Advances in Bioscience and Biotechnology
Vol.3 No.3(2012), Article ID:20036,6 pages DOI:10.4236/abb.2012.33041
Bioremediation of lead by lead-resistant microorganisms, isolated from industrial sample
![]()
Department of Biotechnology, The University of Burdwan, Burdwan, India
Email: *sabsach@rediffmail.com
Received 8 February 2012; revised 22 March 2012; accepted 12 April 2012
Keywords: Bioremediation; Lead Resistant Bacteria; Industrial Bioremediation; Industrial Effluent Toxicity
ABSTRACT
Lead contamination in water is a widespread problem throughout the world and results from industrial use and processing of lead ore. Bio-availability of lead can be hazardous for children and causes mental retardation. The use of lead free petrol is one measure to check this pollution, but this heavy metal is also present in industrial effluents and need to be removed before these effluents are discharged to natural land or water and as well as to the environment. Using bioremediation, bacteria could render lead nonbioavailable would provide an alternative option for detoxifying this contaminant in the environment. The property of some species of bacteria and algae, to extract metals from their surroundings, has been utilized to purify industrial effluents. The first step in devising a bioremediation strategy is to identify candidate bacterial strains capable of modifying the contaminant. Biotechnological approaches are recommended for extraction of metal forms can be grown in ponds where effluents (rich in heavy metals) are discharged. The microbes will extract the heavy metals and sequester them inside their cell membranes. The goal of the present study was to examine the capacity of lead resistant bacteria and bioremediation of lead contaminated water.
1. INTRODUCTION
Living system requires special transport and handling mechanisms to keep them from toxic metals [1]. The toxicity occurs in humans due to environmental pollution via soil or water contamination or due to occupational exposure. Some of these metals are useful to us in low concentrations but are highly toxic in higher concentrations [2]. Bioremediation processes are very attractive in comparison with physicochemical methods such as electrochemical treatment, ion exchange, precipitation, reverse osmosis, evaporation, and sorption for heavy metal removal techniques because they can have lower cost and higher efficiency at low metal concentrations [3,4]. There are a number of bio materials that can be use to remove metal from waste water, such molds, yeasts, bacteria, and seaweeds [5,6]. The ability of microbial stains to grow in the presence of heavy metals would be helpful in the waste water treatment where microorganisms are directly involved in the decomposition of organic matter in biological processes for waste water treatment [7,8], because often the inhibitory effect of heavy metals is a common phenomenon that occurs in the biological treatment of waste water and sewage [9]. Mechanisms of metal resistance in microbes include precipitation of metals as phosphates, carbonates and/or sulfides; volatilization via methylation or ethylation; physical exclusion of electronegative components in membranes and extra cellular polymeric substances (EPS); energy-dependent metal efflux systems; and intra cellular sequestration with low molecular weight, cysteine-rich proteins [10,11]. Growth rate of the sewage isolates in the presence of heavy metal (Cd, Ni, As and Pb) were consistently slower than that of the control similar results have been reported earlier [12,13]. In most of the studies, metal resistance has been reported to hold an association with antibiotic resistance [14-16]. Lead (Pb), a major pollutant that is found in soil, water and air. This metal is also hazardous waste and highly toxic to human, animals, plants and microbes [17]. There are some yeast like Rhodotorula mucilaginosa which is efficient in lead bioadsorption [18]. Hexavalent chromium (Cr (VI)) and trivalent chromium (Cr (III)) are the most prevalent species of chromium in the natural environment [19]. In the present study high degree of heavy metals resistance associated with multiple antibiotics resistance was detected in sewage bacteria. The present study focuses on two lead resistant bacterial strains isolated from sewage water, have metal and antibiotics resistance property and also produce exopolysaccharide. These identified heavy metal resistant bacteria could be useful for the bioremediation of heavy metal contaminated sewage and waste water.
2. MATERIALS AND METHODS
2.1. Sample Collection and Isolation of Lead Resistant Bacteria
Samples were collected from Tamlanala Khal, Durgapur Steel Plant (D.S.P.), West Bengal, India, where effluent discharged from the above industry. Samples were inoculated in nutrient agar (pH ±7.0) plates along with lead acetate with increasing concentrations as 1 mM to 7 mM respectively and incubated at 37˚C for 24 h.
2.2. Microbial Assay, Biochemical Test and Morphological Characterization
After 24 h of incubation 60 colonies were found in plate containing 1 mM lead acetate and around 24 colonies in plate containing 2 mM lead acetate concentration. From both plates, one colony had been selected and designated as A1 and A2. There were no colonies in the plates containing higher concentration of lead ions. Morphological characteristics were observed (Table 1) by bright field microscopy (Olympus GB 89954). Colonies were characterized by Gram’s staining, endospore staining and scanning electron microscopy (SEM). Biochemical tests were performed with isolated bacteria (Table 2, Figure 1).
