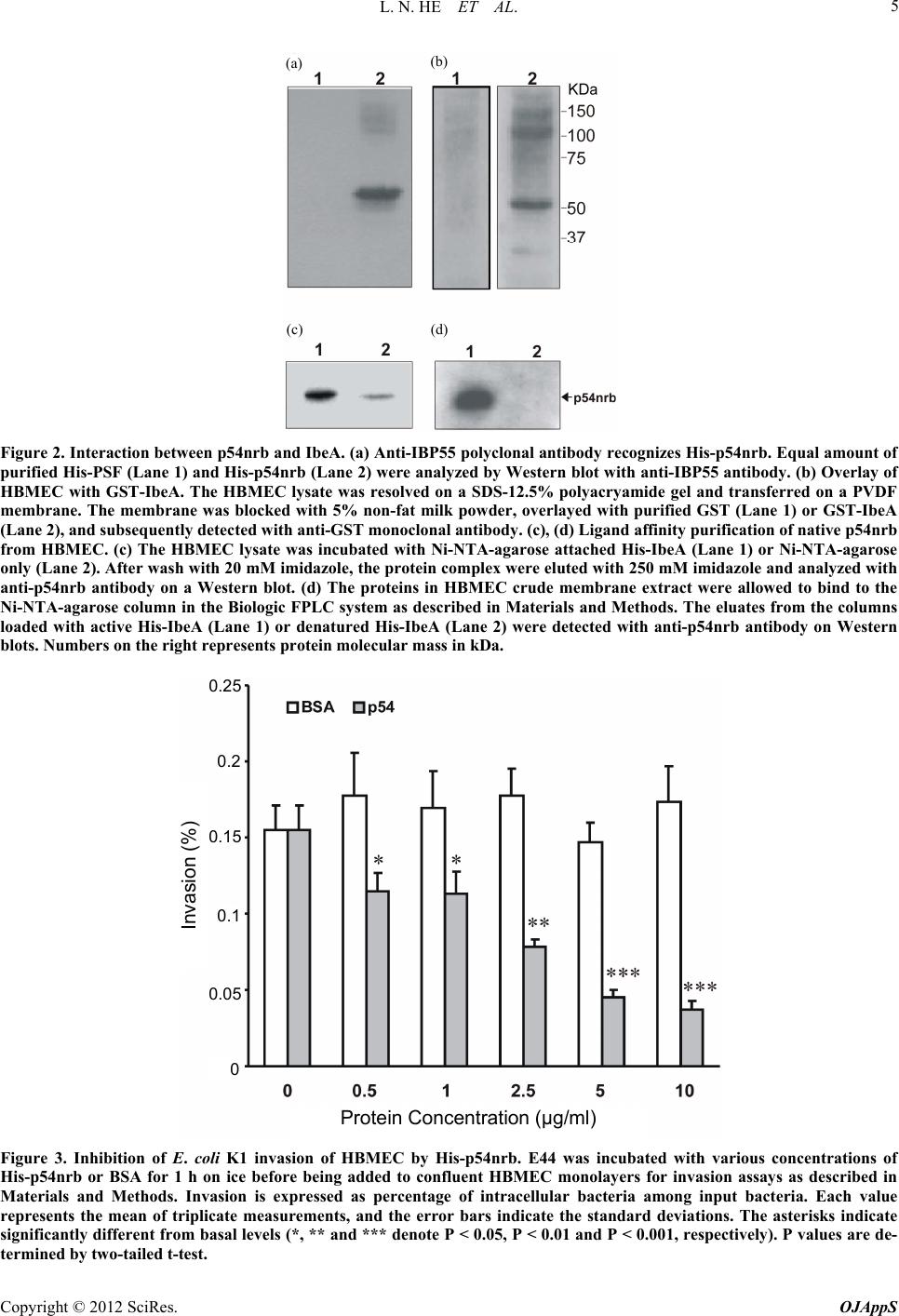 L. N. HE ET AL. 5 (a) (b) (c) (d) Figure 2. Interaction between p54nrb and IbeA. (a) Anti-IBP55 polyclonal antibody recognizes His-p54nrb. Equal amount of purified His-PSF (Lane 1) and His-p54nrb (Lane 2) were analyzed by Western blot with anti-IBP55 antibody. (b) Overlay of HBMEC with GST-IbeA. The HBMEC lysate was resolved on a SDS-12.5% polyacryamide gel and transferred on a PVDF membrane. The membrane was blocked with 5% non-fat milk powder, overlayed with purified GST (Lane 1) or GST-IbeA (Lane 2), and subsequently de te cted with anti-GST monoclonal antibody. (c), (d) Ligand affinity purific ation of native p54nr b from HBMEC. (c) The HBMEC lysate was incubated with Ni-NTA-agarose attached His-IbeA (Lane 1) or Ni-NTA-agarose only (Lane 2). After wash with 20 mM imidazole, the protein complex were eluted with 250 mM imidazole and analyzed with anti-p54nrb antibody on a Western blot. (d) The proteins in HBMEC crude membrane extract were allowed to bind to the Ni-NTA-agarose column in the Biologic FPLC system as described in Materials and Methods. The eluates from the columns loaded with active His-IbeA (Lane 1) or denatured His-IbeA (Lane 2) were detected with anti-p54nrb antibody on Western blots. Numbers on the right represents protein molecular mass in kDa. 0.25 0.2 0.15 0.1 0.05 0 0 Protein Concentration (μg/ml) Invasion (%) Figure 3. Inhibition of E. coli K1 invasion of HBMEC by His-p54nrb. E44 was incubated with various concentrations of His-p54nrb or BSA for 1 h on ice before being added to confluent HBMEC monolayers for invasion assays as described in Materials and Methods. Invasion is expressed as percentage of intracellular bacteria among input bacteria. Each value represents the mean of triplicate measurements, and the error bars indicate the standard deviations. The asterisks indicate significantly different from basal levels (*, ** and *** denote P < 0.05, P < 0.01 and P < 0.001, respectively). P values are de- termined by two-tailed t-test. Copyright © 2012 SciRes. OJAppS 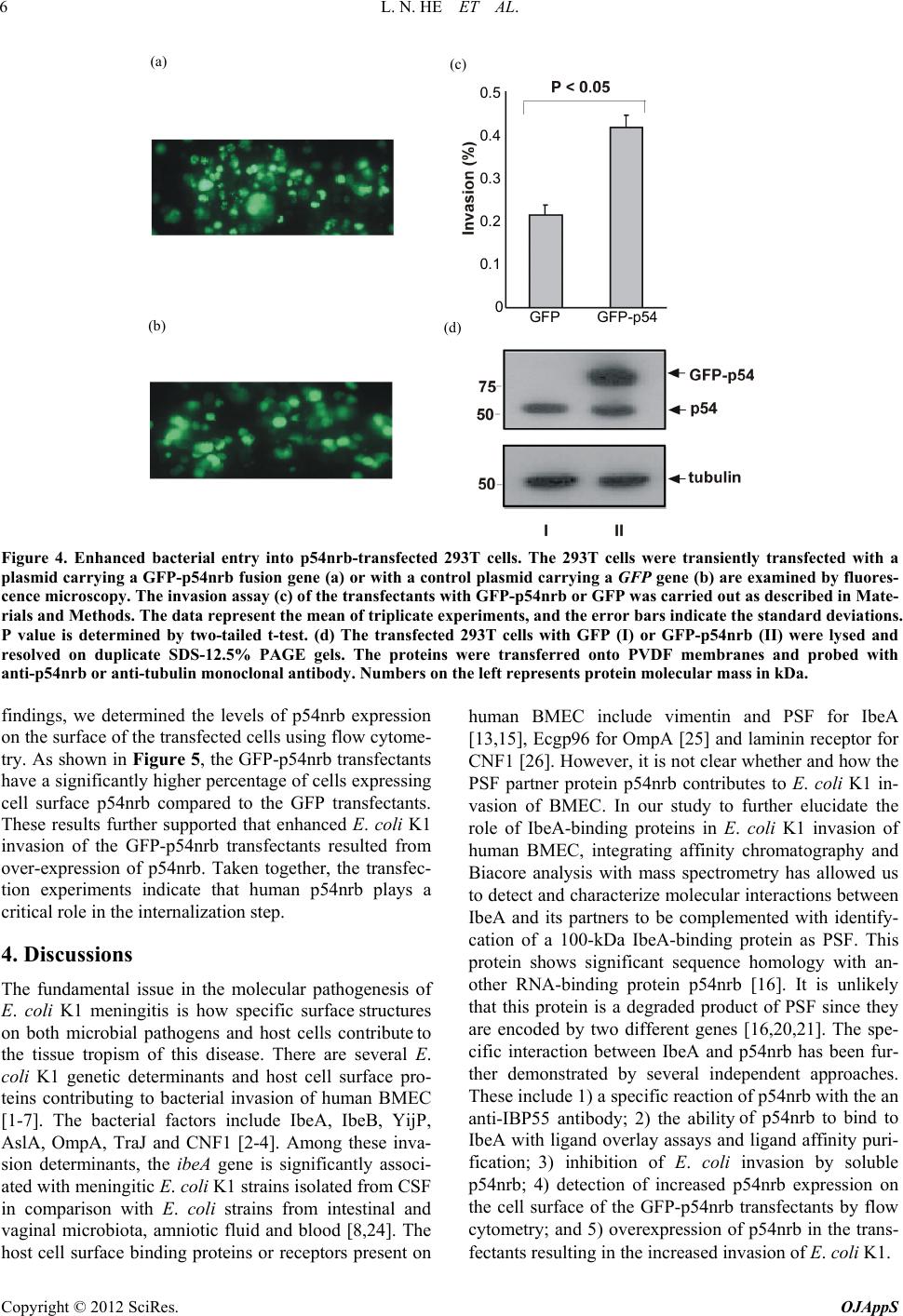 L. N. HE ET AL. 6 GFP GFP-p54 0.5 0.4 0.3 0.2 0.1 0 (a) (b) (c) (d) Figure 4. Enhanced bacterial entry into p54nrb-transfected 293T cells. The 293T cells were transiently transfected with a plasmid carrying a GFP-p54nrb fusion gene (a) or with a control plasmid carrying a GFP gene (b) are examined by fluores- cence microscopy. The invasion assay (c) of the transfectants with GFP-p54nrb or GFP was carried out as described in Mate- rials and Methods. The data represent the mean of triplicate experiments, and the error bars indicate the standard deviations. P value is determined by two-tailed t-test. (d) The transfected 293T cells with GFP (I) or GFP-p54nrb (II) were lysed and resolved on duplicate SDS-12.5% PAGE gels. The proteins were