Advances in Microbiology
Vol.2 No.4(2012), Article ID:25931,10 pages DOI:10.4236/aim.2012.24069
Description of a Putative Oligosaccharyl:S-Layer Protein Transferase from the Tyrosine O-Glycosylation System of Paenibacillus alvei CCM 2051T
Department of NanoBiotechnology, NanoGlycobiology Unit, Universität für Bodenkultur Wien, Wien, Austria
Email: *christina.schaeffer@boku.ac.at
Received August 16, 2012; revised September 22, 2012; accepted October 8, 2012
Keywords: Bacterial Glycosylation; S-Layer; Oligosaccharyl Transferase; Tyrosine-O-Glycosylation; Trans-Membrane Topology
ABSTRACT
Surface (S)-layer proteins are model systems for studying protein glycosylation in bacteria and simultaneously hold promises for the design of novel, glyco-functionalized modules for nanobiotechnology due to their 2D self-assembly capability. Understanding the mechanism governing S-layer glycan biosynthesis in the Gram-positive bacterium Paenibacillus alvei CCM 2051T is necessary for the tailored glyco-functionalization of its S-layer. Here, the putative oligosaccharyl:S-layer protein transferase WsfB from the P. alvei S-layer glycosylation gene locus is characterized. The enzyme is proposed to catalyze the final step of the glycosylation pathway, transferring the elongated S-layer glycan onto distinct tyrosine O-glycosylation sites. Genetic knock-out of WsfB is shown to abolish glycosylation of the S-layer protein SpaA but not that of other glycoproteins present in P. alvei CCM 2051T, confining its role to the S-layer glycosylation pathway. A transmembrane topology model of the 781-amino acid WsfB protein is inferred from activity measurements of green fluorescent protein and phosphatase A fused to defined truncations of WsfB. This model shows an overall number of 13 membrane spanning helices with the Wzy_C domain characteristic of O-oligosaccharyl:protein transferases (O-OTases) located in a central extra-cytoplasmic loop, which both compares well to the topology of OTases from Gram-negative bacteria. Mutations in the Wzy_C motif resulted in loss of WsfB function evidenced in reconstitution experiments in P. alvei ΔWsfB cells. Attempts to use WsfB for transferring heterologous oligosaccharides to its native S-layer target protein in Escherichia coli CWG702 and Salmonella enterica SL3749, which should provide lipid-linked oligosaccharide substrates mimicking to some extent those of the natural host, were not successful, possibly due to the stringent function of WsfB. Concluding, WsfB has all features of a bacterial O-OTase, making it the most probable candidate for the oligosaccharyl:S-layer protein transferase of P. alvei, and a promising candidate for the first O-OTase reported in Gram-positives.
1. Introduction
Bacterial oligosaccharyl:protein transferases (OTases) play a key role in the biosynthesis of glycoproteins, which, in turn, are frequent mediators of interactions between bacterial cells and their environments. A wellknown example is the highly reduced ability of Campylobacter jejuni to colonize mouse intestines when it is deficient in its general protein N-glycosylation system by mutation of either the N-OTase PglB or the prominent glycosylation target PglE [1]. In recent years, interest in biotechnological applications of bacterial protein glycosylation systems has been arising with the aim of efficiently producing recombinant bio-active glycoconjugates for vaccination or drug targeting purposes [2,3].
Bacterial OTases have been characterized in several Gram-negative organisms [4]. PglB has been identified as the N-OTase of Campylobacter jejuni, glycosylating more than 50 proteins at the Asp/Glu-X-Asn-X-Ser/Thr consensus site [5]. PilO and PglL have been found to be the O-OTases responsible for pilin glycosylation in Pseudomonas aeruginosa and Neisseria meningitidis, respectively [6,7]. Recently, homologues of PglL in Vibrio cholerae and Burkholderia thailandensis have been shown to be O-OTases [8]. In all of these cases, glycosylation is a membrane-associated process, with the involvement of a lipid-linked oligosaccharide substrate anchored to the cytoplasmic membrane and an OTase as an integral membrane protein. In most bacteria, similar to eukarya, it appears that O-glycosylation sites are not defined by any primary sequence. However, recent data support the presence of a consensus sequence for Oglycosylation in Bacteriodetes species [9,10].
In Gram-positives, the best studied protein glycosylation systems are those responsible for the O-glycosylation of S-layer proteins in Firmicutes. These organisms are completely covered by a monolayer of self-assembled S-layer glycoproteins, providing a selection advantage to the bacterium in its natural habitat.
