Advances in Bioscience and Biotechnology
Vol.2 No.4(2011), Article ID:6859,7 pages DOI:10.4236/abb.2011.24044
Purification of enzymatically inactive peptidylarginine deiminase type 6 from mouse ovary that reveals hexameric structure different from other dimeric isoforms
![]()
1Department of Internal Medicine, Graduate School of Medicine and Pharmaceutical Sciences, University of Toyama, Toyama, Japan;
2Life Science Research Center, University of Toyama, Toyama, Japan;
3Department of Chemistry, The Scripps Research Institute, Jupiter, USA;
4Institute Ruder Boskovic, Division of Molecular Medicine, Zagreb, Croatia;
5Department of Molecular Neuroscience, Graduate School of Medicine and Pharmaceutical Sciences, University of Toyama, Toyama, Japan.
†Present address: Hiroshima University Hospital, Hiroshima, Japan.
E-mail: *hogawa@med.u-toyama.ac.jp
Received 12 May 2011; revised 7 July 2011; accepted 20 July 2011.
Keywords: Peptidylarginine Deiminase; Isoform; Dimer; Hexamer; Mouse Oocytes; Cytoplasmic Sheets
ABSTRACT
The murine peptidylarginine deiminase (PAD) has five isoforms encoded by different genes and participates in a variety of cellular functions through the citrullination of target proteins. The crystal structure of human PAD4 with a dimeric form was previously solved because of the enzyme’s relevance to rheumatoid arthritis. PAD6, abundant in mouse oocytes and eggs, is believed to take part in early events of embryogenesis, but its biochemical properties are little understood. Here we have purified and characterized a recombinant PAD6. A PAD6 cDNA was cloned from mouse ovary RNA and expressed in Escherichia coli through pET29 and pGEX vectors. When benzoyl-L-arginine ethyl ester was used as a substrate, no appreciable activity was detected with a cell homogenate under conditions where a human PAD4 cDNA caused significant activity. Both proteins were affinity-purified to near homogeneity. The circular dichroism spectra of PAD6 and human PAD4 were similar in the far ultraviolet region. On molecular sieving, PAD6 was eluted faster than human PAD4. The cross-linking of PAD6 with dimethyl suberimidate clearly showed six bands on an sodium dodecyl sulfate-polyacrylamide gel. These results indicate that PAD6 can constitute a hexameric structure. The purified PAD6 still showed no enzymatic activity. This unique structure and loss in enzymatic activity is strongly suggested to favor the formation of egg cytoplasmic sheets as the architectural protein.
1. INTRODUCTION
Peptidylarginine deiminases (PAD) (protein-arginine deiminase; E.C. 3.5.3.15) are an enzyme that catalyzes the removal of the imino-moiety of peptidylarginine to yield peptidylcitrulline and ammonia in the presence of Ca2+. PADs have been demonstrated to play a variety of roles in cell signaling pathways, apoptosis, differentiation and transcriptional regulation. Dysregulation of PAD activity is considered to be responsible for various diseases such as rheumatoid arthritis (RA), cancer and colitis. Therefore, development of agents regulating the PAD activity is required for reducing the disease severity [1].
In humans and rodents, the enzyme has five isoforms differing in distribution: PAD1, epidermis and uterus; PAD2, various tissues; PAD3, hair follicles; PAD4, blood cells; PAD6, egg and ovary (for reviews see, e.g., [2-4]). The genes for these PADs are clustered on chromosome 4 at D3 in mice and on chromosome 1 at p36.13 in humans [5,6], and PADs share 40% - 55% sequence homology. PAD4 has been studied extensively in relation to the pathogenesis of RA, because PAD4 gene expression is prominent in the synovial tissue of RA patients, and levels of an anti-cyclic citrullinated peptide antibody (anti-CCP antibody) are significantly augmented in their sera [7]. Therefore, the anti-CCP antibody has recently become a diagnostic marker of RA. Moreover, Suzuki et al. [8] found that mutations of the PAD4 gene are closely linked to RA in an SNP analysis. For these reasons, the crystal structure of human PAD4 (hPAD4) has been solved [9]. hPAD4 has a dimeric structure with a head-to-tail shape whose Nand C-terminal regions function in Ca2+-binding and catalysis, respectively. The binding of Ca2+ to an apo-hPAD4 induces a gross conformational change resulting in a catalytically active form. Cys645 near the C-terminal is necessary for catalysis. However, only one PAD occurs in prokaryotes [10]; the Porphyromonas gingivalis PAD also carries a reactive Cys residue that acts as a nucleophile essential for catalysis despite no requirement of Ca2+ [11,12].