2.3. Genomic DNA Isolation and PCR Analysis
Genomic DNA was isolated followed by the modification of Vincent and Janarthanan’s protocol [20]. Overnight bacterial cells were cultured in nutrient broth at 37˚C and 3.0 ml of each culture were transferred in a micro centrifuge tube respectively and centrifuged at 10,000
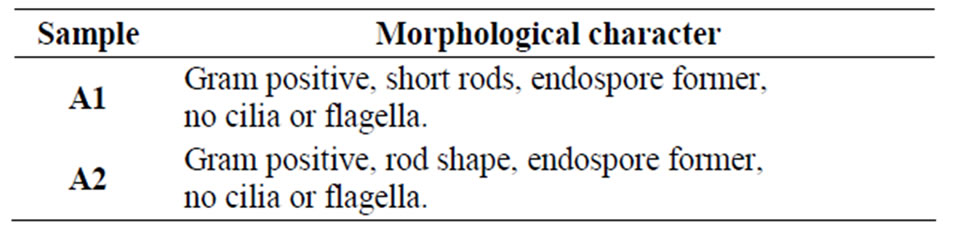
Table 1. Morphological characteristics of isolates obtained by staining and SEM.
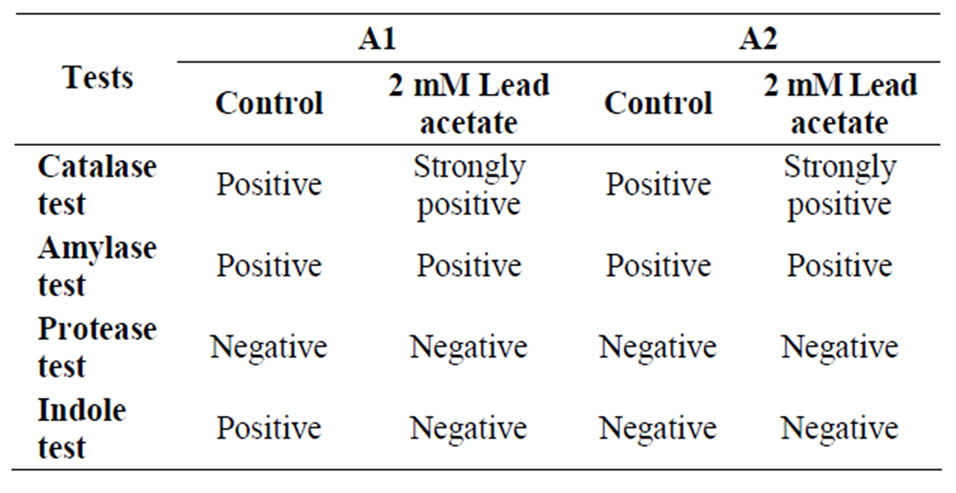
Table 2. Different biochemical tests of isolates.
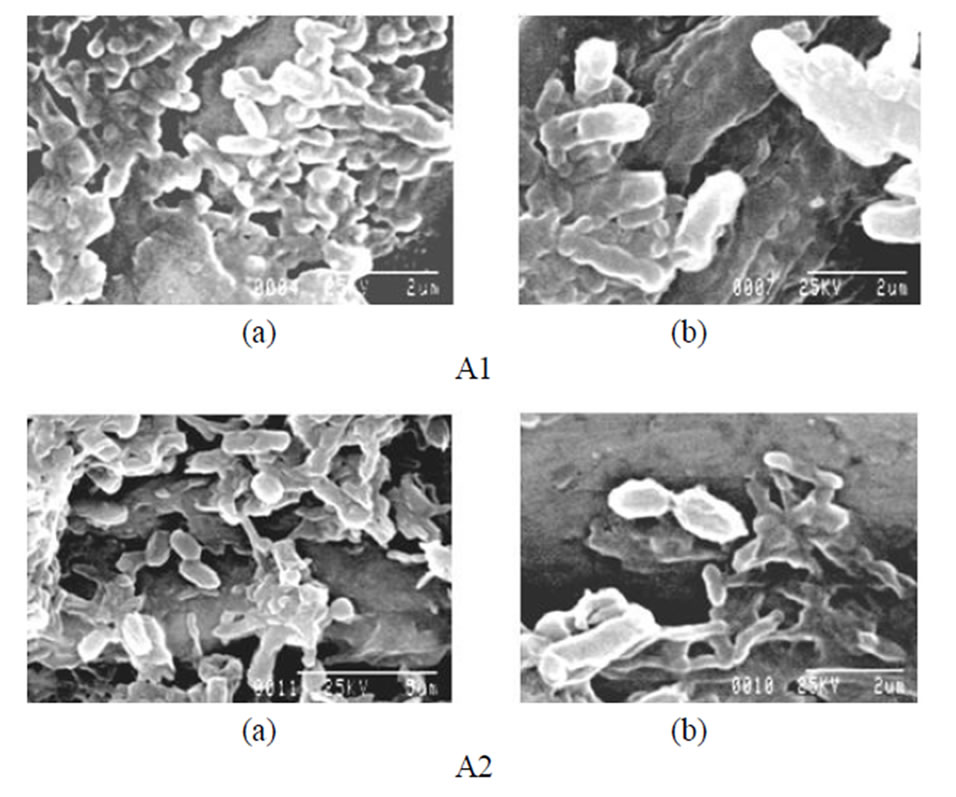
Figure 1. Scanning electron microscopic view of lead resistant bacteria: A1 (a) showing SEM pattern for presence of lead and (b) for absence of lead; A2 (a) showing SEM pattern for presence of lead and (b) for absence of lead.
