American Journal of Plant Sciences
Vol.5 No.5(2014), Article ID:43781,7 pages DOI:10.4236/ajps.2014.55082
Analysis of Genetic Diversity among Wild and Cultivated Chickpea Genotypes Employing ISSR and RAPD Markers
Pramod Kumar Singh1, Himanshu Sharma2, Nidhi Srivastava3, Sameer S. Bhagyawant1*
1School of Studies in Biotechnology, Jiwaji University, Gwalior, India
2Institute of Himalayan Bioresource Technology Palampur, Palampur, India
3Department of Biotechnology, Banasthali University, Banasthali, India
Email: pramod.raju21@gmail.com, himanshusharm@gmail.com, nidhiscientist@gmail.com, *sameerbhagyawant@yahoo.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 26 December 2013; revised 12 February 2014; accepted 1 March 2014
ABSTRACT
Chickpea (Cicer airetinum L.) is an important and most preferred food legume in many parts of the world especially in the Indian sub-continent. Molecular analysis of chickpea using DNA technology has been carried out to identify the diverse genetic base of the cultivars for selected preferential introductions as efficient edible elements. A few of these genetic stocks have been documented here to tap their genetic diversity. The status shows that the level of polymorphism in this species is low. Using PCR based markers, marker assisted selection of polymorphy is one of the established techniques. Here, this procedure has been employed to expedite gene/QTL pyramiding in the chickpea. The study presented here includes analysis of 12 germplasms of chickpea. Standard CTAB method has been performed, with certain modifications, to get better intensity and resolution of DNA bands. Extracted DNA, amplified with 41 RAPD and 21 ISSR primers are thereby tested to determine genetic diversity. The presentation discusses the results of chickpea germplasm diversity on the basis of these observations.
Keywords:Chickpea; Randomly Amplified Polymorphic DNA and Inter Simple Sequence Repeats; Genetic Diversity

1. Introduction
Ever increasing population pressures an urgent need to catalogue the available genotype of the majority of food crops; this includes chickpea. Chickpea being self-pollinated crop hence is supposed to display little genetic variation. Thus seeds exhibiting genetic variation among diverse genotypes of Cicer could be a practical way to select parents to be crossed [1] . DNA based markers are available for understanding genetic variations in plant genomes. Thus, DNA finger printing for cultivar or varietal identification has become an important parameter for genetic signature in plant breeding and germplasm management. Such DNA markers take fingerprints of a species in revealing polymorphism at molecular level. Polymorphisms could be cause either due to differences of a single-nucleotide sequence at the priming sites such as point mutations, or by structural arrangements within the amplified sequence e.g., insertions, deletions, inversions. RAPD detects nucleotide sequence polymorphisms in DNA using a single primer of arbitrary nucleotide sequence. In this reaction, a single species of primer anneals the genomic DNA at two different sites on complementary strands of DNA template. Inter simple sequence repeat (ISSR) analysis involves the polymerase chain reaction (PCR) amplification of regions between adjacent, inversely oriented microsatellites, using a single simple sequence repeat (SSR) motifs. The motifs can be di-, tri-, tetra-, or penta-nucleotides. These techniques are useful in genetic typing and mapping [2] [3] .
In the present investigation therefore, polymorphic data are generated, by employing ISSR and RAPD markers to analyze the inter genetic relatedness of chickpea germplasm. This polymorphic marker data can hence be converted into discrete character data and analyzed subsequently using bioinformatics software for taping inter genetic relationships. Therefore, the aimed genome analysis using such PCR-based markers may help to produce a detailed linkage map of chickpea. The present study conducted ascertain the genetic variability between 12 chickpea cultivar seeds collected from different regions of Madhya Pradesh using Random Amplified Polymorphic DNA (RAPD) and ISSR markers.
2. Materials and Methods
2.1. Plant Material
Total twelve chickpea (Cicer arietinum L.) cultivars were selected for analysis. The mature and dry seed material was procured from R. A. K. College, Sehore Madhya Pradesh and wild genotypes were obtained from ICRISAT Hyderabad, India as shown in the Table 1.
DNA was isolated using a modified cetyl trimethyl ammonium bromide (CTAB) procedure of Khan et al. [4] . CTAB is a cationic detergent which solubilizes membrane and forms a complex with DNA. After cell disruption and incubation with hot CTAB isolation buffer, proteins were extracted using chloroform isoamyl-alcohol, and the CTAB-DNA complex was precipitated with isopropanol. The DNA pellet was washed, dried and re-dissolved in TE buffer.