The most detailed data on S-layer protein glycosylation is available for Geobacillus stearothermophilus NRS 2004/3a and Paenibacillus alvei CCM 2051T [11-13]. In both organisms, a single S-layer protein species is multiply decorated with a distinct type of polysaccharide, which consists of an adaptor saccharide and an elongated glycan chain composed of a distinct number of repeating units. The repeating structure of the P. alvei CCM 2051T S-layer glycan is [-3)-β-D-Galp-(1[α-D-Glcp-(1,6)]-4)- β-D-ManpNAc-(1-]n=22-25, which is linked via the adapter- [GroA-2-OPO2-4-β-D-ManpNAc-(1,4)]-3)-α-L-Rhap-(1, 3)-α-L-Rhap-(1,3)-α-L-Rhap-(1,3)-β-D-Galp-(1- to specific tyrosine residues of the S-layer protein SpaA. The glycan from G. stearothermophilus is less complex being a poly-L-rhamnan which is linked to Thr 590, Thr 620 and Ser 794 of the S-layer protein SgsE via an adaptor resembling that of P. alvei without the branching part [14,15]. The required enzymatic machinery is encoded in a single S-layer glycosylation (slg) gene cluster [16]. In G. stearothermophilus NRS 2004/3a, each step of the S-layer glycan assembly line could be attributed to a distinct enzyme from its slg gene cluster using in vitro experiments [12]. Finally, an OTase is required for the transfer of the complete glycan chain onto the protein. In the case of P. alvei CCM 2051T, the corresponding enzyme is assumed to be the predicted membrane protein WsfB, the only protein from the slg gene locus, to which (in addition to WsaA, which is encoded in a very short upstream coding sequence) no distinct function could be attributed in the glycan biosynthesis so far. The architecture of the slg gene locus of P. alvei is similar to that of G. stearothermophilus, even though it is not a polycistronic cluster as is the case for G. stearothermophilus [17]. Sequence based annotation of the slg gene locus-enzymes identified WsfB as the putative O-OTase in this organism. Both, WsfB and the G. stearothermophilus ortholog WsaB, contain a Wzy_C domain, which is characteristic of O-OTases as well as O-antigen ligases and polymerases. However, only the former function is required for S-layer protein glycosylation in these organisms.
WsfB function is of special interest, because the glycosylation sites in the P. alvei S-layer protein SpaA are exclusively tyrosine residues, while the vast majority of O-glycosylation in prokarya and eukarya is targeted to serine and threonine residues. Tyrosine glycosylation is only prominent in glycogen, where glycogenin catalyses the transfer of glucose from UDP-glucose to its own Tyr 194 residue [18], and in the insect humoral factor D-glucosyl-L-tyrosine [19]. The O-glycosylation system of P. alvei might, thus, serve as a tool-box for uncommon glycoengineering, complementing the huge potential of known Ser/Thr O-glycosylation systems.
In this work, the possibility to genetically manipulate P. alvei CCM 2051T is exploited to introduce mutations in the Wzy_C domain of WsfB and to study the functional outcome. Further, a transmembrane topology model of WsfB is built on the activity measurements of translationally fused alkaline phosphatase A (PhoA) and green fluorescence protein (GFP), which have complementary activity in the cytoplasm and periplasm, respectively. Following up reports on relaxed substrate specificity of the O-OTases PilO and PglL, transfer of heterologous glycans onto SpaA using WsfB was attempted in distinct Escherichia coli and Salmonella enterica strains that would provide defined lipid-linked sugar substrates for the WsfB enzyme.
2. Materials and Methods
2.1. Genetic Constructs
Point mutations and deletions were introduced into WsfB by overlap extension PCR using genomic DNA of P. alvei CCM 2051T (Czech Collection of Microorganisms, CCM). Two parts of the WsfB sequence were amplified separately, one comprising the part upstream of the site to mutate, the other being the respective downstream part. The reverse primer of the upstream part and the forward primer of the downstream part were overlapping and included the mutation or deletion that was consequently introduced in both stretches. In a second round of PCR, these two amplicons were mixed with overall forward and reverse primers and a product was obtained that contained the desired mutation or deletion. All mutated WsfB sequences were cloned into the Bacillus/E. coli shuttle and expression vector pEXALV [13] via SphI/ KpnI. The primer pair WsfB_SphI_for/WsfB_KpnI_rev was used as outermost primers and the inner primers are named according to the introduced mutation or deletion. The point mutations introduced were R353A, D359A, H395A, Q400A, and E404A. The deletions were WsfB- Δ353-362 and WsfBΔ383-405. A truncated form WsfB1- 714 was obtained by using the respective reverse primer WsfB_K714_KpnI_rev.
WsfB was translationally fused to PhoA at residues E147, S184, R224, F257, E292, T342, K362, E385, L402, T431, F461, S489, E610, G679, and the native C-terminal E781. WsfB-PhoA fusions were cloned by inserting WsfB amplified with the primer pair WsfB_XhoI_for/ WsfB_KpnI_rev upstream of PhoA into the expression vector pHA1-PhoA via XhoI/KpnI. WsfB was translationally fused to GFP at residues E147, S184, R224, F257, E292, T342, E385, L402, T431, F461, S489, E711, and E781. For GFP fusions, WsfB amplified using the primer pair WsfB_NcoI_for/WsfB_KpnI_rev and was inserted into the fusion insertion vector pET28-GFP [20] upstream of GFP via NcoI/KpnI.
SpaA including its native signal peptide was cloned into the expression vector pMLBAD using the primer pair spaA_KpnI_for/spaA_PstI_rev and the restriction sites KpnI/PstI. WsfB was cloned into the expression vector pEXT20 using the primer pair WsfB_BamHI_for/ WsfB_HindIII_rev and the restriction sites BamHI/HindIII.
Cloning was performed according to standard procedures using E. coli DH5α. Restriction enzymes, T4 DNA ligase and DNA preparation kits were purchased from Fermentas and used according to the manufacturer’s instructions.
The primers used in this study are listed in Table 1.
2.2. Mutational Analysis
The constructs containing mutated WsfB variants in pEXALV were transformed into P. alvei CCM 2051T WsfB:L1.LtrB cells (henceforth abbreviated ΔWsfB) by electroporation [13]. The transformed bacteria were cultivated in LB medium containing 10 µg/mL chloramphenicol at 37˚C and 200 rpm to an OD600 1.