transferred onto PVDF membranes and probed with anti-p54nrb or anti-tubulin monoclonal antibody. Numbers on the left represents protein molecular mass in kDa. findings, we determined the levels of p54nrb expression on the surface of the transfected cells using flow cytome- try. As shown in Figure 5, the GFP-p54nrb transfectants have a significantly higher percentage of cells expressing cell surface p54nrb compared to the GFP transfectants. These results further supported that enhanced E. coli K1 invasion of the GFP-p54nrb transfectants resulted from over-expression of p54nrb. Taken together, the transfec- tion experiments indicate that human p54nrb plays a critical role in the internalization step. 4. Discussions The fundamental issue in the molecular pathogenesis of E. coli K1 meningitis is how specific surface structures on both microbial pathogens and host cells contribute to the tissue tropism of this disease. There are several E. coli K1 genetic determinants and host cell surface pro- teins contributing to bacterial invasion of human BMEC [1-7]. The bacterial factors include IbeA, IbeB, YijP, AslA, OmpA, TraJ and CNF1 [2-4]. Among these inva- sion determinants, the ibeA gene is significantly associ- ated with meningitic E. coli K1 strains isolated from CSF in comparison with E. coli strains from intestinal and vaginal microbiota, amniotic fluid and blood [8,24]. The host cell surface binding proteins or receptors present on human BMEC include vimentin and PSF for IbeA [13,15], Ecgp96 for OmpA [25] and laminin receptor for CNF1 [26]. However, it is not clear whether and how the PSF partner protein p54nrb contributes to E. coli K1 in- vasion of BMEC. In our study to further elucidate the role of IbeA-binding proteins in E. coli K1 invasion of human BMEC, integrating affinity chromatography and Biacore analysis with mass spectrometry has allowed us to detect and characterize molecular interactions between IbeA and its partners to be complemented with identify- cation of a 100-kDa IbeA-binding protein as PSF. This protein shows significant sequence homology with an- other RNA-binding protein p54nrb [16]. It is unlikely that this protein is a degraded product of PSF since they are encoded by two different genes [16,20,21]. The spe- cific interaction between IbeA and p54nrb has been fur- ther demonstrated by several independent approaches. These include 1) a specific reaction of p54nrb with the an anti-IBP55 antibody; 2) the ability of p54nrb to bind to IbeA with ligand overlay assays and ligand affinity puri- fication; 3) inhibition of E. coli invasion by soluble p54nrb; 4) detection of increased p54nrb expression on the cell surface of the GFP-p54nrb transfectants by flow cytometry; and 5) overexpression of p54nrb in the trans- fectants resulting in the increased invasion of E. coli K1. Copyright © 2012 SciRes. OJAppS 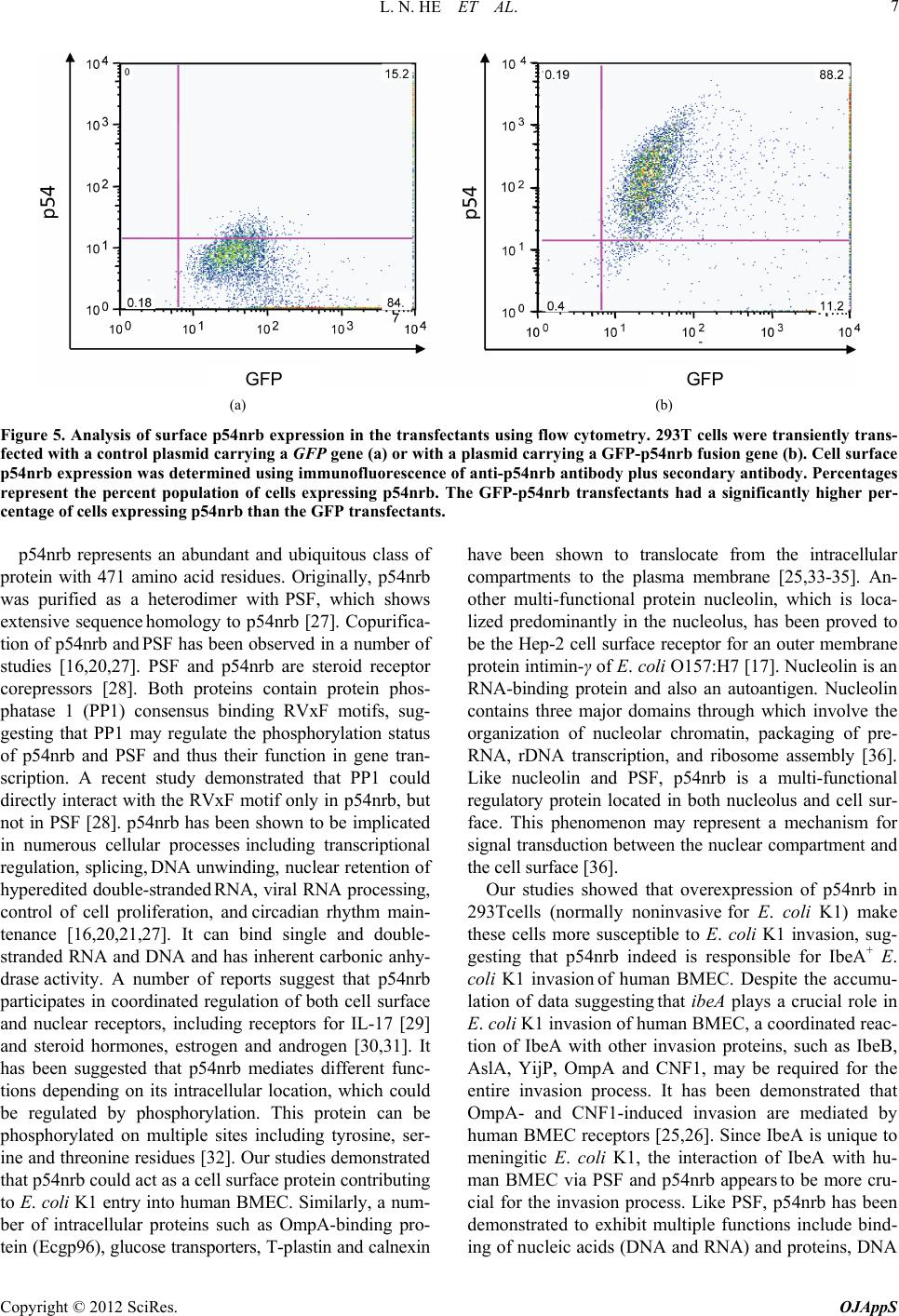 L. N. HE ET AL. 7 10 GFP GFP (a) (b) Figure 5. Analysis of surface p54nrb expression in the transfectants using flow cytometry. 