Table 1. Details of chickpea germplasm used for genetic diversity analysis.
Taq DNA polymerase and dNTPs were obtained from Perkin Elmer Inc. USA. RAPD and ISSR primers were obtained from Operon Technologies Ltd., (Alameda, California). The lyophilized DNA primers were dissolved in sterile TE buffer at a concentration of 10 pm·µl−1.
2.2. RAPD-PCR Amplification
PCR amplifications were carried out using a Bio-Rad 3.03 version thermo-cycler. Fourty one primers were used. Out of these 22 primers did not give satisfactory results (Table 2). Amplifications were performed for 35 cycles with denaturation at 94˚C for 1 min, annealing at 37˚C for 1 minute and extension at 72˚C for 2 minutes. The reaction mixture (20 µl) contained 20 ng of DNA template, 10 mM of Tris-Hcl (pH 9.0), 2 mM MgCl2, 50 mM KCl, 200 mM dNTP mix, 0.25 mM primer, 5 mM of spermidine and 0.8 units of Taq DNA polymerase. Master mixes of each reaction were overlaid with 10 µl of mineral oil to prevent evaporation.
2.3. ISSR-PCR Amplification
Twenty one ISSR primers were used in this study; only six primers gave the products (Table 3). PCR amplification was done using a Bio-Rad 3.03 version thermo-cycler. Amplification were performed for 35 cycles with denaturation at 94˚C for 1 min, annealing at 56˚C for 1 min. and extension at 72˚C for 2 minutes. Initial denaturation was done at 94˚C for 5 min and a final extension step of 5 min. at 72˚C was also included. The reaction mixture (25 µl) contained 20 ng of DNA template, 10 mM Tris-HCl pH 9.0; 50 mM KCl; 0.1% Triton X-100; 1.5 mM MgCl2; 0.1 mM dNTP; 2 mM primer; 0.5 unit of Taq DNA polymerase. Master mixes of each reaction were overlaid with 10 µl of mineral oil to prevent evaporation. Amplification products were separated on 1.2% agarose gel in 1X TAE (Tris base, acetic acid and EDTA) buffer.
2.4. Data Analysis
RAPD and ISSR bands were manually scored as present (1) or absent (0) for estimation the similarity among all tested samples. The matrix of similarity (Jaccard) was obtained by clustering according to the Unweighted Pair-Group Method with Arithmetic averages (UPGMA) using, Infor-Bio (version 5.26) software. Polymorphism percentage was estimated by dividing the number of polymorphic bands over the total number of bands.
Table 2. Statistics of RAPD primers used to detect polymorphism, in chickpea.
Table 3. Statistics of ISSR primers used to detect polymorphism, in chickpea.
3. Result
In the present study, both RAPD and ISSR markers have been used in combination for genetic diversity estimation. Both RAPD and ISSR showed same patterns of clustering of genotypes. Forty two RAPD primers were screened against twelve selected chickpea DNA to identify potential primers producing higher number of polymorphic and repeatable fragments. RAPD analysis revealed a good polymorphism among chickpea cultivars. Of the 41 primers used 22 primers did not give satisfactory results. This causes majority of the amplicons were found to be monomorphic. Such kind of non amplifying primers were also reported earlier in chickpea [5] . The remaining 19 primers generated reproducible RAPD patterns were used for subsequent analysis.
Each primer-template yielded distinct, easily detectable bands of variable intensities. Light, dense and sharp bands are considered for the consideration of genetic diversity. The monomorphic bands of low degree of similarity indicated high divergence between the genotypes evaluated. Assaying RAPD variation in accession of Cicer cultivars with these primers yielded total of 73 bands ranging molecular weight between 100 - 3000 bp. Number of bands produced ranged from 3 to 9 with an average of 3.84 bands per genotype. The total polymorphic bands in 19 primers are 57 and therefore, reproducible polymorphic band are 3 averages per primer and percentage polymorphism are 71.10%. Maximum number of 9 bands was shown by primer OPA 06, while least number of single bands was shown by OPA8, OPA9, OPC18. Out of 42 potential primers, 19 RAPD primers were thus chosen for fingerprinting logically based on the high number of polymorphic fragments generated and then concomitant reproducibly. The sequences of primers, the number of RAPD fragments they amplified and their approximate sizes are presented in Table 2.