In contrast to PAD1 to 4, the biochemical properties of PAD6 are poorly understood. Interestingly, the crystallographic data on PAD4 along with a sequence alignment among five PADs imply that the acidic residues involved in Ca2+-binding and a reactive Cys in the Cterminal region are not conserved in PAD6 [9]. These findings provide the possibility that PAD6 lacks enzymatic activity. To learn more about structure-function relationships among PADs, it is necessary to purify and characterize PAD6. Since the crystal structure of hPAD4 is available, and hPAD4 and mouse PAD4 have high amino acid sequence homology, we have compared the properties of PAD6 and hPAD4, and present data for a unique structure of PAD6.
2. MATERIALS AND METHODS
2.1. Materials
The biochemical reagents and materials used were obtained as follows: murine reverse transcriptase, Takara Bio (Kyoto, Japan); KOD+ DNA polymerase, Toyobo (Shiga, Japan); Sephacryl S300, Ni2+ Sepharose, PreScissionTM endoprotease, GE Health care; pET28 and pET29, Novagen/Merck; Luria-Bertani (LB) broth, Terrific broth (TB), dimethyl suberimidate, Nacalai tesque (Kyoto, Japan); DE52, Whatman (NJ, USA) and the Amicon UltraTM spin column (cut-off Mr ~ 10,000), Millipore (MA, USA). Other materials were also obtained commercially and used without further treatment.
2.2. cDNA Cloning
cDNA cloning and the construction of expression vectors were performed using general PCR-based procedures as described previously [13]. Total RNA was isolated from mouse ovaries. cDNA to PAD6 was synthesized with random hexamer/oligo(dT) primers by reverse transcriptase. An aliquot of the cDNA library was amplified by KOD+ DNA polymerase with custom-made primers in a thermal cycler (ABI 2720) (Figure 1(a)). The reaction profile was 30 cycles of denaturing at 94˚C for 30 sec, annealing at 55˚C for 30 sec, and elongation at 68˚C for 2 min. The product, once treated with phenol/chloroform, was digested with restriction endonucleases and ligated to a pET29 or pGEX6 vector precut with the corresponding restriction enzymes. These DNAs were used to transform E. coli BL21 (DE3) or BL21, and antibiotic-resistant colonies were subjected to DNA sequencing in a Hitachi-ABI sequencer 3100.
2.3. Purification of PAD6
Proteins were purified at 0˚C - 4˚C. Herein, 10 mM Tris-HCl (pH 8.0) is referred to as “Tris”, unless otherwise stated. A one-fiftieth volume of overnight culture of E. coli harboring pET29/PAD6 cDNA (Figure 1(b)) was inoculated into a 2L-flask containing 500 ml of TB/ kanamycin, and shaken at 37˚C (95 shuttles/min) until the cell density reached 0.7 (absorbance at 600 nm). Then, isopropyl 1-thio-β-D-galactoside (IPTG) was added to make a concentration of 0.15 mM, and the culture was continued at 25˚C overnight. The cells collected by centrifugation were stored in four 50-ml tubes at –80˚C until used. The thawed cells were suspended in 20 ml of Tris, 1 mM EDTA, lysozyme (5 mg), 10 mM 2-mercaptoethanol and 1mM phenylmethylsulfonyl fluoride, and frozen again at –80˚C for 30 min. The thawed sample was sonicated at 200-W at three intervals over a pe-
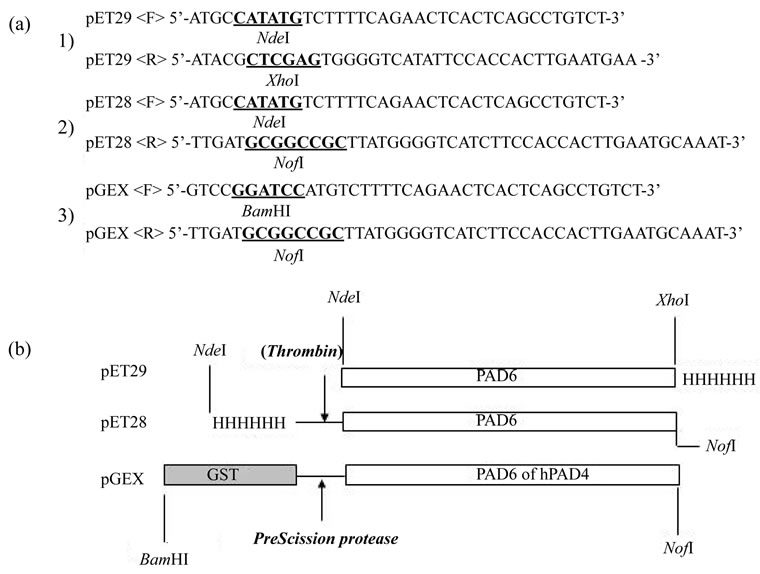
Figure 1. Primers and vectors used. (A) Forward (F) and reverse (R) primer sets, a, b, c, were used for the amplification of PAD6 cDNA from a mouse ovary cDNA library. The PCR products were cleaved with NdeI + XhoI, NdeI + NotI, or BamHI + NotI restriction endonucleases, and introduced into a pET29, pET28, or pGEX vector previously cut with the corresponding endonucleases.