293T cells were transiently trans- fected with a control plasmid carrying a GFP gene (a) or with a plasmid carrying a GFP-p54nrb fusion gene (b). Cell surface p54nrb expression was determined using immunofluorescence of anti-p54nrb antibody plus secondary antibody. Percentages represent the percent population of cells expressing p54nrb. The GFP-p54nrb transfectants had a significantly higher per- centage of cells expressing p54nrb than the GFP transfectants. p54nrb represents an abundant and ubiquitous class of protein with 471 amino acid residues. Originally, p54nrb was purified as a heterodimer with PSF, which shows extensive sequence homology to p54nrb [27]. Copurifica- tion of p54nrb and PSF has been observed in a number of studies [16,20,27]. PSF and p54nrb are steroid receptor corepressors [28]. Both proteins contain protein phos- phatase 1 (PP1) consensus binding RVxF motifs, sug- gesting that PP1 may regulate the phosphorylation status of p54nrb and PSF and thus their function in gene tran- scription. A recent study demonstrated that PP1 could directly interact with the RVxF motif only in p54nrb, but not in PSF [28]. p54nrb has been shown to be implicated in numerous cellular processes including transcriptional regulation, splicing, DNA unwinding, nuclear retention of hyperedited double-stranded RNA, viral RNA processing, control of cell proliferation, and circadian rhythm main- tenance [16,20,21,27]. It can bind single and double- stranded RNA and DNA and has inherent carbonic anhy- drase activity. A number of reports suggest that p54nrb participates in coordinated regulation of both cell surface and nuclear receptors, including receptors for IL-17 [29] and steroid hormones, estrogen and androgen [30,31]. It has been suggested that p54nrb mediates different func- tions depending on its intracellular location, which could be regulated by phosphorylation. This protein can be phosphorylated on multiple sites including tyrosine, ser- ine and threonine residues [32]. Our studies demonstrated that p54nrb could act as a cell surface protein contributing to E. coli K1 entry into human BMEC. Similarly, a num- ber of intracellular proteins such as OmpA-binding pro- tein (Ecgp96), glucose transporters, T-plastin and calnexin have been shown to translocate from the intracellular compartments to the plasma membrane [25,33-35]. An- other multi-functional protein nucleolin, which is loca- lized predominantly in the nucleolus, has been proved to be the Hep-2 cell surface receptor for an outer membrane protein intimin-γ of E. coli O157:H7 [17]. Nucleolin is an RNA-binding protein and also an autoantigen. Nucleolin contains three