The second aspect of investigation was to ascertain the most followed ISSR patterns to estimate diversity with anchored primer at 3’ end. ISSRs will have an important role in securing plant variety rights by virtue of its unique efficiency in distinguishing even closely related germplasm. Twelve Cicer genotypes were screened using 21 ISSR primers for the present PCR investigation. The primers tested were six dinucleotide (AG)8C, (AG)8T, (AG)8YT, (AG)8YC, (TG)8YC, (CA)7BDB and their anchored repeat primers were included in the ISSR-PCR studies. Of the 21 primers used for PCR studies, 15 primers did not amplify DNA of any chickpea genotype.
Among the dinucleotide repeats tested, (AG)8C, (AG)8T, (AG)8YT, (CA)7BDB produced better amplification patterns (Figure 1). 06 primers on an average produced 3 bands across 12 genotype, of which 3 were polymorphic accounting for 88%. The primer sequences (AG) and (AG) 8YC amplified least number of bands viz. 2 respectively, whereas, primer (AG) 8C and (AG) 8YT, amplified a maximum number of 4 bands. The percent polymorphism as shown by these ISSR primer range from 50% by primer (AG) 8T to 100% by (AG)8YT, (AG)8YC, (TG)8YC and (AC)7BDB primer. An average polymorphism of 88% across all genotype is observed. The dendogram separated the genotypes into two major clusters. The dendogram displayed one major cluster containing subgroups of cultivated and wild accessions. While ICC7554 came as an out group. In the major cluster I, BG1053 exist as an independent strain type. While accessions JAKI 9218, JG16, JG218, JGG1, JGK1 along with one wild ICC5434 grouped into one sub group. In this JG11 exhibits separate existence. The clade is formed amongst ICC13441, ICC1180, and ICC6537. This clade also shows separate existence in dendogram as being correlated in the PCRproducts (Figure 2).
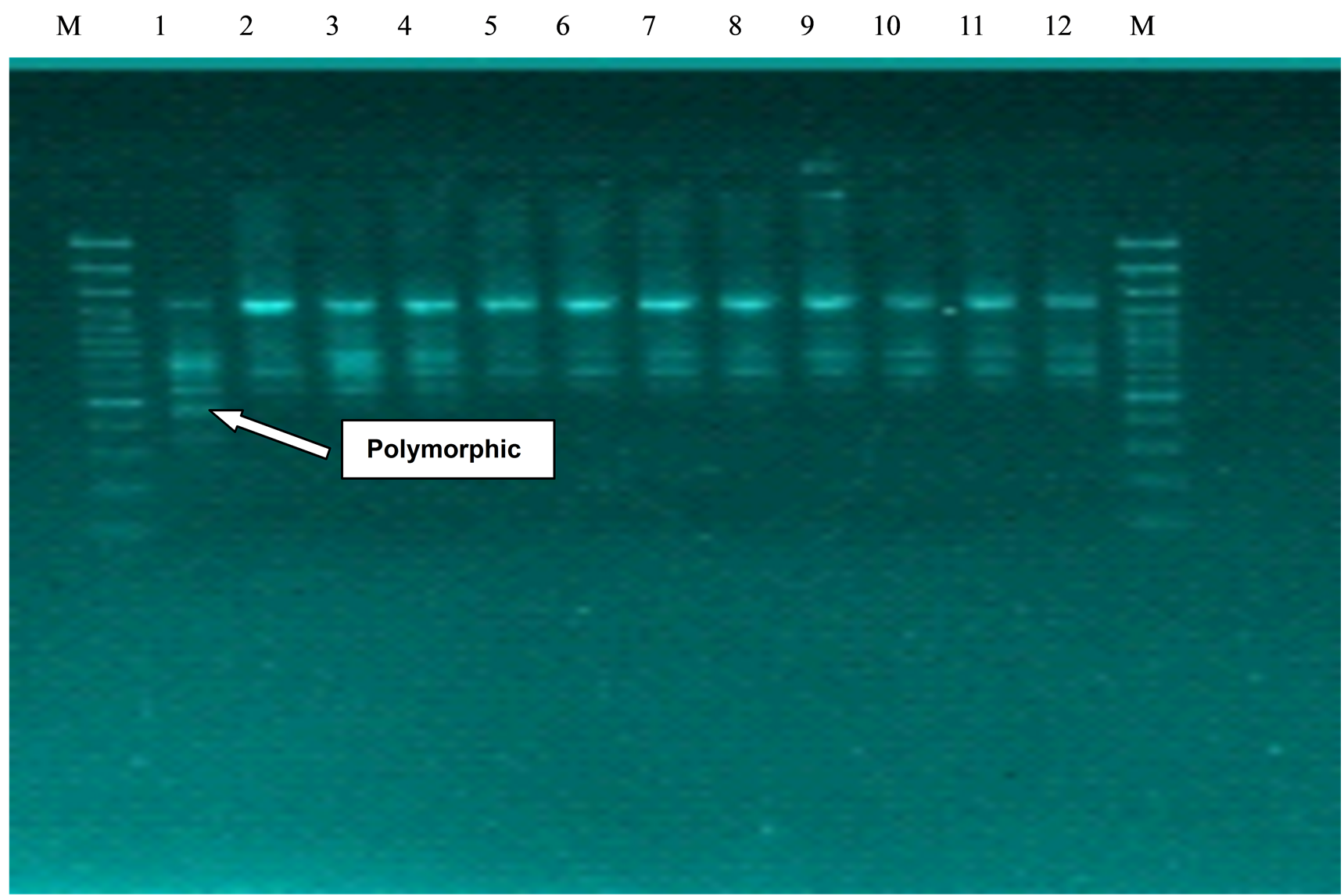
Figure 1. Amplification of 12 accessions of Cicer with primer OPC 12. Lane identification (from left to right)— M: 3Kb markers Lane 1-12: BG1053, JAKI 9218, JG11, JG16, JG218, JGK1, JGG1, ICC6537, ICC 5434, ICC 13441, ICC 7554 and ICC 1180.
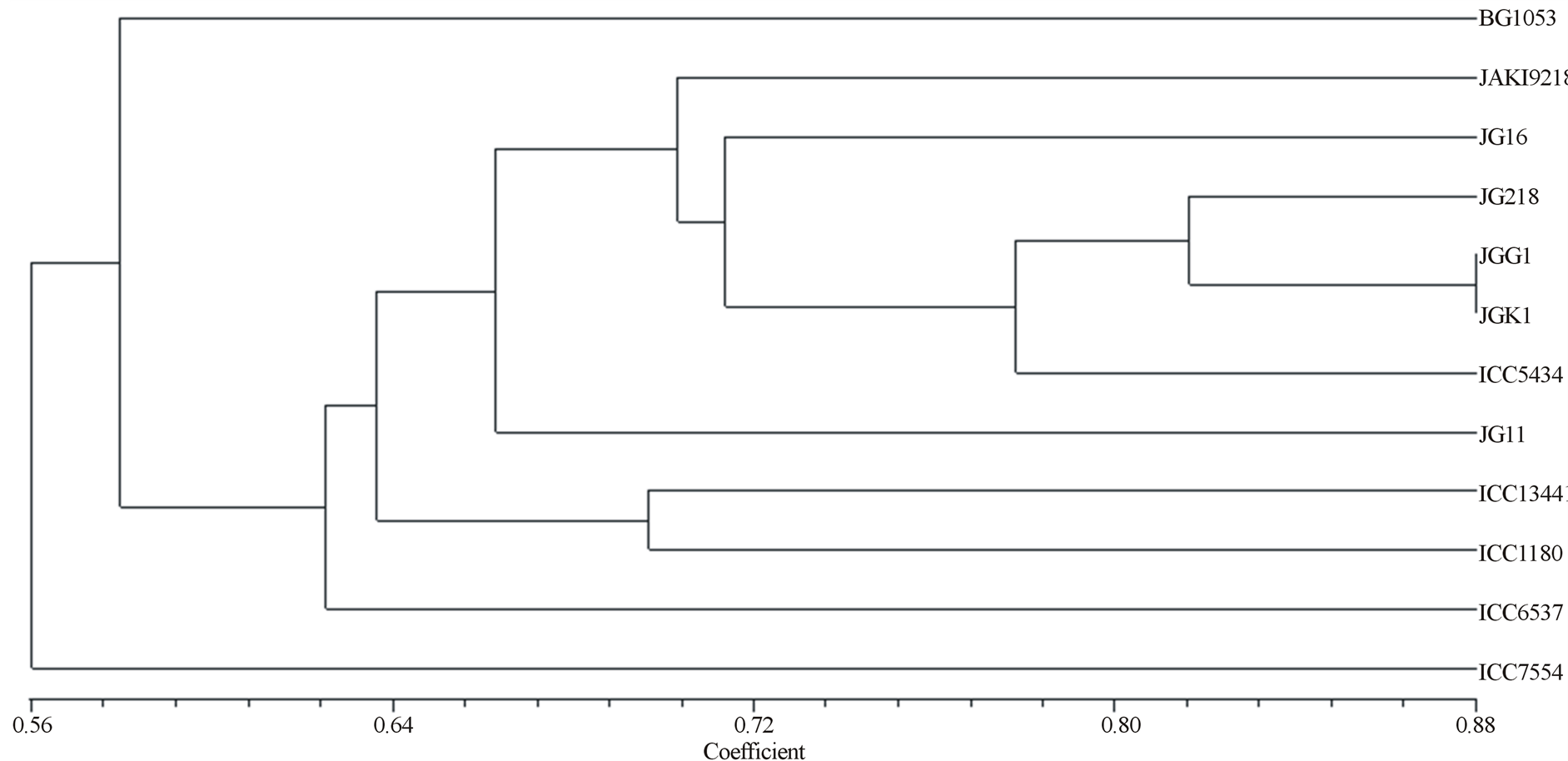
Figure 2. Dendogram as generated by RAPD and ISSR primers in Cicer.