riod of 1 min. After centrifugation at 12,000× g for 15 min, the supernatant was diluted 2-fold with Tris, and applied to a DE52 column (2-cm diameter X 4-cm height) prewashed with Tris containing 1 mM EDTA and 10 mM 2-mercaptoethanol (Buffer A). After a wash with 60 ml of Buffer A, PAD6 was eluted with 90 ml of Tris containing 5 mM mercaptoethanol and 0.2 M NaCl (Buffer B). The eluent was adjusted to make 0.5 M NaCl and 20 mM imidazole. To this solution in 50-ml tubes was added 5 ml of Ni2+ Sepharose (50% v/v). The mixture was added was gently rotated for 1 h, and centrifuged at 1000× g for 2 min. The precipitate was washed five times with Tris containing 0.5 M NaCl and 20 mM imidazole. Finally, PAD6 was released by adding triturated imidazole to make 0.3 M. The eluent concentrated on a spin column was applied to a Sephacryl S300 column (1.5 cm × 98 cm) prewashed with Buffer B, and 5-ml portions were collected. The purity of each fraction was checked by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE), concentrated by a spin column again and kept in aliquots at –80˚C. Alternatively, Ni2+ Sepharose chromatography was preceded by gel chromatography; a DE52-eluted preparation was concentrated with 70% ammonium sulfate and applied to the Sephacryl column. Each fraction (5 ml) was purified by the Ni2+ Sepharose treatment as above.
2.4. Purification of hPAD4 and PAD6 Derived from pGEX6
hPAD4 and PAD6 were obtained through the vector pGEX6 (Figure 1(b)). E. coli cells harboring pGEX6/ hPAD4 or PAD6 were cultured overnight in TB supplemented with ampicillin and IPTG as described above. The cell homogenate was spun, and the supernatant was applied to a DE52 column equilibrated with Tris. The column was extensively washed with Tris containing 0.15 M NaCl and 10 mM 2-mercaptoethanol. The washout solution was brought to a concentration of 0.5 M with respect to NaCl. This preparation received 5 ml of GST-Sepharose (50% v/v), and was gently mixed by rotation for 2 h. The gels were washed 5 times with Tris containing 5 mM 2-mercaptoehanol and 0.5 M NaCl, and then the 10-ml suspension received 30 µl of PreScission protease. The digestion was continued for 24 h with slow rotation. The supernatant after centrifugation was further concentrated on a spin column and applied to the Sephacryl S300 column. Aliquots of the 5-ml fraction were subjected to SDS-PAGE and active fractions were used for experiments.
2.5. Chemical Cross-Linking Experiment
The chemical cross-linking experiment was carried out as described by Davies and Stark [14]. The reaction mixture (20 µl) consisted of 0.1 M triethanolamine-HCl (pH 8.5), 2 - 5 µg of protein, and a 10- to 30-fold excess of dimethyl suberimidate-HCl. The reaction was allowed to proceed for 2 h at 20˚C, and stopped by adding an equal volume of 2% SDS, 20 mM sodium phosphate (pH 7.2), 5% 2-mercaptoethanol, 0.05% bromophenol blue and 50% glycerol. After being denatured at 95˚C for 2 min, the samples were subjected to 3.5% SDS-polyacrylamide disc gel electrophoresis at 8 mA/disc, and stained with Coomassie Brilliant Blue.