major domains through which involve the organization of nucleolar chromatin, packaging of pre- RNA, rDNA transcription, and ribosome assembly [36]. Like nucleolin and PSF, p54nrb is a multi-functional regulatory protein located in both nucleolus and cell sur- face. This phenomenon may represent a mechanism for signal transduction between the nuclear compartment and the cell surface [36]. Our studies showed that overexpression of p54nrb in 293Tcells (normally noninvasive for E. coli K1) make these cells more susceptible to E. coli K1 invasion, sug- gesting that p54nrb indeed is responsible for IbeA+ E. coli K1 invasion of human BMEC. Despite the accumu- lation of data suggesting that ibeA plays a crucial role in E. coli K1 invasion of human BMEC, a coordinated reac- tion of IbeA with other invasion proteins, such as IbeB, AslA, YijP, OmpA and CNF1, may be required for the entire invasion process. It has been demonstrated that OmpA- and CNF1-induced invasion are mediated by human BMEC receptors [25,26]. Since IbeA is unique to meningitic E. coli K1, the interaction of IbeA with hu- man BMEC via PSF and p54nrb appears to be more cru- cial for the invasion process. Like PSF, p54nrb has been demonstrated to exhibit multiple functions include bind- ing of nucleic acids (DNA and RNA) and proteins, DNA Copyright © 2012 SciRes. OJAppS  L. N. HE ET AL. 8 pairing, promotion of pre-mRNA splicing, and transcript- tional regulation [16,20,21]. Due to its dual RNA/ DNA-binding properties and the ability to interact with several protein counterparts, p54nrb might be regarded as a “sticky” protein that can link multiple gene regulation processes, which may play a crucial role in the E. coli K1 invasion process. Recently, we have demonstrated that vimentin and PSF could cooperatively contribute to IbeA+ E. coli K1-induced NF-κB signaling, bacterial invasion and leukocyte transmigration across the BBB [37], which are three hallmark features of bacterial meningitis [38]. Thus, it is tempting to speculate that IbeA/vimentin/ PSF/p54nrb interactions may represent an important mechanism responsible for a coordinated modulation of both bacterial and host factors that contribute to the inva- sion process. In summary, the data presented in this report establish that p54nrb is the 55-kDa IbeA-binding protein, IBP55, which mediates E. coli K1 invasion of BMEC. Further work is needed to examine how vimentin, PSF and p54nrb coordinately contribute to IbeA-induced signal transduction and subsequently E. coli K1 entry into the host cells. 