Cells were harvested at 14,000 × g for 1 min and the pellet was resuspended in 20 µL of Laemmli buffer per OD unit and incubated for 5 min at 95˚C. Samples were analyzed by SDS-PAGE (10% gel) using Coomassie Brilliant Blue G250 and periodic acid Schiff (PAS) staining [21] for proteins and glycoproteins, respectively. Protein bands corresponding to non-glycosylated and glycosylated SpaA appear at 109 kDa and 155 kDa/240 kDa, respectively [17].
2.3. PhoA Activity Assay
PhoA and GFP activity assays were performed as described previously [22]; (D. Daley, personal communication). Cells of the PhoA deficient strain E. coli CC118 were transformed with the WsfB-PhoA fusion constructs. Transformed cells and untransformed control cells were cultivated in LB medium at 37˚C overnight, diluted 1:100 in fresh medium, and grown for 2.5 h under the same conditions. Ampicillin was added at a concentration of 100 µg/mL, when appropriate. Expression of WsfB-PhoA fusion proteins was induced with L-arabinose at a final concentration of 0.16% and cells were incubated for additional 1.5 h. Iodoacetamide was added to a final concentration of 0.8 mM from a 200 mM iodoacetamide stock solution in 10 mM Tris-HCl, pH 8.0 (buffer A). Cells were incubated for 5 min followed by harvesting from a 1-mL culture by centrifugation at 5000 × g (4˚C, 15 min). Pellets were washed with buffer A, containing 10 mM MgSO4 and 1 mM iodoacetamide. Pellets were resuspended in 800 µL of the same buffer without MgSO4. OD600 of the resuspension was recorded. 100 µL of this suspension were added to 900 µL of 1 M Tris-HCl, pH 8.0, containing 0.1 mM ZnCl2 and 1 mM iodoacetamide. 4 µL of a 0.1% SDS solution and 4 µL of chloroform were added, samples were mixed thoroughly and incubated at 37˚C for 5 min with shaking. Samples were subsequently cooled on ice for 5 min, followed by addition of 100 µL of 0.4% p-nitrophenyl phosphate in 1 M Tris-HCl, pH 8.0, substrate stock solution. Samples were mixed and incubated for 1 h at 37˚C without shaking. Absorbance of the reactions was measured at 405 nm (OD405) to assess PhoA activity and at 550 nm (OD550) to correct for the background. The activity of PhoA was calculated in terms of activity units (AU) according to
 with 0.1 being the dilution factor when transferring the cell suspension to the activity buffer.
with 0.1 being the dilution factor when transferring the cell suspension to the activity buffer.
2.4. GFP Activity Assay
E. coli BL21 cells were transformed with the WsfB-GFP fusion constructs. Transformed cells were grown to OD600 0.4 at 37˚C in LB medium. Expression was induced with IPTG at a final concentration of 0.4 mM and incubation was continued for 2 h. Cells were harvested by centrifugation at 5000 × g (4˚C, 15 min). Cell pellets from 1-mL culture aliquots were resuspended in 200 µL of 0.2 M Tris-HCl, pH 8.0, containing 15 mM NaCl and 50 mM EDTA, and incubated in the dark at 25˚C for 2 h.
Fluorescence was measured at 512 nm from excitiation at 485 nm in a TECAN Infinite F200 plate reader. Obtained values were normalized by sample OD600 to account for variations in cell density.
2.5. Location of Fusion Sites Respective to the Cytoplasmic Membrane
Signals from positions, for which both a PhoA fusion and a GFP fusion was available, were used to define the position of the fusion with respect to the cytoplasmic membrane. A normalized activity ratio (NAR) was calculated from both signals according to Islam [23] by normalizing PhoA and GFP signals to their respective maximum and


Table 1. PCR primers used in this studya.
calculating the ratio of the normalized PhoA and the normalized GFP signal intensity. NAR values above 1 indicate periplasmic location and values below 1 indicate cytoplasmic location.
2.6. Western Blotting
Expression of plasmid encoded proteins (WsfB, SpaA, WsfB-GFP and WsfB-PhoA fusion proteins) was confirmed by Western-blotting as described previously [24], using rabbit antibodies against WsfB and SpaA, and mouse anti-GFP antibody (Roche) as well as mouse antiPhoA antibody (Invitrogen). The secondary antibodies were IRDye 680LT goat anti-rabbit or IRDye 800CW goat anti-mouse (LI-COR), respectively, and detection was performed accordingly at 700 nm or 800 nm using the Odyssey imaging system (LI-COR).
2.7. Heterologous Co-Expression of WsfB and SpaA
Cells of E. coli CWG702 and S. enterica SL3749 were transformed with both pMLBAD-SpaA and pEXT20- WsfB simultaneously. Cells were grown in LB-medium, containing 50 µg/mL trimethoprim and 100 µg/mL ampicillin, at 37˚C and shaking at 200 rpm. At an OD600 0.6 expression was induced by the addition of L-arabinose to a final amount of 0.02% and IPTG to a final concentration of 1 mM. Cells were harvested after 4 h of expression.