2.6. Enzyme Assay
PAD activity was measured in a 700-µl volume with benzoyl-L-arginine ethyl ester (BAEE) as the substrate as described previously [15], in which 10 mM dithiothreitol was included. After incubation at 37˚C for an appropriate period, the reaction was terminated by adding 100 µl of 8 N perchloric acid. Citrulline in the 333- µl supernatant was reacted with 417 µl of 2.25 M H3PO4/4.5 M H2SO4, 167 µl of 20 mM diacetyl monoximine, and 43 µl each of 30 mM thiosemicarbazide and NH4Fe(SO4)2 at 99˚C for 22 min. The absorbance of the reactant was recorded at 540 nm. The molar extinction coefficient for citrulline was 5 × 104. One unit of activity was defined as the amount of enzyme catalyzing the formation of 1 µmol of product per min.
2.7. Other Methods
Circular dichroism (CD) spectra of purified proteins were recorded in a JASCO J-805 as described previously [13]. Prior to analysis, the samples were dialyzed against 10 mM potassium phosphate (pH 7.2) and 0.2 M NaCl for 15 h. Protein was measured by the Lowry method using bovine serum albumin (BSA) as a standard [16]. Site-directed mutagenesis of PAD4 was performed as described [13].
3. RESULTS
3.1. Expression of PAD6 cDNA
A PAD6 cDNA was cloned from mouse ovary RNA, ligated to a pET29 expression vector, and expressed in E. coli in the presence of IPTG. For the PAD enzyme assay, BAEE was used as a substrate throughout the experiment. As shown in Figure 2, the cell homogenate of hPAD4 showed timeand dose-dependent activity, whereas that of PAD6 did not. The latter result verifies a predicttion by X-ray analysis that PAD6 may not bear enzymatic activity [9].
3.2. Purification of PAD6
PAD6 protein was purified to near homogeneity from the supernatant of the E. coli homogenate, because an intense band of PAD6 with a Mr of 76,777 was not seen in the insoluble fraction upon SDS-PAGE. A single band of

Figure 2. hPAD4 and PAD6 activity in cell homogenates. (A) The time course of PAD activity was determined colorimetrically with 50 ml of cell homogenate (~ 20 mg/ml) as described in Materials and Methods. Open and closed circles denote the activity of the hPAD4 and PAD6 homogenates, respectively. (B) Concentration-dependence of the hPAD4 (○) and PAD6 (●) activity of the cell homogenates. The reaction time was 30 min. The acid soluble fraction (333-ml) of the reaction mixture was subjected to color development and measured at 540 nm (the ordinate absorbance).
protein was recovered from the SDS gel and assigned as PAD6 by peptide sequencing (Figure 3). Ser2 was found to be the N-terminal residue instead of the translation initiator Met. The yield of PAD6 was extremely low (below 0.5 mg from 1L-cutured cells), whereas that of hPAD4 exceeded 3 mg. Since hPAD4 was isolated from a GST-fused polypeptide by cleavage with PreScission protease, the N-terminal GST segment might be useful for translation efficiency, and/or protein stability. To test this possibility, PAD6 was expressed through pGEX (Figure 1(b)). However, the yield of PAD6 was still comparable to, or less than, that through pET29. A pET28 vector having a long N-terminal tag (20 residues) was
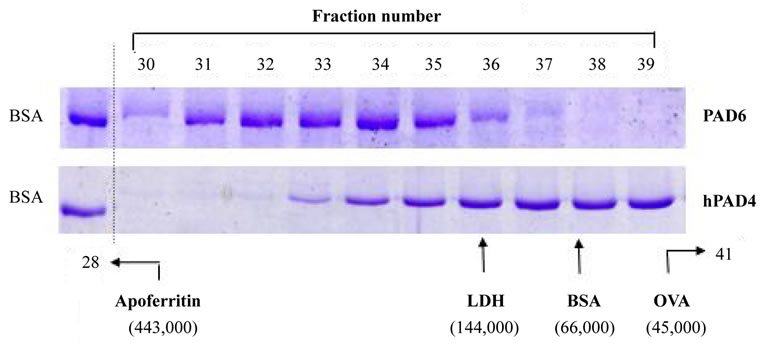
Figure 3. Elution profiles of PAD6 and hPAD4 obtained by Sephacryl S300 gel chromatography. (Upper panel) An ammonium sulfate-enriched preparation (~50 mg) of an eluent from a DE52 column was applied to a Sephacryl S300 column, and fraction numbers 30 to 39 were subjected to Ni2+ Sepharose chromatography as described in Materials and Methods. Aliquots of the imidazole-eluted samples were analyzed by 8% SDS-PAGE. (Lower panel) An hPAD4 preparation (~5 mg) released through PreScission protease digestion was applied to a Sephacryl S300 column, and aliquots of the eluent were analyzed by 8% SDS-PAGE. Separately, authentic standards were run to locate the elution position as monitored at 280 nm: LDH, lactate dehydrogenase; BSA, bovine serum albumin; OVA, ovalbumin.