5. Acknowledgements We thank Dr. Kwang S Kim for providing brain endothe- lial cells, Dr. Prasadarao V. Nemani for the polyclonal antibody against IBP55, Dr. Yanming Zou for Western blot and ligand overlay assays, Dr. Zhi-Hua Xie (Pro- teinX Lab) for protein purification, Dr. Yu-Zhou Yang for anti-His and anti-GST monoclonal antibodies, and Dr. James G. Patton for p54nrb and PSF constructs. This project is financially supported by Public Health Service grants R01-AI40635 (S.H.H.) and R01-NS047599 (A.J.), and China Natural Science Foundation grant 81070514 (L.W). REFERENCES [1] S. H. Huang, C. A. Wass, Q. Fu, N. V. Prasadarao, M. F. Stins and K. S. Kim, “Escherichia coli Invasion of Brain Microvascular Endothelial Cells in Vitro and in Vivo: Molecular Cloning and Characterization of Invasion Gene ibe10,” Infection and Immunity, Vol. 63, No. 11, 1995, pp. 4470- 4475. [2] S. H. Huang, M. Stins and K. S. Kim, “Bacterial Penetra- tion across the Blood-Brain Barrier during the Develop- ment of Neonatal Meningitis,” Microbes and Infection, Vol. 2, No. 10, 2000, pp. 1237-1244. doi:10.1016/S1286-4579(00)01277-6 [3] S. H. Huang and A. Jong, “Cellular Mechanisms of Micro- bial Proteins Contributing to Invasion of the Blood-Brain Barrier,” Cellular Microbiology, Vol. 3, No. 5, 2001, pp. 277-287. doi:10.1046/j.1462-5822.2001.00116.x [4] K. S. Kim, “Pathogenesis of Bacterial Meningitis: From Bacteraemia to Neuronal Injury,” Nature Reviews Neuro- science, Vol. 4, No. 5, 2003, pp. 376-385. doi:10.1038/nrn1103 [5] S. H. Huang, Z. S. Wan, Y. H. Chen, A. Y. Jong and K. S. Kim, “Further Characterization of Escherichia coli Brain Microvascular Endothelial Cell Invasion Gene ibeA by Deletion, Complementation, and Protein Expression,” Journal of Infectious Diseases, Vol. 183, No. 7, 2001, pp. 1071-1078. doi:10.1086/319290 [6] S. H. Huang, Y. H. Chen, G. Y. Kong, S. H. Chen, M. Borodovsky, J. Besemer and A. Y. Jong, “A Novel Ge- netic Island of Meningitic E. coli K1 Containing the ibeA Invasion Gene (GimA): Functional Annotation and Car- bon Source-Regulated Invasion of Human Endothelial Cells,” Functional & Integrative Genomics, Vol. 1, No. 5, 2001, pp. 312-322. doi:10.1007/s101420100039 [7] S. H. Huang, P. Germon and A. Y. Jong, “The Invasion- Associated Genetic Island GimA in Meningitic E. coli K1,” In: F. Columbus, Ed., Focus on Meningitis Research, Nova Science Publisher, New York, 2004, pp. 97-114. [8] S. Bonacorsi, O. Clermont, V. Houdouin, C. Cordevant, N. Brahimi, A. Marecat, C. Tinsley, X. Nassif, M. Lange and E. Bingen, “Molecular Analysis and Experimental Virulence of French and North American Escherichia coli Neonatal Meningitis Isolates: Identification of a New Virulent Clone,” Journal of Infectious Diseases, Vol. 188, No. 7, 2003, pp. 1895-1906. doi:10.1086/375347 [9] Y. H. Chen, S. H. Chen, A. Jong, Z. Y. Zhou, W. Li, K. Suzuki and S. H. Huang, “Enhanced Escherichia coli In- vasion of Human Brain Microvascular Endothelial Cells Is Associated with Alterations in Cytoskeleton Induced by Nicotine,” Cellular Microbiology, Vol. 4, No. 8, 2002, pp. 503-514. doi:10.1046/j.1462-5822.2002.00209.x [10] F. Chi, T. Jong, L. Wang, Y. Ouyang, C. Wu, W. Li and S. H. Huang, “Vimentin-Mediated Signaling Is Required for IbeA+ E. coli K1 Invasion of Human Brain Microvas- cular Endothelial Cells,” Biochemical Journal, Vol. 427, No. 1, 2010, pp. 79-90. doi:10.1042/BJ20091097 [11] F. Chi, L. Wang, X. Zheng, A. Jong and S. H. Huang, “Recruitment of α7 Nicotinic Acetylcholine Receptor to Caveolin-1-enriched Lipid Rafts is Required for Nico- tine-Enhanced Escherichia coli K1 Entry into Brain En- dothelial Cells,” Future Microbiology, Vol. 6, No. 8, 2011, pp. 953-966. [12] T. Kihara, S. Shimohama, H. Sawada, K. Honda, T. Na- kamizo and H. Shibasaki, “Alpha 7 Nicotinic Receptor Transduces Signals to Phosphatidylinositol 3-Kinase to Block A Beta-Amyloid-Induced Neurotoxicity,” Journal of Biological Chemistry, Vol. 276, No. 17, 2001, pp. 13541-13546. [13] Y. Zou, L. He and S. H. Huang, “Identification of a Sur- face Protein on Human Brain Microvascular Endothelial Cells as Vimentin Interacting with Escherichia coli Inva- sion Protein IbeA,” Biochemical and Biophysical Research Communications, Vol. 351, No. 3, 2006, pp. 625-630. doi:10.1016/j.bbrc.2006.10.091 [14] N. V. Prasadarao, C. A. Wass, S. H. Huang and K. S. Kim, “Identification and Characterization of a Novel Ibe10 Binding Protein that Contributes to Escherichia coli In- Copyright © 2012 SciRes. OJAppS  L. N. HE ET AL. 9 vasion of Brain Microvascular Endothelial Cells,” Infec- tion and Immunity, Vol. 67, No. 3, 1999, pp. 1131-1138. [15] Y. Zou, L. He, C. H. Wu, H. Cao, Z. H. Xie, Y. Wang, A. Y. Jong and S. H. Huang, “PSF Is an IbeA-Binding Pro- tein Contributing to Meningitic Escherichia coli K1 Inva- sion of Human Brain Microvascular Endothelial Cells,” Medical Microbiology and Immunology, Vol. 196, No. 3, 2007, pp. 135-143. doi:10.1007/s00430-006-0034-x [16] C. L. Bladen, D. Udayakumar, Y. Takeda and W. S. Dy- nan, “Identification of the Polypyrimidine Tract Binding Protein-Associated Splicing Factor p54(nrb) Complex as a Candidate DNA Double-Strand Break Rejoining Fac- tor,” Journal of Biological Chemistry, Vol. 280, No. 7, 2005, pp. 5205-5210. doi:10.1074/jbc.M412758200 [17] J. F. Sinclair and A. D. O'Brien, “Cell Surface-Localized Nucleolin Is a Eukaryotic Receptor for the Adhesin In- timin-Gamma of Enterohemorrhagic Escherichia coli O157:H7,” Journal of Biological Chemistry, Vol. 277, No. 4, 2002, pp. 2876-2885. doi:10.1074/jbc.M110230200 [18] S. F. Altschul, T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, “Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs,” Nucleic Acids Research, Vol. 25, No. 7, 1997, pp. 3389-3402. doi:10.1093/nar/25.17.3389 [19] K. Guan and J. E. Dixon, “Eukaryotic Proteins Expressed in Escherichia coli: An Improved Thrombin Cleavage and Purification Procedure of Fusion Proteins with Glu- tathione S-Transferase,” Analytical Biochemistry, Vol. 