3. Results
3.1. Conserved Domains and Functional Association
WsfB is a 781-amino acids protein (calculated molecular mass, 87.5 kDa) containing a conserved Wzy_C domain (pfam 04932) between the amino acid residues 351 and 414. This domain is typically found in a class of sugar transferring enzymes including O-antigen polymerases, O-antigen ligases and OTases. To our current understanding, the S-layer glycosylation pathway of P. alvei requires only the latter. When comparing PAS-stained SDS gels of whole cell extracts of wild-type P. alvei CCM 2051T and DWsfB cells, numerous glycoprotein bands are visible. The distinct S-layer glycoprotein bands appearing at 155 kDa and 240 kDa [17] are the only glyco-stained bands missing in the knock-out strain (Figure 1). Therefore, the function of WsfB is confined to the S-layer protein glycosylation pathway, as was expected from its coding sequence being located within the slg gene locus. Together, the unique function within P. alvei and the presence of the conserved Wzy_C motif are clear indications of the suggested O-OTase function of WsfB.
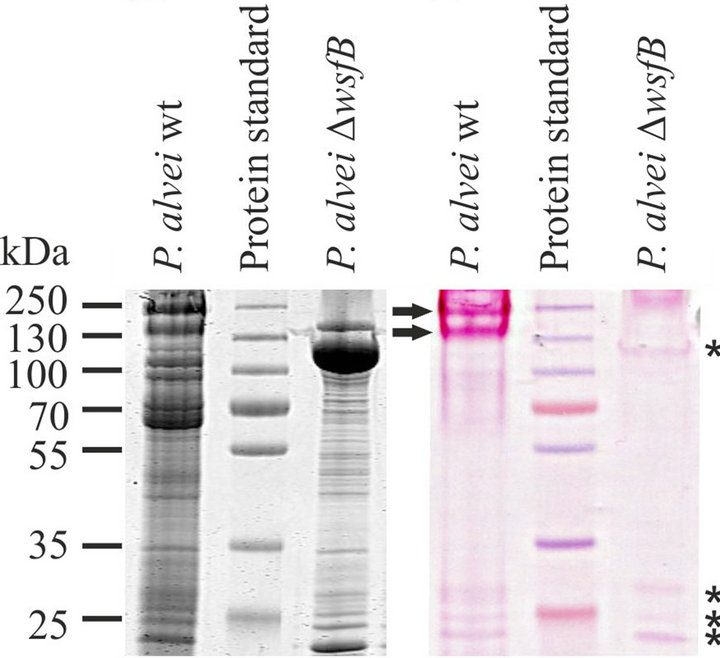 (a) (b)
(a) (b)
Figure 1. Restriction of WsfB function to S-layer protein glycosylation. SDS-PAGE analysis of the glycoprotein profile of P. alvei CCM 2051T wild-type and DWsfB cells upon Coomassie Brilliant Blue (a) and PAS (b) staining. Knockout of WsfB in P. alvei results in loss of S-layer glycosylation while the other glycoproteins remain unaffected. Slayer glycoproteins are indicated by arrows while other glycoproteins are indicated by asterisks.
3.2. Transmembrane Topology
A transmembrane topology model of WsfB was developed based on the translational fusion of PhoA and GFP [22], respectively, to defined C-terminal truncations of WsfB. The fusion constructs with PhoA were expressed in the PhoA-deficient strain E. coli CC118. The activity of PhoA, being only functional when located in the periplasm of the host cells, was assessed photometrically using p-nitrophenyl phosphate as a substrate. Fusions to GFP were expressed in E. coli BL21 cells. The activity of GFP, which is restricted to the cytoplasm, was measured by fluorometry. Using both, PhoA and GFP signals, from twelve fusion sites, a rough pattern of cytoplasmically and extra-cytopasmically located regions of WsfB was obtained. The experimentally determined locations were used as a set of restrictions in the transmembrane topology prediction program HMMTOP [25]. The resulting topology model for WsfB is shown in Figure 2.
The model shows a clear orientation of WsfB towards the extra-cytoplasmic space. Four loops of considerable size are found at the outer side of the membrane, while the cytoplasmic loops are not larger than 22 amino acid residues and probably serve only a structural purpose.
The model contains a 79-residue cytoplasmic tail, which might be of functional importance. The conserved Wzy_C region is found to encompass the majority of the central extra-cytoplasmic loop. The location of this domain at the exterior is consistent with finding of the glycosylation reaction to occur outside the cytoplasm (B. Janesch, unpublished data). A large loop is present at the very C-terminus of WsfB from amino acid 515 to 734. Within this loop, a tetratricopeptide repeat (TPR) motif, which is a mediator of protein-protein interaction [26], is found between residues 519 and 625. The topology
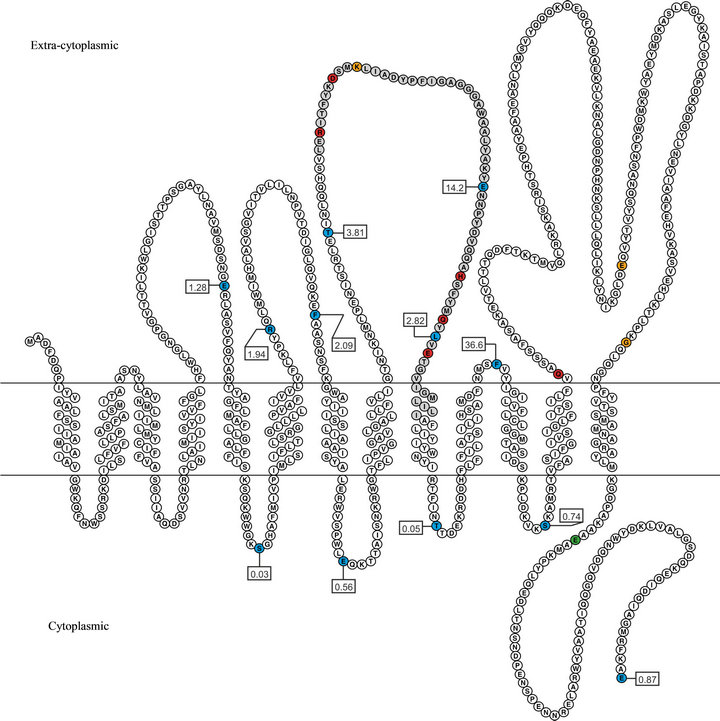
Figure 2. Transmembrane topology model of WsfB. The model was calculated with HMMTOP using the results from PhoA and GFP activity measurements as constraints. Coloured residues indicate fusion to both PhoA and GFP (blue), only to PhoA (orange), only to GFP (green) as well as the Wzy_C motif (gray) and sites of studied mutations (red). The boxed labels contain the normalized activity ratios (NAR) obtained for the PhoA/GFP fusion sites.