also without effect (Figure 1), suggesting that the Cterminal His tag does not interfere with the expression. Therefore, the secondary structure of a local nucleotide sequence in the PAD6 mRNA may impede translation, or the codon usage of the PAD6 mRNA may be inadequate in prokaryotic cells.
3.3. Elution Profile of PAD6 on Sephacryl S300
PAD6 and hPAD4 were subjected to molecular sieving, and each fraction was visualized by staining after SDSPAGE (Figure 3). PAD6 appeared with a broad peak at Mr ~ 200,000, while hPAD4 was recovered much slower than PAD6. A major band of hPAD was seen around the position of BSA, which differed from the expected elution position of about Mr 150,000 (dimer) [9].
3.4. Chemical Cross-Linking
The results of size-exclusion chromatography were slightly ambiguous. Thus, we estimated the quaternary structure based on results of chemical cross-linking experiments. RNase A, creatine kinase, lactate dehydrogenase and glutamate dehydrogenase were used as a monomer or oligomeric proteins. The protein bands were visualized by staining after disc SDS-PAGE. Unexpectedly, PAD6 revealed six bands (Figure 4). hPAD4 produced a dimer although its population was slightly small compared with a parent band. PAD6 via pGEX also showed a hexamer (data not shown). These results, together with those of the molecular sieving experiment, imply that hPAD4 constitutes in part a monomeric
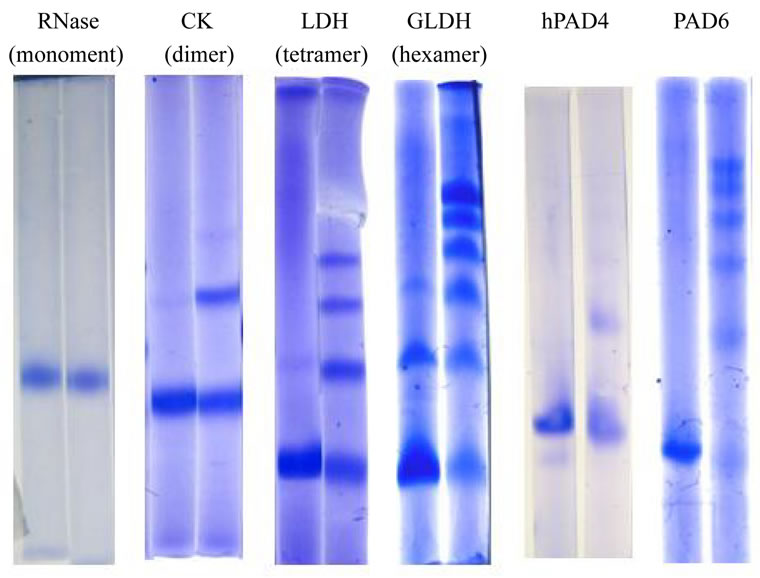
Figure 4. Chemical cross-linking of PAD6 and hPAD4 with dimethyl suberimidate. Chemical cross-linking experiments were carried out as described in Materials and Methods. RNase A (Mr 14,000), creatine kinase (CK, subunit Mr 41,000), lactate dehydrogenase (LDH, subunit Mr 35,000), and bovine glutamate dehydrogenase (GLDH, subunit Mr 52,000) were used as standards. The gel concentration was 3.5% except for RNase A (8%) and CK (6%).
population under the present conditions, and PAD6 can form a hexamer (calculated Mr ~ 456,000) despite that little PAD6 was seen at the elution position of apoferritin (calculated Mr 443,000). The shape of PAD6 is considered to be different from that of apoferritin.
3.5. CD Spectra
CD spectra of PAD6 and hPAD4 were measured in the far ultraviolet region (Figure 5). Their patterns seemed to be similar; the α-helix content was calculated to be 26.0% and 27.2% for hPAD4 and PAD6, respectively, from the [θ]obs value based on the equation of Chen and Yang [17]. The CD spectrum of mouse PAD2 indicated an α-helix content of 29.1% [18]. An X-ray analysis revealed that hPAD4 is comprised of 22.4% α-helix and 39.1% β-strand [9]. It is conceivable that PAD6 and hPAD4 have a similar secondary structure.