gx 2 min. Supernatant was removed and pellets were collected and resuspended in 450 µl of TE buffer. 45 µl of 10% SDS was added and incubated for 1 hour at 37˚C. 500 µl phenol-chloroform mixtures was added and centrifuged at 10,000 rpm for 2 minutes. The upper layer was transferred into a new tube and 50 µl of sodium acetate was added and mixed. 300 µl of isopropanol was added to precipitate the DNA and washed by dipping the end of the loop into 1 ml of 70% ethanol for 30 seconds and centrifuged briefly. The DNA was re-suspended in 100 - 200 µl TE buffer. Presence of genomic DNA was confirmed by 0.8% agarose gel electrophoresis [20]. PCR amplification was done using 16s rDNA universal primer as 8F 5’AGA GTT TGA TCC TGG CTC AG3’ and 1492R 5’ACG GCT ACC TTG TTA TTA CGA CTT3’ (Primer delivered by Chromous Biotech Ltd., Bengaluru, India). Thermal cycling was performed with Gene Amp PCR System 2400. Amplification reactions were performed in 25 uL × 4 tube, (total 100 uL volume), each 25 uL tube containing 20 mmol/L TrisHCl (pH = 8.4), 50 mmol/L KCl, 2.0 mmol/L MgCl2, 200 µmol/L of dNTPs, 1 μmol/L of each primer, 30 ng of genomic DNA and 1.5 U of Taq DNA polymerase (0.5 uL for each 25 uL reaction mixture). The temperature profile was followed by initial denaturation at 95˚C × 3 min; 35 cycles of denaturation at 94˚C × 1 min, annealing at 55˚C × 1 min, and extension at 72˚C × 2 min; and final extension at 72˚C × 3 min. The PCR products were analyzed by electrophoresis on 1.5% agarose gel and stained with ethidium bromide (Sigma). 100 bp ladder was used for evaluating the size of amplicons. DNA bands were visualized and documented using a Gel documentation system (Biorad Gel Doc. 2000 system) (Figure 2). Amplicons were sent for sequencing in GCC Biotech Pvt. Ltd., Kolkata, West Bengal, India.
2.4. Determination of Growth Profile
The growth pattern was observed by performing bacterial growth curve experiment. Two sets of 30 ml nutrient broth (in which 1 conical flask contain 2 mM lead acetate and another one was control) were taken and inoculated with 5 ml of activated culture of isolates respectively and kept in shaker incubator at 180 rpm, 30˚C. After every 60 minutes interval optical density was taken (OD620) (Figure 3).
2.5. Polysaccharide Test
Four different cultures (A1 with metal, A1 without metal, A2 with metal and A2 without metal) were taken in different centrifuge tube and centrifuge it for 10 min at 10,000 rpm and supernatant were separated in different test tubes and washed the pellet with water. Those tubes (containing pellet with water) kept in hot water bath for 15 min at 70˚C. Then 2 - 3 drops of 1(N) HCl were added into pellet and heated those tubes again for 3 - 4 min in hot water bath and centrifuge those mixtures and separate the supernatant. Then double volume of 70% ethanol was added with those tubes and previously separated supernatant and pellet treated tubes and all tubes were kept inrefrigerator for overnight (Table 3). White precipitate was observed after overnight incubation which was further confirmed by Dubois method [21].
2.6. Dubois Method
200 µl of white precipitate were taken in different testtubes which were formed after polysaccharide test. Then 2 ml distilled water were added in each tube respectively.
1 ml of 5% phenol solution was added in each tube. There after 5 ml of conc. H2SO4 were added in each tube & kept the tubes in ice-bath till the tubes comes in room temperature. After 30 minutes incubation at room temperature OD was taken (OD480) (Table 4, Figure 4).