model contains 13 membrane spanning helices, which is a reasonably high number related to both, the size of the protein and to similar models of other Wzy_C-like enzymes [27,28].
Activity measurements from one additional fusion site (E711), for which only a WsfB-GFP fusion construct was obtained and three additional sites with only a WsfBPhoA fusion (K362, E610, G679) were not included for calculating the model. However, the absolute values of these signals were amongst the highest of their respective kind and, thus, confirmed the locations of these sites in the model.
3.3. Mutational Analysis
To identify amino acids within the Wzy_C region directly involved in the function of WsfB, point mutations to alanine were introduced between residues 353 and 404. The mutation sites were selected based on their conservation between the Wzy_C domains of WsfB, PilO, PglL and the Wzy_C HMM consensus sequence, excluding Pro and Gly residues (Figure 3). Mutated residues are also indicated in Figure 2.
None of the mutated WsfB sequences was able to reconstitute WsfB function upon homologous expression in P. alvei CCM 2051T ΔWsfB cells (Figure 4). Also deletions of residues R353-K362 and K383-T405 within the Wzy_C region rendered WsfB non-functional. This confirms the Wzy_C domain as a critical region for WsfB function, however, this does not allow for the conclusion of either of the selected residues being of particular importance.
It has to be noted that the truncation of the C-terminal tail of WsfB at K715 also resulted in loss of function. A WsfB variant containing a spontaneous mutation of Glu 516 to His, which is located at the start of the large fourth extra-cytoplasmic loop in the topology model, was able to fully reconstitute the wild-type glycosylation phenoltype in ΔWsfB cells. As a control, non-mutated native WsfB is shown to be able to fully reconstitute the glycosylation when expressed in the knock out strain (Figure 4).
3.4. Substrate Specificity of WsfB
Assuming that WsfB would have a relaxed substrate specificity in heterologous hosts comparable to what has been reported to other OTases from Gram-negative bacteria [8,29,30], WsfB and its native target protein SpaA were co-expressed in E. coli CWG702 and S. enterica SL3749, intending to directly confirm its catalytic function. The native S-layer signal peptide has been shown previously to allow for export of an S-layer protein to the periplasm of E. coli cells [31] and it was employed here to direct SpaA to the periplasm of the Gram-negative host cells, where protein glycosylation is known to occur. Both host strains are deficient in their O-antigen ligase and, thus, accumulate lipid-linked oligosaccharides in the cytoplasm, which makes them suitable to check for utilization of these sugar substrates to be transferred by WsfB. The S. enterica SL3749 O-glycan has the reducing end structure L-Rha-β1,3-D-Gal, which besides the anomeric configuration of the Gal residue matches the reducing end of the P. alvei CCM 2051T S-layer glycan adaptor saccharide L-Rha-a1,3-D-Gal. The repeating unit structure of the E. coli CWG702 O-glycan is (D-Man)n-a1,3-D-GlcNAc, thus providing a rather non-natural substrate for WsfB to be recognized. To analyse for the transfer of these glycans onto SpaA by rWsfB, Western blots of whole cell extracts of the respective expression cultures with anti-SpaA antibody were performed and the glycosylation status of SpaA was checked by PAS staining after separation by SDS-PAGE. However, neither approach for assaying WsfB enzyme activity was successful (data not shown), pointing to more specific requirements of the WsfB/SpaA glycosyla-

Figure 3. Multiple alignment of the Wzy_C hidden Markov model (HMM) consensus sequence and the Wzy_C regions of WsfB and the O-OTases PilO (P. aeruginosa) and PglL (N. meningitidis). Conserved residues are indicated with asterisks. The alignment was performed using Clustal X v2.1.
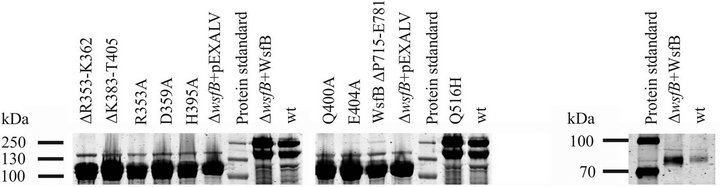 (a) (b)
(a) (b)
Figure 4. Mutational analysis. (a) Whole cell extracts of P. alvei ΔWsfB expressing mutated WsfB variants. Samples were separated by SDS-PAGE and protein bands were visualized by Coomassie Brilliant Blue staining. Mutations within the conserved Wzy_C region as well as truncation of the C-terminal tail resulted in loss of glycosylation, indicated by the appearance of a strong S-layer protein band at 109 kDa and no band at 240 kDa. WsfB Q516H and wild-type WsfB are fully functional. (b) Western-blot of whole cell extracts of P. alvei CCM 2051T expressing native WsfB and P. alvei ΔWsfB expressing plasmid encoded WsfB. WsfB was detected by rabbit anti-WsfB antibody.
tion system than those reported for the Gram-negative OTase systems studied so far.