3.6. Enzymatic Activity of Purified Preparations
The specific activity of purified hPAD4 was 1.2 µmol/ mg/min, whereas purified PAD6 exhibited no detectable activity even with 50 µg and 15-h incubation. hPAD4 Cys645 is essential for catalysis because a Cys645Ala mutant did not show activity [9]. We also confirmed the Cys645Ala mutant to be enzymatically inactive. In PAD6, the corresponding residue is Ala664. It is of interest whether an Ala664Cys protein has activity. The mutant, however, did not exhibit enzymatic activity, indicating that there exist critical residues other than Ala664.
4. DISCUSSION
The structure of mammalian eggs is fortified with cytoplasmic sheets. An Mr ~ 69,000 protein, occurring in relatively high abundance in eggs, was found to associate with the fibrous network of intermediate filaments [19]. Wright et al. separated this soluble protein by two-
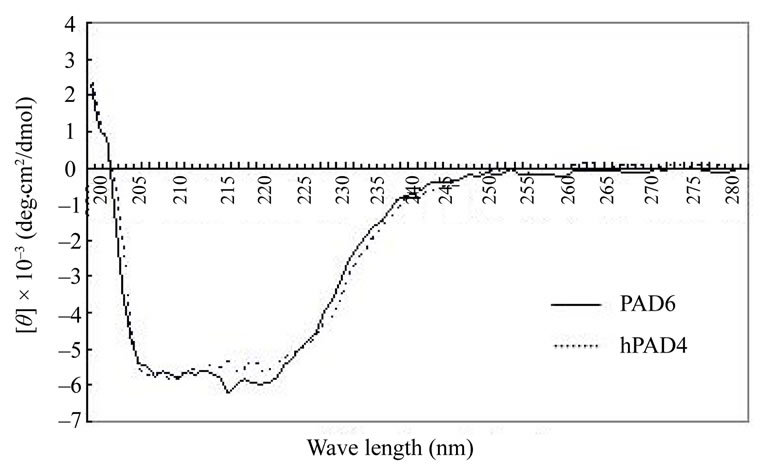
Figure 5. CD spectral profiles of PAD6 and hPAD4. About 0.3 mg/ml each of PAD6 and hPAD4 were recorded in the far ultraviolet region. The measurement conditions were: cell, 10- mm light path; response, 2 sec; band width, 1 nm, scan speed, 20 nm/min.
dimensional gel electrophoresis, identified it as an isoform of PAD by mass spectrometry, and named it ePAD [20]. An ortholog was found in humans, and ePAD was renamed PAD6 [6]. Esposito et al. [21] created PAD6- null mice lacking cytoplasmic sheets and female fertility, and showed an absence of positive signals in these animals when ovarian sections were stained with an antiH4Cit3 antibody. They concluded that this was due to the lack of PAD6 activity. On the other hand, an X-ray analysis of hPAD4 along with a sequence alignment of PADs suggested that enzymatic activity may not be inherent to PAD6 [9]. In the present study, we first showed that the purified PAD6 does not exhibit enzymatic activity when BAEE is used as a substrate. A microbial PAD occurs in P. gingivalis whose primary sequence differs greatly from that of PAD6 [22]. Rodriguez et al. [11,12] showed with purified a monomeric P. gingivalis PAD that Cys351 acts as a nucleophile in catalysis and no Ca2+ cofactor is required. These results suggest that a particular Cys residue in the active site is a prerequisite for deimination, but Ca2+ is not necessarily required in the microbial enzyme. Our preliminary finding that an Ala664Cys mutant did not exhibit enzymatic activity shows the necessity of acidic residues for Ca2+ binding. Among 19 residues involved in the binding of Ca2+, 13 are acidic. In human PAD6, 7 Glu and Asp residues are not conserved [9]. Human and mouse PADs are similar in primary structure. Accordingly, the PAD6 Ala664Cys mutant is expected to be incapable of binding to Ca2+. Whether bound Ca2+ exists in PAD6 remains to be determined. It is tempting to examine whether the replacement of these non-acidic residues with acidic residues could restore the enzymatic activity. The creation of a chimera between hPAD4 and PAD6 may be another approach.