4. Discussion
The present investigation used a total of 41 RAPD and 21 ISSR primers. In combinations displayed complex banding pattern with minor bands showing weak amplification and hence were not considered for further analysis. Such comparative studies in legumes involving RAPD and ISSR markers have been successfully used by few researchers in crops like black gram [6] , vigna species [7] and rice [8] .
Literature perusal suggests that PCR can successfully forecast the growth habitats and geographic origin. Studies of Iruela et al. [9] in the genus Cicer and cultivated chickpea using combination of RAPD and ISSR markers of 26 accessions including kabuli and desi types were employed. A dendogram based on the Jaccard’s similarity index showed that a distribution pattern of variability between species was related to both growth habitats and geographical origin and also revealed differentiation amongst kabuli and desi types. In our investigation a taped relationship amongst the Cicer accessions BG1053 and ICC 7554 displayed unique banding patterns. Such banding trend was furthered supported by the dendogram. This could infer that the extra band/ fingerprinting pattern btained may be because of resistant to the pod borer and/or fungal diseases. In 2007, Rao et al. [10] developed RAPD and ISSR fingerprinting in cultivated chickpea and its wild progenitor Cicer reticulatum L. They concluded ISSR analysis as a reliable attributes for estimation of genetic diversity than of RAPD.
In present study, we report a strong multi-locus association between molecular and morphological traits. RAPD markers could establish a morphological divergence as a means of indicator. The identification of genetic variation, using PCR markers generated data may be applied in breeding programs, to reshuffle the alleles of recombination, having better chances for appearance of transgressive segregation and thus beneficial traits in the germplasm can be selected to extract high yielding lines with desirable trait combination. Bhagyawant and Srivastava [5] reported the applications of ISSR marker in identifying genetic relationships in chickpea and ascertain that the unique band produced by (GGAGA) primers employed in the BCP- 15 genotypes may be attributed to temperature tolerance phenotype. In present studies also the bands produced by BG1053 and ICC7554 may attribute to a fungal resistant phenotype. However, this needs to be further characterized using more number of primers and the banding pattern produced by such accessions needs to be converted into more reliable a SCAR marker. In similar studies, Talebi et al. 2008 [11] revealed a strong multilocus association between molecular and morphological traits chickpea cultivars. Hence, they concluded their studies that RAPD markers were good indicators of morphological divergence. The gene differentiation of chickpea into desi and kabuli types during their evolution was successfully taped by Agrawal and Srivastava using RAPD based on dendogram [12] . Recently Fatemeh at al. 2013 [13] employed ISSR markers to fingerprint genetic diversity and conservation of land race chickpea from north-west of Iran.
The present investigation therefore demonstrates in detecting polymorphism amongst the 12 chickpea cultivars and their wild accession using RAPD and ISSR technique. The genetic diversity in the wild annual Cicer germplasm overall is narrow. [14] [15] may therefore, be exploited constructively for mapping studies. It can further be employed for introgression of disease and insect resistance in cultivated genotypes [16] .