4. Discussion
The utilization of bacterial protein glycosylation systems is aimed at the production of tailor-made glycoproteins in bacterial cell factories [32,33]. The capability of such production systems requires the detailed understanding of the involved carbohydrate-active enzymes, with OTases being key modules. Here we focused on the predicted O-OTase WsfB from the P. alvei S-layer glycosylation system that allows for the formation of a rare tyrosine O-glycosidic linkage. Our results strongly suggest that WsfB indeed transfers the elongated S-layer glycan onto the tyrosine glycosylation sites.
The O-OTase-characteristic Wzy_C motif of WsfB shares a range of conserved amino acids with PilO and PglL (Figure 3). Mutations of the conserved residues as well as the truncation of the C-terminal tail resulted in inactivation of WsfB. Similarly, in PilO the mutation of conserved R281 to alanine, matching R353A in WsfB, was reported to disable this enzyme [27]. Also, a C-terminal truncation led to inactivation of PilO; however the PilO “tail” is located in the periplasm in contrast to the cytoplasmic orientation in the WsfB model.
A transmembrane topology model for WsfB was derived from measuring the activity of PhoA and GFP fused to several sites in WsfB. The model shows the Wzy_C region being located outside the cytoplasm, which matches the current model of S-layer glycosylation taking place in the extra-cytoplasmic space. The location of the Wzy_C region within WsfB just C-terminal of the protein sequence centre is comparable to the location in PilO and PglL, respectively. Also, the number of 13 transmembrane helices predicted for WsfB is consistent with 12 and 13 helices in the models for PilO and PglL [28], respectively. An additional large extra-cytoplasmic loop is found at the very C-terminus of WsfB, which, like the extra-cytoplasmic tail of PglL, contains a tetratricopeptide repeat (TPR) region [26]. This region probably mediates protein-protein interaction to other enzymes of the S-layer glycosylation pathway. Potential interaction partners would have to be located on the external face of the membrane, which precludes them to the ABC-transporter and the enzymes WsfD and WsfH, which are proposed to be involved in the formation and addition of a branching glucose residue on the S-layer glycan [17].
One remarkable feature of other OTases is their relaxed specificity towards the glycan substrate. It was therefore a worthwhile approach to use readily available though not entirely matching substrates to assess WsfB function in heterologous hosts. However, neither the O-antigen structure from E. coli CWG702 nor the glycan from S. enterica SL3749 where transferred to SpaA by recombinantly expressed WsfB. While this is not yet a proof of a restriction of WsfB function to S-layer glycan transfer in heterologous host, this observation is in line with the confinement of WsfB to the S-layer glycosylation system (Figure 1), which contrasts the diverse native functionalities of other OTases, such as PglB.
Bacterial OTases that have been identified and characterised originate from Gram-negative organisms. The presence of similar enzymes in Gram-positives has not been shown so far [34]. WsfB has all features of a bacterial O-OTase, making it the most probable candidate for the O-OTase of P. alvei CCM 2051T, and a promising candidate for the first O-OTase reported in Gram-positives.
5. Acknowledgements
The authors gratefully acknowledge the kind gifts from Daniel O. Daley, Department of Biochemistry and Biophysics, Stockholm University, Sweden (pHA1-PhoA), from Mario F. Feldman, Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada (pMLBAD, pEXT20, and S. enterica SL3749), and from Chris Whitfield, Department of Molecular and Cellular Biology, University of Guelph, ON, Canada (E. coli CWG702).
Financial support came from the Austrian Science Fund, projects P19047-B12 and P21954-B20 (to C.S.) and P20745-B11 and P22791-B11 (to P. M.).