2.7. Effect of Temperature, pH, Salt and Minimal Media on Isolates
To study the effect of temperature, pH, salt and minimal media two sets of tubes were prepared. One set containing 2 mM lead acetate with inoculums & other was control without inoculums. The tubes were kept for incubation at different temperatures like 0˚C, 28˚C, 37˚C, 45˚C, 60˚C for 24 h and OD was taken at 620 nm (Figure 5). To study the effect of pH one control set and one 2 mM lead acetate containing set were prepared and the pH was adjusted from 1 - 9 respectively with the help of 1N HCl and 1N NaOH. Tubes were inoculated with A1 and A2 respectively, incubated at 37˚C for 24 hrs and OD was taken at 620 nm (Figure 6). To study the effect of salt one control set (without inoculam) and one 2 mM lead acetate containing set (with inoculam) were prepared with different concentrations of NaCl (1 M, 2 M, 3 M, 4 M),
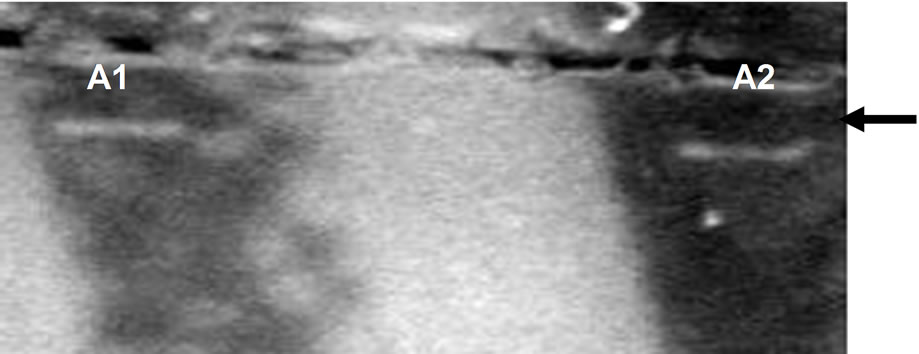
Figure 2. Agarose gel electrophoresis of genomic DNA. 1.5% Agarose gel electrophoresis showing lead resistant isolates, where two DNA bands were found. Band 1 for sample A1 and Band 2 for sample A2.
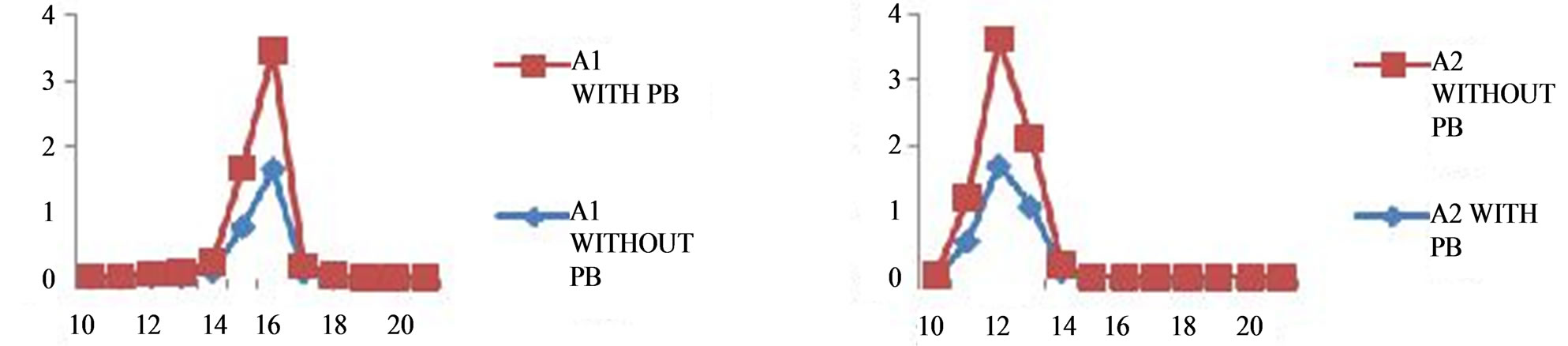
Figure 3. Bacterial growth curve of isolates.
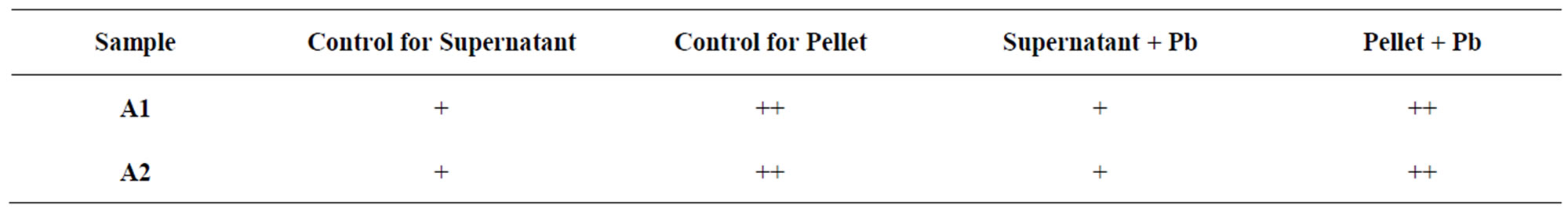
Table 3. Polysaccharide test result of isolates, where A1: sample 1, A2: sample 2, (++): More precipitate, (+): Less precipitate.