192, No. 2, 1991, pp. 262-267. doi:10.1016/0003-2697(91)90534-Z [20] R. Peng, B. T. Dye, I. Perez, D. C. Barnard, A. B. Thom- pson and J. G. Patton, “PSF and p54nrb Bind a Conserved Stem in U5 snRNA,” RNA, Vol. 8, No. 10, 2002, pp. 1334-1347. doi:10.1017/S1355838202022070 [21] B. T. Dye and J. G. Patton, “An RNA Recognition Motif (RRM) Is Required for the Localization of PTB Associ- ated Splicing Factor (PSF) to Subnuclear Speckles,” Ex- perimental Cell Research, Vol. 263, No. 1, 2001, pp. 131- 144. doi:10.1006/excr.2000.5097 [22] L. Braun, B. Ghebrehiwet and P. Cossart, “gC1q-R/p32, a C1q-Binding Protein, Is a Receptor for the InIB Invasion Protein of Listeria monocytogenes,” EMBO Journal, Vol. 19, No. 7, 2000, pp. 1458-1466. doi:10.1093/emboj/19.7.1458 [23] R. E. Kingston, C. A. Chen and H. Okayama, “Calcium Phosphate Transfection,” In: J. S. Bonifacino, M. Dasso, J. B. Harford, J. L. Schwartz and K. M. Yamada, Eds., Current Protocols in Cell Biology, John Wiley & Sons, New York, 2003, pp. 2031-2038. doi:10.1002/0471143030.cb2003s19 [24] S. Watt, P. Lanotte, L. Mereghetti, M. M. Schouleur, B. Picard and R. Quentin, “Escherichia coli Strains from Pregnant Women and Neonates: Intraspecies Genetic Dis- tribution and Prevalence of Virulence Factors,” Journal of Clinical Microbiology, Vol. 41, No. 5, 2003, pp. 1929- 1935. doi:10.1128/JCM.41.5.1929-1935.2003 [25] N. V. Prasadarao, P. K. Srivastava, R. S. Rudrabhatla, K. S. Kim, S. H. Huang and S. K. Sukumaran, “Cloning and Expression of the Escherichia coli K1 Outer Membrane Protein A Receptor, a gp96 Homologue,” Infection and Immunity, Vol. 71, No. 4, 2003, pp. 1680-1688. doi:10.1128/IAI.71.4.1680-1688.2003 [26] K. J. Kim, J. W. Chung and K. S. Kim, “67-kDa Laminin Receptor Promotes Internalization of Cytotoxic Necrotiz- ing Factor 1-Expressing Escherichia coli K1 into Human Brain Microvascular Endothelial Cells,” Journal of Bio- logical Chemistry, Vol. 280, No. 2, 2005, pp. 1360-1368. doi:10.1074/jbc.M410176200 [27] A. H. Fox, C. S. Bond and A. I. Lamond, “P54nrb Forms a Heterodimer with PSP1 That Localizes to Paraspeckles in an RNA-Dependent Manner,” Molecular Biology of the Cell, Vol. 16, No. 11, 2005, pp. 5304-5315. doi:10.1091/mbc.E05-06-0587 [28] L. Liu, N. Xie, P. Rennie, J. R. Challis, M. Gleave, S. J. Lye and X. Dong, “Consensus PP1 Binding Motifs Regu- late Transcriptional Corepression and Alternative RNA Splicing Activities of the Steroid Receptor Coregulators, p54nrb and PSF,” Molecular Endocrinology, Vol. 25, No. 7, 2011, pp. 1197-1210. doi:10.1210/me.2010-0517 [29] K. Thiele, D. Riemann, S. A. Navarrete, J. Langner and A Kehlen, “Cell-Cell Contact of Human T Cells with Fibro- blasts Changes Lymphocytic mRNA Expression: In- creased mRNA Expression of Interleukin-17 and Inter- leukin-17 Receptor,” European Cytokine Network, Vol. 11, No. 1, 