REFERENCES
- C. M. Szymanski, D. H. Burr and P. Guerry, “Campylobacter Protein Glycosylation Affects Host Cell Interactions,” Infection and Immunity, Vol. 70, No. 4, 2002, pp. 2242-2244. doi:10.1128/IAI.70.4.2242-2244.2002
- C. Lizak, Y. Y. Fan, T. C. Weber and M. Aebi, “NLinked Glycosylation of Antibody Fragments in Escherichia coli,” Bioconjugate Chemistry, Vol. 22, No. 3, 2011, pp. 488-496. doi:10.1021/bc100511k
- J. D. Valderrama-Rincon, A. C. Fisher, J. H. Merritt, Y. Y. Fan, C. A. Reading, K. Chhiba, C. Heiss, P. Azadi, M. Aebi and M. P. DeLisa, “An Engineered Eukaryotic Protein Glycosylation Pathway in Escherichia coli,” Nature Chemical Biology, Vol. 8, No. 5, 2012, pp. 434-436. doi:10.1038/nchembio.921
- H. Nothaft and C. M. Szymanski, “Protein Glycosylation in Bacteria: Sweeter than Ever,” Nature Reviews in Microbiology, Vol. 8, No. 11, 2010, pp. 765-778. doi:10.1038/nrmicro2383
- N. E. Scott, B. L. Parker, A. M. Connolly, J. Paulech, A. V. Edwards, B. Crossett, L. Falconer, D. Kolarich, S. P. Djordjevic, P. Hojrup, N. H. Packer, M. R. Larsen and S. J. Cordwell, “Simultaneous Glycan-Peptide Characterization Using Hydrophilic Interaction Chromatography and Parallel Fragmentation by Cid, Higher Energy Collisional Dissociation, and Electron Transfer Dissociation MS Applied to the N-Linked Glycoproteome of Campylobacter jejuni,” Molecular Cell Proteomics, Vol. 10, No. 2, 2011, pp. M000031-MCP201. doi:10.1074/mcp.M000031-MCP201
- A. Faridmoayer, M. A. Fentabil, D. C. Mills, J. S. Klassen and M. F. Feldman, “Functional Characterization of Bacterial Oligosaccharyl Transferases Involved in OLinked Protein Glycosylation,” Journal of Bacteriology, Vol. 189, No. 22, 2007, pp. 8088-8098. doi:10.1128/JB.01318-07
- M. D. Hartley, M. J. Morrison, F. E. Aas, B. Borud, M. Koomey and B. Imperiali, “Biochemical Characterization of the O-Linked Glycosylation Pathway in Neisseria gonorrhoeae Responsible for Biosynthesis of Protein Glycans Containing N,N’-Diacetylbacillosamine,” Biochemistry, Vol. 50, No. 22, 2011, pp. 4936-4948. doi:10.1021/bi2003372
- C. Gebhart, M. V. Ielmini, B. Reiz, N. L. Price, F. E. Aas, M. Koomey and M. F. Feldman, “Characterization of Exogenous Bacterial Oligosaccharyl Transferases in Escherichia coli Reveals the Potential for O-Linked Protein Glycosylation in Vibrio cholerae and Burkholderia thailandensis,” Glycobiology, Vol. 22, No. 7, 2012, pp. 962- 974. doi:10.1093/glycob/cws059
- C. M. Fletcher, M. J. Coyne, O. F. Villa, M. ChatzidakiLivanis and L. E. Comstock, “A General O-Glycosylation System Important to the Physiology of a Major Human Intestinal Symbiont,” Cell, Vol. 137, No. 2, 2009, pp. 321-331. doi:10.1016/j.cell.2009.02.041
- G. Posch, M. Pabst, L. Brecker, F. Altmann, P. Messner and C. Schäffer, “Characterization and Scope of S-Layer Protein O-Glycosylation in Tannerella forsythia,” Journal of Biological Chemistry, Vol. 286, No. 44, 2011, pp. 38714-38724. doi:10.1074/jbc.M111.284893
- R. Ristl, K. Steiner, K. Zarschler, S. Zayni, P. Messner and C. Schäffer, “The S-Layer Glycome: Adding to the Sugar Coat of Bacteria,” International Journal of Microbiology, Vol. 2011, No. 2011, 2011, Article ID: 127870.
- K. Steiner, R. Novotny, D. B. Werz, K. Zarschler, P. H. Seeberger, A. Hofinger, P. Kosma, C. Schäffer and P. Messner, “Molecular Basis of S-Layer Glycoprotein Glycan Biosynthesis in Geobacillus stearothermophilus,” Journal of Biological Chemistry, Vol. 283, No. 30, 2008, pp. 21120-21133. doi:10.1074/jbc.M801833200
- K. Zarschler, B. Janesch, S. Zayni, C. Schäffer and P. Messner, “Construction of a Gene Knockout System for Application in Paenibacillus alvei CCM 2051T, Exemplified by the S-Layer Glycan Biosynthesis Initiation Enzyme WsfP,” Applied and Environmental Microbiology, Vol. 75, No. 10, 2009, pp. 3077-3085. doi:10.1128/AEM.00087-09
- C. Schäffer, T. Wugeditsch, H. Kählig, A. Scheberl, S. Zayni and P. Messner, “The Surface Layer (S-Layer) Glycoprotein of Geobacillus stearothermophilus NRS 2004/ 3a. Analysis of Its Glycosylation,” Journal of Biological Chemistry, Vol. 277, No. 8, 2002, pp. 6230-6239. doi:10.1074/jbc.M108873200
- P. Messner, R. Christian, C. Neuninger and G. Schulz, “Similarity of ‘Core’ Structures in Two Different Glycans of Tyrosine-Linked Eubacterial S-Layer Glycoproteins,” Journal of Bacteriology, Vol. 177, No. 8, 1995, pp. 2188- 2193.
- R. Novotny, A. Pfoestl, P. Messner and C. Schäffer, “Genetic Organization of Chromosomal S-Layer Glycan Biosynthesis Loci of Bacillaceae,” Glycoconjugate Journal, Vol. 20, No. 7-8, 2004, pp. 435-447.