Studies in vitro and in vivo have evoked a great discrepancy as to why an inactive PAD6 can function as an enzyme in the egg and ovary. Several possibilities exist: 1) a true proteinous substrate of PAD6 occurs in vivo; 2) an unknown protein such as a chaperone is needed for the activity; 3) Ca2+ is not the correct cofactor; and 4) the recombinant PAD6 differs from the naturally occurring form due to a lack of post-translational modification. Concerning the second possibility, Snow et al. [23] reported the necessity of tyrosine/tryptophan monooxygenase activation protein (YWHA or 14-3-3 protein) for interaction with PAD6 via phosphorylation at the stage encompassing oocytes to early embryos.
The principle finding of this study is that PAD6 is a hexamer unlike hPAD4, although the two resemble each other in secondary structure. This was not noted previously. PAD6 may be in a state of equilibrium regarding the association-dissociation of the protomer depending on the protein concentration or the solvent. Under chemical cross-linking conditions, the PAD6 concentration was relatively high (about 0.5 mg/ml), whereas about 0.1 mg/ml of PAD6 was used for a Sephacryl S300 chromatography, and so the protein would be gradually diluted during development; this may explain why PAD6 migrated so far. Arita et al. [9] in their X-ray analysis queried whether the dimeric form is specific to PAD4 or common to all PADs although the residues involved in the dimerization are partially conserved. Unfortunately, we could not determine which residues play a role in the subunit-subunit interaction of hPAD4. The residues responsible for the hexamer of PAD6 are expected to be different from those in the case of hPAD4. Further study is needed to clarify whether the unique structure of PAD6 favors the formation of cytoplasmic sheets in eggs.
In conclusion, we have first demonstrated that PAD6 has no enzymatic activity when BAEE is used as a substrate, and is a hexameric protein unlike other dimeric PADs. This unique structure is suggested to favor the formation of egg cytoplasmic sheets.
REFERENCES
- Slack, J.L., Jones, L.E., Bhatia, M.M. and Thompson, P.R. (2011) Autoimination of protein arginine deiminase 4 alters protein-protein interactions but not activity. Biochemistry, 50, 3997-4010. doi:10.1021/bi200309e
- Jones, J.E., Causey, C., Knuckley, B., Slack-Noyes, J.L. and Thompson, P.R. (2009) Protein arginine deiminase 4 (PAD4): Current understanding and future therapeutic potential. Current opinion in drug discovery & development, 12, 616-627.
- Vossenaar, E.R., Zendman, A.J.W., Venrooij, W.J. and Pruijn, G.J.M. (2003) PAD, a growing family of citrullinating enzymes: Genes, features and involvement in disease. BioEssays, 25, 1106-1118. doi:10.1002/bies.10357
- György, B., Tóth, E., Tarcsa, E., Falus, A. and Buzás, E.I. (2004) Citrullination: A posttranslational modification in health and disease. The International Journal of Biochemistry & Cell Biology, 38, 1662-1677. doi:10.1016/j.biocel.2006.03.008
- Zhang, J., Dai, J., Zaho, E., Lin, Y., Zeng, L., Chen, J., Zhen, H., Wang, Y., Li, X., Ying, K., Xie, Y. and Mao, Y. (2004) cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type VI. Acta Biochimica Polonica, 51, 1051-1058.
- Chavanas, S., Méchin, M.-C., Takahara, H., Kawada, A., Nachat, R., Serre, G. and Simon, M. (2004) Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene, 330, 19-27. doi:10.1016/j.gene.2003.12.038
- Venrooij, W.J., Zendman, A.J. and Pruijn, G.J.M. (2006) Autoantibodies to citrullinated antigens in (early) rheumatoid arthritis. Autoimmune Review, 6, 37-41. doi:10.1016/j.autrev.2006.03.008
- Suzuki, A., Yamada, R., Chang, X., Tokuhiro, S., Sawada, T., Suzuki, M., Nagasaki, M., Nakayama-Hamada, M., Kawaida, R., Ono, M., Ohtsuki, M., Furukawa, H., Yoshino, S., Yukioka, M., Tohma, S., Matsubara, T., Wakitani, S., Teshima, R., Nishioka. A. Sekine, Y., Iida, A., Takahashi, A., Tsunoda., T., Nakamura, Y. and Yamamoto, K. (2003) Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nature Genetics, 34, 395-402. doi:10.1038/ng1206
- Arita, K., Hashimoto, H., Shimizu, T., Nakashima, K., Yamada, M. and Sato, M. (2004) Structural basis for Ca2+-induced activation of human PAD4. Nature Structural & Molecular Biology, 11, 777-783. doi:10.1038/nsmb799
- Mangat, P., Wegner, N., Venables, P.J. and Potempa, J. (2010) Bacterial and human peptidylarginine deiminases: Targets for inhibiting the autoimmune response in rheumatoid arthritis? Arthritis Research & Therapy, 12, 209- 218.