2000, pp. 53-58. [30] K. Ishitani, T. Yoshida, H. Kitagawa, H. Ohta, S. Nozawa and S. Kato, “p54nrb Acts as a Transcriptional Coactiva- tor for Activation Function 1 of the Human Androgen Receptor,” Biochemical and Biophysical Research Com- munications, Vol. 306, No. 3, 2003, pp. 660-665. doi:10.1016/S0006-291X(03)01021-0 [31] O. Adegbola and G. R. Pasternack, “A pp32-Retinoblas- toma Protein Complex Modulates Androgen Recep- tor-Mediated Transcription and Associates with Compo- nents of the Splicing Machinery,” Biochemical and Bio- physical Research Communications, Vol. 334, No. 2, 2005, pp. 702-708. doi:10.1016/j.bbrc.2005.06.153 [32] A. Proteau, S. Blier, A. L. Albert, S. B. Lavoie, A. M. Traish and M. Vincent, “The Multifunctional Nuclear Protein p54nrb Is Multiphosphorylated in Mitosis and In- teracts with the Mitotic Regulator Pin1,” Journal of Mo- lecular Biology, Vol. 436, No. 4, 2005, pp. 1163-1172. doi:10.1016/j.jmb.2004.12.034 [33] T. Adam, M. Arpin, M. C. Prevost, P. Gounon and P. J. Sansonetti, “Cytoskeletal Rearrangements and the Func- tional Role of T-Plastin during Entry of Shigella flexneri into HeLa Cells,” Journal of Cell Biology, Vol. 192, No. 2, 1995, pp. 367-381. doi:10.1083/jcb.129.2.367 [34] D. Zhou, M. S. Mooseker and J. E. Galan, “An Inva- sion-Associated Salmonella Protein Modulates the Ac- tin-Bundling Activity of Plastin,” Proceeding of the Na- tional Academy of Sciences of the United States of Amer- ica, Vol. 96, No. 18, 1999, pp. 10176-10181. [35] N. Manel, F. J. Kim, S. Kinet, N. Taylor, M. Sitbon and J. L. Battini, “The Ubiquitous Glucose Transporter GLUT-1 Is a Receptor for HTLV,” Cell, Vol. 115, No. 4, 2003, pp. 449-459. doi:10.1016/S0092-8674(03)00881-X [36] M. Srivastava and H. B. Pollard, “Molecular Dissection Copyright © 2012 SciRes. OJAppS  L. N. HE ET AL. Copyright © 2012 SciRes. OJAppS 10 of Nucleolin’s Role in Growth and Cell Proliferation: New Insights,” FASEB Journal, Vol. 13, No. 14, 1999, pp. 1911-1922. [37] F. Chi, T. Bo, C. H. Wu, A. Jong and S. H. Huang, “Vimentin and PSF Act in Concert to Regulate IbeA+ Escherichia coli K1-Induced Activation and Nuclear Translocation of NF-κB in Human Brain Microvascular Endothelial Cells,” PLoS One, in Press, 2012. [38] X. Che, F. Chi, L. Wang, T. Jong, C. H. Wu, X. Wang and S. H. Huang, “Involvement of IbeA in Meningitic Escherichia coli K1-Induced Polymorphonuclear Leuko- cyte Transmigration across Brain Endothelial Cells,” Brain Pathology, Vol. 21, No. 4, 2011, pp. 389-404. doi:10.1111/j.1750-3639.2010.00463.x Abbreviations PSF, polypyrimidine tract-binding protein (PTB)-asso- ciated splicing factor; BMEC, brain microvascular en- dothelial cells; p54nrb, 54-kDa nuclear RNA-binding protein; CSF, cerebrospinal fluid; BBB, blood-brain bar- rier; IBP55, 55-kDa IbeA-binding protein; IBP100, 100-kDa IbeA-binding protein.
|