- K. Zarschler, B. Janesch, M. Pabst, F. Altmann, P. Messner and C. Schäffer, “Protein Tyrosine O-Glycosylation: A Rather Unexplored Prokaryotic Glycosylation System,” Glycobiology, Vol. 20, No. 6, 2010, pp. 787-798. doi:10.1093/glycob/cwq035
- C. Smythe and P. Cohen, “The Discovery of Glycogenin and the Priming Mechanism for Glycogen Biogenesis,” European Journal of Biochemistry, Vol. 200, No. 3, 1991, pp. 625-631. doi:10.1111/j.1432-1033.1991.tb16225.x
- P. Lu, K. J. Kramer, P. A. Seib, D. D. Mueller, R. Ahmed and T. L. Hopkins, “β-D-Glucopyranosyl-O-L-Tyrosine: Synthesis, Properties and Titre during Insect Development,” Insect Biochemistry, Vol. 12, No. 4, 1982, pp. 377-381. doi:10.1016/0020-1790(82)90034-8
- B. Kainz, K. Steiner, M. Möller, D. Pum, C. Schäffer, U. B. Sleytr and J. L. Toca-Herrera, “Absorption, SteadyState Fluorescence, Fluorescence Lifetime, and 2D SelfAssembly Properties of Engineered Fluorescent S-Layer Fusion Proteins of Geobacillus stearothermophilus NRS 2004/3a,” Biomacromolecules, Vol. 11, No. 1, 2010, pp. 207-214. doi:10.1021/bm901071b
- C. Hart, B. Schulenberg, T. H. Steinberg, W. Y. Leung and W. F. Patton, “Detection of Glycoproteins in Polyacrylamide Gels and on Electroblots Using Pro-Q EMERALD 488 Dye, a Fluorescent Periodate Schiff-Base Stain,” Electrophoresis, Vol. 24, No. 4, 2003, pp. 588- 598. doi:10.1002/elps.200390069
- D. O. Daley, M. Rapp, E. Granseth, K. Melen, D. Drew and G. von Heijne, “Global Topology Analysis of the Escherichia coli Inner Membrane Proteome,” Science, Vol. 308, No. 5726, 2005, pp. 1321-1323. doi:10.1126/science.1109730
- S. T. Islam, V. L. Taylor, M. Qi and J. S. Lam, “Membrane Topology Mapping of the O-Antigen Flippase (Wzx), Polymerase (Wzy), and Ligase (WaaL) from Pseudomonas aeruginosa PAO1 Reveals Novel Domain Architectures,” MBio, Vol. 1, No. 3, 2010, Article ID: e00189.
- K. Zarschler, B. Janesch, B. Kainz, R. Ristl, P. Messner and C. Schäffer, “Cell Surface Display of Chimeric Glycoproteins via the S-Layer of Paenibacillus alvei,” Carbohydrate Research, Vol. 345, No. 10, 2010, pp. 1422- 1431. doi:10.1016/j.carres.2010.04.010
- G. E. Tusnady and I. Simon, “The HMMTOP Transmembrane Topology Prediction Server,” Bioinformatics, Vol. 17, No. 9, 2001, pp. 849-850. doi:10.1093/bioinformatics/17.9.849
- G. L. Blatch and M. Lassle, “The Tetratricopeptide Repeat: A Structural Motif Mediating Protein-Protein Interactions,” Bioessays, Vol. 21, No. 11, 1999, pp. 932-939. doi:10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N
- M. Qutyan, M. Paliotti and P. Castric, “PilO of Pseudomonas aeruginosa 1244: Subcellular Location and Domain Assignment,” Molecular Microbiology, Vol. 66, No. 6, 2007, pp. 1444-1458.
- P. M. Power, K. L. Seib and M. P. Jennings, “Pilin Glycosylation in Neisseria meningitidis Occurs by a Similar Pathway to wzy-Dependent O-Antigen Biosynthesis in Escherichia coli,” Biochemical and Biophysical Research Communications, Vol. 347, No. 4, 2006, pp. 904-908. doi:10.1016/j.bbrc.2006.06.182
- M. F. Feldman, M. Wacker, M. Hernandez, P. G. Hitchen, C. L. Marolda, M. Kowarik, H. R. Morris, A. Dell, M. A. Valvano and M. Aebi, “Engineering N-Linked Protein Glycosylation with Diverse O Antigen Lipopolysaccharide Structures in Escherichia coli,” Proceeding of the National Academy of Sciences USA, Vol. 102, No. 8, 2005, pp. 3016-3021.
- A. Faridmoayer, M. A. Fentabil, M. F. Haurat, W. Yi, R. Woodward, P. G. Wang and M. F. Feldman, “Extreme Substrate Promiscuity of the Neisseria Oligosaccharyl Transferase Involved in Protein O-Glycosylation,” Journal of Biological Chemistry, Vol. 283, No. 50, 2008, pp. 34596-34604. doi:10.1074/jbc.M807113200
- K. Steiner, A. Hanreich, B. Kainz, P. G. Hitchen, A. Dell, P. Messner and C. Schäffer, “Recombinant Glycans on an S-Layer Self-Assembly Protein: A New Dimension for Nanopatterned Biomaterials,” Small, Vol. 4, No. 10, 2008, pp. 1728-1740. doi:10.1002/smll.200701215
- S. A. Brooks, “Appropriate Glycosylation of Recombinant Proteins for Human Use: Implications of Choice of Expression System,” Molecular Biotechnology, Vol. 28, No. 3, 2004, pp. 241-255. doi:10.1385/MB:28:3:241
- J. Pandhal and P. C. Wright, “N-Linked Glycoengineering for Human Therapeutic Proteins in Bacteria,” Biotechnology Letters, Vol. 32, No. 9, 2010, pp. 1189-1198. doi:10.1007/s10529-010-0289-6
- A. Dell, A. Galadari, F. Sastre and P. Hitchen, “Similarities and Differences in the Glycosylation Mechanisms in Prokaryotes and Eukaryotes,” International Journal of Microbiology, Vol. 2011, 2011, Article ID: 148178.
NOTES
*Corresponding author.