References
- Ahmad, F., Khan, A.I., Awan, F.S., Sadia, B., Sadaqat, H.A. and Bahadur, S. (2010) Genetic Diversity of Chickpea (Cicer arietinum L.) Germplasm in Pakistan as Revealed by RAPD Analysis. Genetics and Molecular Research, 9, 1414-1420. http://dx.doi.org/10.4238/vol9-3gmr862
- William, J.G.K., Kubelik, A.R., Livak, K.J., Rafalski, J.A. and Tingey, S.V. (1990) DNA Polymorphisms Amplified by Arbitrary Primers Are Useful as Genetic Markers. Nucleic Acids Research, 18, 6531- 6535. http://dx.doi.org/10.1093/nar/18.22.6531
- Welsh, J. and Mcclelland, M. (1990) Fingerprinting Genomes Using PCR with Arbitrary Primers. Nucleic Acids Research, 18, 7213-7218. http://dx.doi.org/10.1093/nar/18.24.7213
- Khan, S., Qureshi, M.I., Kamaluddin, T.A. and Abdin, M.Z. (2007) Protocol for Isolation of Genomic DNA from Dry and Fresh Roots of Medicinal Plants Suitable for RAPD and Restriction Digestion. African Journal of Biotechnology, 6, 175-178.
- Bhagyawant, S.S. and Srivastava, N. (2008) Genetic Fingerprinting of Chickpea (Cicer arietinum L.) Germplasm Using ISSR Markers and Their Relationships. African Journal of Biotechnology, 7, 4428-4431.
- Souframanien, J. and Gopalakrishna, T.G. (2004) A Comparative Analysis of Genetic Diversity in Black Gram Genotypes Using RAPD and ISSR Markers. Theoretical and Applied Genetics, 109, 1687-1693. http://dx.doi.org/10.1007/s00122-004-1797-3
- Dikshit, H.K., Jhang, T., Singh, N.K., Koundal, K.R. Bansal, K.C., Chandra, N., Tickoo, J.L. and Sharma, T.R. (2007) Genetic Differentiation of Vigna Species by RAPD, URP and SSR Markers. Biologia Plantarum, 3, 451-457. http://dx.doi.org/10.1007/s10535-007-0095-8
- Ravi, M., Geethanjali, S., Sammeyafarheen, F. and Maheswaran, M. (2003) Molecular Marker Based Genetic Diversity Analysis in Rice (Oryza sativa L.) Using RAPD and SSR Markers. Euphytica, 133, 243-253. http://dx.doi.org/10.1023/A:1025513111279
- Iruela, M., Rubio, J., Cubero, J.I., Gil, J. and Millán, T. (2002) Phylogenetic Analysis in the Genus Cicer and Cultivated Chickpea Using RAPD and ISSR Markers. Theoretical and Applied Genetics, 104, 643-651. http://dx.doi.org/10.1007/s001220100751
- Rao, L., Usha, R.P., Deshmukh, P., Kumar, P. and Panguluri, S. (2007) RAPD and ISSR Finger-Printing in Cultivated Chickpea (Cicer arietinum L.) and Its Wild Progenitor Cicer reticulatum Ladizinsky. Genetic Resources and Crop Evolution, 54, 1235-1244. http://dx.doi.org/10.1007/s10722-006-9104-6
- Talebi, R., Naji, A.M. and Fayaz, F. (2008) Geographical Patterns of Genetic Diversity in Cultivated Chickpea (Cicer arietinum L.) Characterized by Amplified Fragment Length Polymorphism. Plant Soil Environment, 54, 447-452.
- Agrawal, P.K. and Srivastava, A. (2010) Assessment of genetic Diversity among Chickpea Cultivars of India Using RAPD Marker. Indian Journal of Human Genetics, 70, 264-270.
- Pakseresht, F., Talebi, R. and Karami, E. (2013) Comprative Assessment of ISSR, DAMD and SCoT Markers for Evaluation of Genetic Diversity and Conservation of Landrace Chickpea (Cicer arietinum L.) Genotypes Collected from North-West of Iran. Physiology and Molecular Biology of Plants, 19, 563-574. http://dx.doi.org/10.1007/s12298-013-0181-7
- Virk, P.S., Newbury, H.J., Jacckson, M.T. and Ford-Lioyd, B.V. (1995) The Identification of Duplicate Accessions within a Rice Germplasm Collection Using RAPD Analysis. Theoretical and Applied Genetics, 90, 1049-1055. http://dx.doi.org/10.1007/BF00222920
- Abbo, S., Berger, J. and Turner, N.C. (2003) Evaluation of Cultivated Chickpea: Four Bottlenecks Limit Diversity and Constrain Adaptation. Functional Plant Biology, 30, 1081-1087.
NOTES
*Corresponding author.