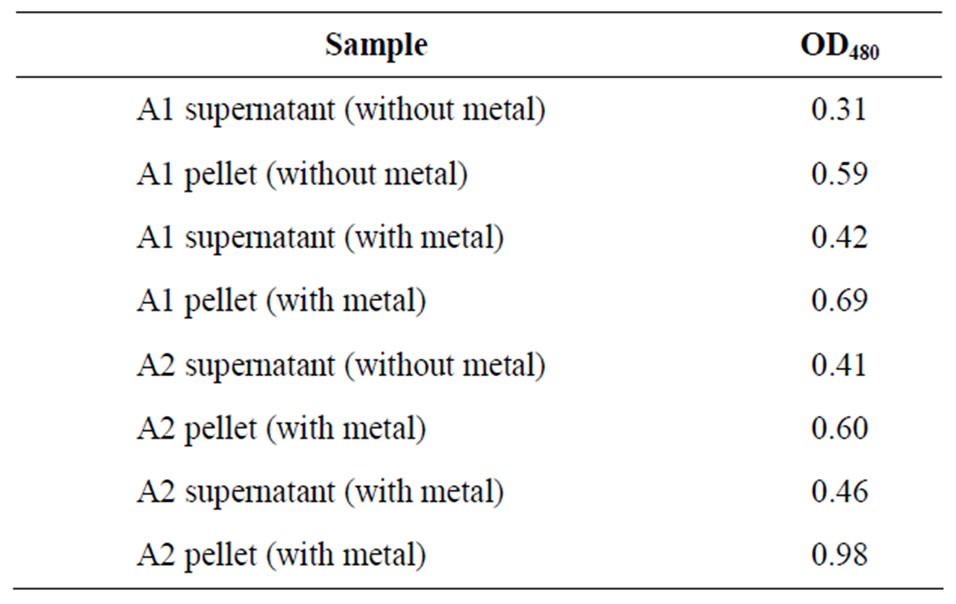
Table 4. Dubois test result of isolates.
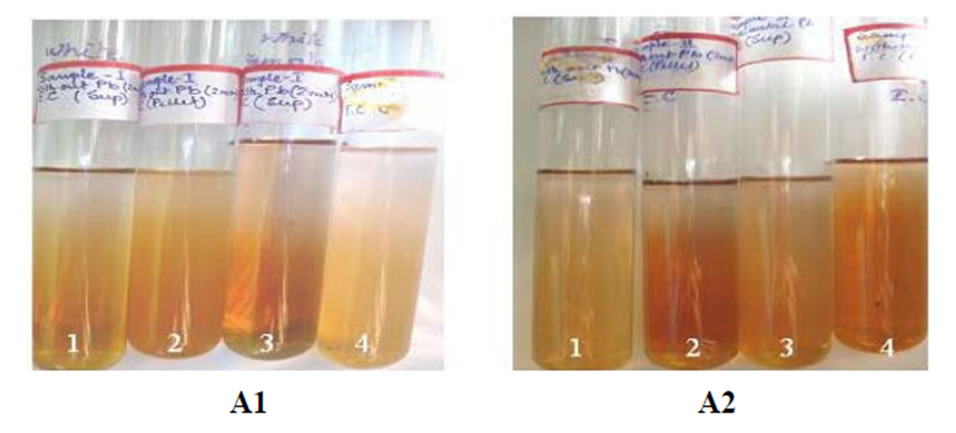
Figure 4. Dubois method. A1: Tube 1 and Tube 2 are without lead, Tube 3 and Tube 4 with lead. 1st and 3rd from supernatant, 2nd and 4th from pellet; A2: Tube 1 and Tube 2 are without lead, Tube 3 and Tube 4 with lead. 1st and 3rd from supernatant, 2nd and 4th from pellet.

Figure 5. Optimum temperature for growth of isolates.

Figure 6. Optimum pH for growth of isolates.
incubated at 37˚C for 24 h and OD was taken at 620 nm (Table 5). For study the effect of minimal media 2 mM lead acetate containing minimal media was prepared, one set was inoculated with isolates and other set was kept as control without inoculum. Both the sets were incubated at 37˚C for 24 h and OD was taken (OD620). Antibiotic resistant tests also done by “agar cup” method with different antibiotics like Tetracyclin, Ampicilin, Kanamycin, Chloramphenicol and Rifampicin.
2.8. Quantification of Lead Accumulation
Atomic Absorption Spectroscopy (AAS) was used for quantification of lead absorption. Isolates were grown in 2 mM lead acetate with nutrient broth and incubated at 37˚C for 72 h. After incubation supernatant were collected. Control was made with 2 mM lead acetate with nutrient broth but without inoculum and sent for AAS at Durgapur Pollution control board (Table 6).