- Rodríguez, S.B., Stitt, B.L. and Ash, D.E. (2009) Expression of peptidylarginine deiminase from Porphyromonas gingivalis in Escherichia coli: Enzyme purification and characterization. Archives of Biochemistry and Biophysics, 488, 14-22. doi:10.1016/j.abb.2009.06.010
- Rodríguez, S.B., Stitt, B.L. and Ash, D.E. (2010) Cysteine 351 is an essential nucleophile in catalysis by Porphyromonas gingivalis peptidylarginine deiminase. Archives of Biochemistry and Biophysics, 504, 190-196. doi:10.1016/j.abb.2010.09.008
- Gomi, T., Takusagawa, F., Nishizawa, M., Agussalim, B., Usui, I., Sugiyama, E., Taki, H., Shinoda, K., Hounoki, H., Miwa, T., Tobe, K., Kobayashi, M., Ishimoto, T., Ogawa, H. and Mori, H. (2008) Cloning, bacterial expression, and unique structure of adenosylhomocysteine hydrolase-like protein 1, or inositol 1,4,5-triphosphate receptor-binding protein from mouse kidney. Biochimica et Biophysica Acta, 1784, 1786-1794.
- Davies, G.E. and Stark, G.R. (1970) Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oliogomeric proteins. Proceedings of National Academy of Sciences USA, 66, 651-656. doi:10.1073/pnas.66.3.651
- Knipp, M. and Vašák, M. (2000) A colorimetric 96-well microtiter plate assay for the determination of enzymatically formed citrulline. Analytical Biochemistry, 286, 257-264. doi:10.1006/abio.2000.4805
- Lowry, O.H., Rosebrugh, N.J., Farr, A.L. and Randall, R.J. (1951) Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry, 193, 265-275.
- Chen, Y.-H., Yang, J.T. and Martinez, H.M. (1972) Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry, 11, 4120-4131. doi:10.1021/bi00772a015
- Ohsugi, I., Takahara, H., Shiraiwa, M. and Sugawara, K. (1995) Expression of mouse uterine peptidylarginine deiminase in Escherichia coli: Construction of expression plasmid and properties of the recombinant enzyme. Archives of Biochemistry and Biophysics, 317, 62-68. doi:10.1006/abbi.1995.1136
- Liu, M., Oh, A., Calarco, P., Yamada, M., Coonrod, S.A. and Talbot, P. (2005) Peptidylarginine deiminase (PAD) is a mouse cortical granule protein that plays a role in preimplantation embryonic development. Reproductive Biology and Endocrinology, 3, 42-63. doi:10.1186/1477-7827-3-42
- Wright, P.W., Bolling, L.C., Calvert, M.E., Sarmento, O.F., Berkeley, E.V., Shea, M.C., Hao, Z., Jayes, F.C., Bush, L.A., Shetty, J., Shore, A.N., Redid, P.P., Tung, K.S., Samy, E., Alietta, M.M., Sherman, N.E., Herr, J.C. and Coonrod, S.A. (2003) ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Developmental Biology, 256, 73-89. doi:10.1016/S0012-1606(02)00126-4
- Esposito, G., Vitale, A.M., Leijten, F.P.J., Strik, A.M., Koonen-Reemst, A.M.C.B., Yurttas, P., Robben, T.J.A.A., Coonrod, S. and Gossen, J.A. (2007) Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Molecular and Cellular Endocrinology, 273, 25-31. doi:10.1016/j.mce.2007.05.005
- McGraw, W.T., Potermpa, J., Farley, D. and Travis, J. (1999) Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infection and Immunity, 67, 3248-3256.
- Snow, A.J., Puri, P., Acker-Palmer, A., Bouwmeester, T., S., Vijayaraghavan, S. and Kline, D. (2008) Phosphorylation-dependent interaction of tyrosine 3-monooxygenase/ tryptophan 5-monooxygenase activation protein (YWHA) with PADI6 following oocyte maturation in mice. Biological Reproduction, 79, 337-347.