3. RESULT AND DISCUSSIONS
From the above study it was conclude that the isolated microbes (i.e. sample A1 and sample A2) grown well at 37˚C - 45˚C and pH 6 - 7, good capability to tolerate toxic concentration of heavy metal present in industrial effluent. Sample A1 was a Gram positive, rod shape, endospore former bacteria. It was amylase, indole positive in presence of 2 mM lead acetate but in absence of lead acetate it was indole negative (Tables 1 and 2). Sample A1 utilized xylose, lactose, galactose, sucrose, dextrose, maltose in presence of 2 mM lead acetate (Table 7). Sample A2 was also a Gram positive, short rod and endospore forming bacteria. A2 was amylase and protease positive in presence of 2 mM lead acetate (Tables 1 and 2). Sample A2 also utilized xylose, galactose, dextrose, maltose in presence of 2 mM lead acetate (Table 7). Sample A2 grown well in minimal media also. After atomic absorption spectroscopy analysis it was observed that concentration of lead ion decreased in supernatant after 72 h comparing with the control (Table 8). The morphology of isolates A1 and A2 were unchanged after growth in medium with 2 mM lead acetate which suggests that 2 mM concentration of lead acetate was not toxic to the isolate (Figures 1(a) and (b)). Preliminary
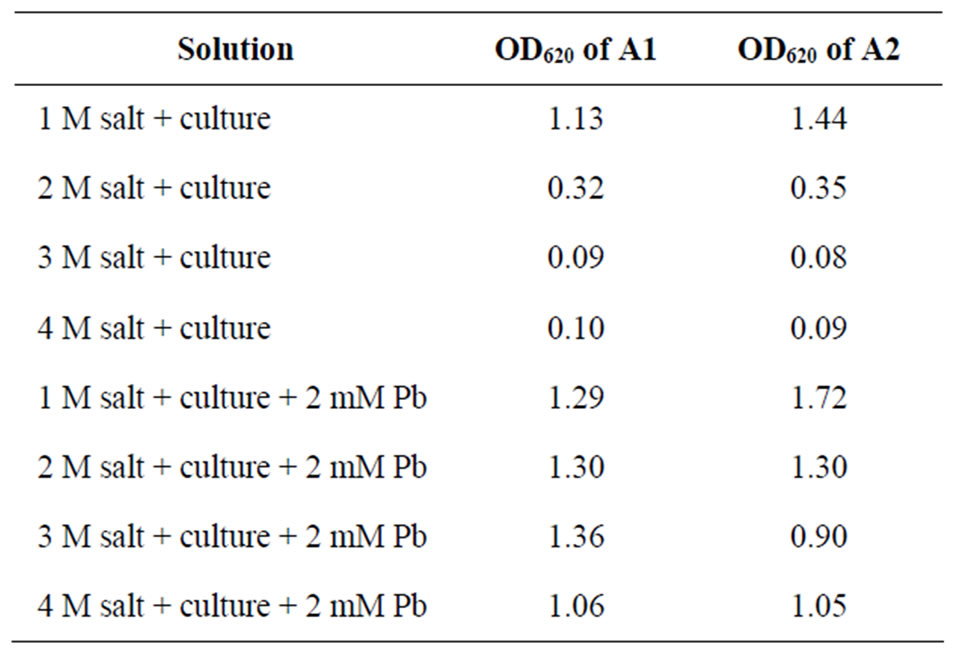
Table 5. Effect of salinity on bacterial growth of isolates.
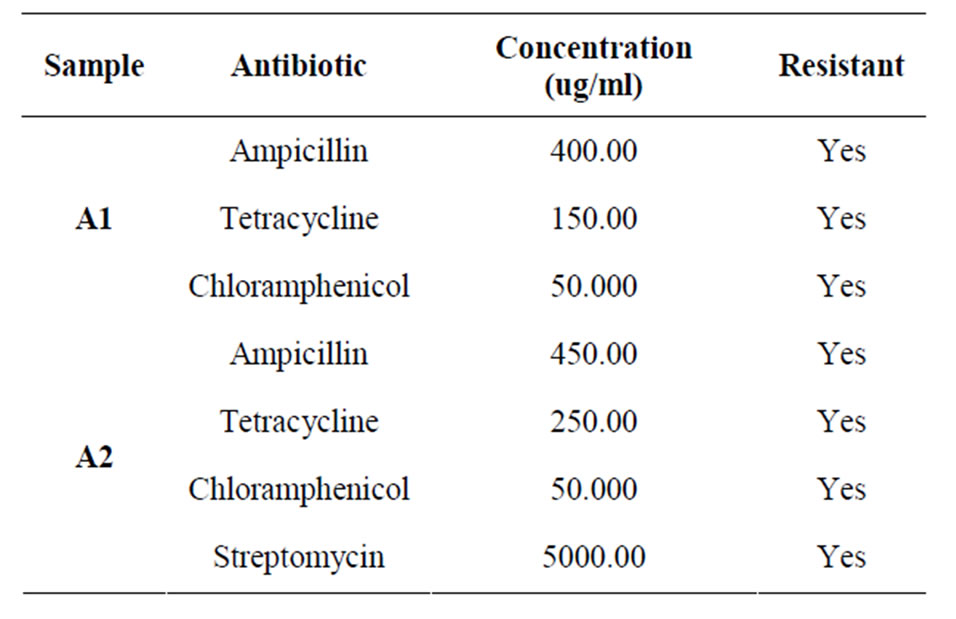
Table 6. Agar cup assay with A1 and A2 of isolates.
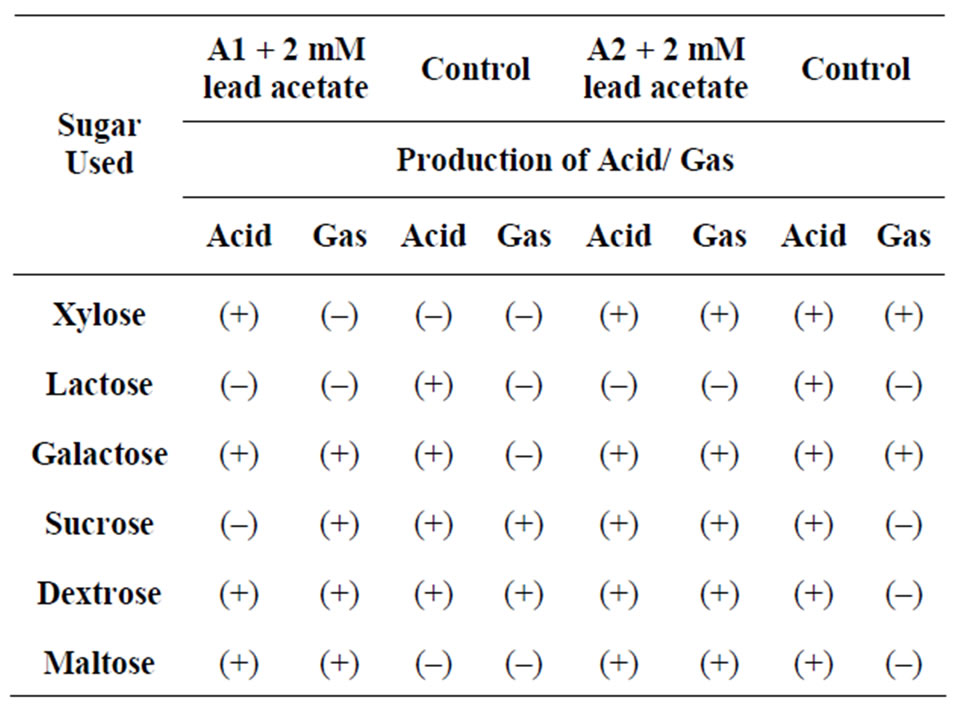
Table 7. Acid-gas production test of isolates, where A1: sample 1, A2: sample 2, (+): positive, (–): negative.

Table 8. Concentration of Pb in sample after Atomic Absorption Spectroscopy, where A1: sample 1, A2: sample 2.
studies showed that the lead resistant bacteria produced large quantities of EPS (Table 3). The Growth curve experiment for A1 showed extended log phase with lead acetate than in absence of lead acetate (Figure 3). A1 and A2 capable of utilizing wide range of carbohydrates in presence of 2 mM lead acetate in media. In both cases when isolated microbes are inoculated in 2 mM lead acetate containing nutrient broth, color of media changed white to brown after 10 days incubation which may indicated the deposition of lead in form of metal complex. These isolated bacteria grown well in nutrient rich media (Nutrient media, Luria burtanii media) as well as in minimal media. Strain A1 grows well at 37˚C - 45˚C and pH 6 - 7. It can tolerate 150 µg/ml of tetracycline, 400 µg/ml of ampicilin, and 50 µg/ml of chloramphenicol. The strain A2 also tolerate 250 µg/ml of tetracycline, 450 µg/ml of ampicilin, 5 mg/ml streptomycin and 50 µg/ml of chloramphenicol.
4. CONCLUSION
From the result of AAS it can be concluded that both isolated lead resistant strains are well efficient and useful for bioremediation of heavy metal contaminated sewage and waste water. These strains are able to grown well in minimal media that could be very helpful in industrial purpose of bioremediation. Strain A1 was more efficient in lead remediation compared to strain A2 though the mechanism of lead remediation is still unknown. Both strains can produce exopolysaccharide. Isolation, purification, structure of the exopolysaccharide and mechanism of lead remediation are further area of interest.
5. ACKNOWLEDGEMENTS
The authors wish to acknowledge Department of Biotechnology, Govt. of India for supporting the research activity. Prof. Pranab Roy wishes to acknowledge Mrigendranath Mondal endowment fund to Burdwan University.
REFERENCES
- Rehman, A., Zahoor, A., Muneer, B. and Hasnain, S. (2008) Chromium tolerance and reduction potential of a Bacillus sp.ev3 isolated from metal contaminated wastewater. Bulletin of Environmental and Contamination Toxicology, 81, 25-29. doi:10.1007/s00128-008-9442-5
- Ge, H.W., Lian, M.F., Wen, F.Z., Yun, Y.F., Jian, F.Y. and Ming, T. (2009) Isolation and characterization of the heavy metal resistant bacteria CCNWRS33-2 isolated from root nodule of Lespedeza cuneata in gold mine tailings in China. Journal of Hazard Materials, 162, 50-56. doi:10.1016/j.jhazmat.2008.05.040
- Bogdanova, E.S., Mindlin, S.Z., Pakrova, E., Kocur, M. and Rouch, D.A. (1992) Mercuric reductase in environmental Gram-positive bacteria sensitive to mercury. FEMS Microbiology Letters, 97, 95-100. doi:10.1111/j.1574-6968.1992.tb05446.x
- Gadd, G.M. and White, C. (1993) Microbial treatment of metal pollution—A working biotechnology. Trends Biotechnology, 11, 353-359. doi:10.1016/0167-7799(93)90158-6
- Vieira, R. and Volesky, B. (2000) Biosorption: A solution to pollution? International Journal of Food Microbialogy, 3, 17-24.
- Waisberg, M., Joseph, P., Hale, B. and Beyersmann, D. (2003) Molecular mechanism of cadmium carcinogenesis. Toxicology, 192, 95-117. doi:10.1016/S0300-483X(03)00305-6
- Munoz, R.A., Munoz, M.T., Terrazas, E., Guieysse, B. and Mattisasson, B. (2006) Sequential removal of heavy metals ions and organic pollutants using an algal-bacterial consortium. Chemosphere, 63, 903-991. doi:10.1016/j.chemosphere.2005.09.062
- Prasenjit, B. and Sumathi, S. (2005) Uptake of chromium by Aspergillus foetidus. Journal of Material Cycles and Waste Management, 7, 88-92. doi:10.1007/s10163-005-0131-8
- Filali, B.K., Taoufik, J., Zeroual, Y., Dzairi, F.Z., Talbi, M. and Blaghen, M. (2000) Waste water bacteria resistant to heavy metals and antibiotics. Current Microbiology, 41, 151-156. doi:10.1007/s0028400
- Gadd, G.M. (1990) Heavy metal accumulation by bacteria and other microorganisms. Experientia, 46, 834-840. doi:10.1007/BF01935534
- Silver, S. (1996) Bacterial resistances to toxic metals—A review. Gene, 179, 9-19. doi:10.1016/S0378-1119(96)00323-X
- Kadirvelu, K. Thamaraiselvi, K. and Namasivayam, C. (2001) Adsorption of nickel (II) from aqueous solution onto activated carbon prepared from coirpith. Seperation and Purification Technology, 24, 477-505. doi:10.1016/S1383-5866(01)00149-6
- Pal, A., Choudhuri, P., Dutta, S., Mukherjee, P.K. and Paul, A.K. (2004) Isolation and characterization of nickel-resistant microflora from serpentine soils of Andaman. World Journal of Microbiology and Biotechnology, 20, 881-886. doi:10.1007/s11274-004-2776-1
- Singh, V., Chauhan, P.K., Kanta, R., Dhewa, T. and Kumer, V. (2010) Isolation and characterization of pseudomonas resistant to heavy metals contaminants. International Journal of Pharmaceutical Sciences Review and Research, 3, 165-168.
- Silver, S. and Misra, T.K. (1988) Plasmid mediated metal resistance. Annual Review of Microbiology, 42, 717-743. doi:10.1146/annurev.mi.42.100188.003441
- Verma, T., Srinath, T., Gadpayle, R.U., Ramtake, P.W., Hans, R.K. and Garg, S.K. (2001) Chromate tolerant bacteria isolated from tannery effluent. Bioresource Technology, 78, 31-35. doi:10.1016/S0960-8524(00)00168-1
- Low, K.S., Lee, C.K. and Liew, S.C. (2000) Sorption of cadmium and lead from aqueous solution by spent grain. Process Biochemistry, 36, 59-64. doi:10.1016/S0032-9592(00)00177-1
- Chatterjee, S., Gupta, D., Roy, P., Chatterjee, N.C., Saha, P. and Dutta, S. (2011) Study of a lead tolerant yeast strain BUSCY1 (MTCC9315). African Journal of Microbiology Research, 5, 5362-5372.
- Chung, J., Nerenberg, R. and Rittmann, B.E. (2006) Bioreduction of soluble chromate using hydrogen based membrane bioflim reactor. Water Research, 40, 1634-1642. doi:10.1016/j.watres.2006.01.049
- Janarthanan, S. and Vincent, S. (2007) Phenol chloroform extraction of DNA. Practical Biotechnology, 29-30.
- Dubois, M., Gilles, K.A., Hamilton, J.K., Rebers, P.A. and Smith, F. (1956) Colorimetric method for determination of sugars and related substances. Analytical Chemistry, 28, 350. doi:10.1021/ac60111a017
NOTES
*Corresponding author.

