Open Journal of Preventive Medicine
Vol.2 No.1(2012), Article ID:17438,7 pages DOI:10.4236/ojpm.2012.21017
Novel variants in the hepatocyte nuclear factor-1-alpha gene in MODY and early onset NIDDM: Evidence for a mutational hotspot in exon 5
![]()
1Department of Biochemistry, Institute of Post Graduate Medical Education and Research, Kolkata, India
2Department of Endocrinology and Metabolism, Institute of Post Graduate Medical Education and Research, Kolkata, India; *Corresponding Author: bbasudev@rediffmail.com
Received 19 October 2011; revised 24 November 2011; accepted 30 December 2011
Keywords: Maturity Onset Diabetes of Young; PCR-SSCP; HNF1-Alpha; Non Insulin Dependant Diabetes Mellitus
ABSTRACT
The disorder, Maturity Onset of Diabetes of the young (MODY) is a monogenic form of Non-Insulin dependent Diabetes Mellitus (NIDDM), characterized by autosomal dominant mode of inheritance and onset is usually before 25 years of age. Clinical studies of subjects with the different forms of MODY indicate that each is associated with a different defect in the normal pattern of glucose stimulated insulin secretion. MODY can result from mutations in any one of the six different genes, one of which encodes the glycolytic enzyme Glucokinase, associated with MODY2 and the other five encode transcription factors HNF4-alpha associated with MODY 1, HNF1- alpha associated with MODY 3, IPF with MODY 4, HNF1-Beta with MODY 5 and NeuroD1 with MODY6. Studies related to mutations in the MODY genes have led to a better understanding of the genetic causes of the Beta cell dysfunction as genetic factors plays a great role in this disorder. Objective: To investigate the mutation pattern in the different transcription factor genes with special reference to HNF1-alpha which are highly penetrant with 63% mutation carriers manifesting clinical diabetes by the age of 25 years. Hence study of mutation pattern in this gene is essential in our population i.e. Eastern Indian population. Our study is focused on HNF1-alpha related to MODY 3, which is the most common one. Methods: In our study enzyme amplification (PCR) of the 10 target exons of the said gene with simultaneous mutation detection in them by PCR-SSCP (Polymerase chain reaction-single strand conformational polymorphism) reaction analysis method was attempted by screening of exon 1 - 10 with respect to normal healthy controls without Diabetes Mellitus. The nature of the specific mutations was also determined by sequencing. Result: It was observed that maximum number of variations exist in exon 5 of HNF1-alpha gene which may be referred to as “Mutational Hotspot” in our Eastern Indian population. Conclusion: Since maximum number of variations exists in exon 5 of the said gene, hence one can initially go for exon 5 followed by other exons, while screening for pathogenic MODY 3 mutations in the responsible gene by PCR-SSCP method.
1. INTRODUCTION
Type 2 Diabetes Mellitus (DM) is an entity of metabolic diseases which is characterized by both impaired Beta cell function and insulin resistance. Type 2 or Non insulin dependent Diabetes Mellitus (NIDDM) is a genetically, metabolically and clinically heterogeneous syndrome of multifactorial etiology. Although in most cases T2DM is a Polygenic disorder, several monogenic forms have also been identified [1].
Maturity onset of diabetes of the young (MODY) is one such monogenic form of non-insulin dependent diabetes Mellitus (NIDDM) characterized by autosomal dominant mode of inheritance and is a disorder with an early age of onset usually before 25 years of age [2]. Genetic Studies have provided evidences for at least 3 unlinked loci that can cause this form of diabetes designated as MODY 1, MODY 2, and MODY 3. Till date, six types, MODY 1 - MODY 6 have been enumerated but more are likely to be added [3]. MODY 2 and MODY 3 are the most common forms. Well defined mode of inheritance with high penetrance and early age of onset makes MODY an attractive model for genetic studies of T2DM. The severity of the different types varies considerably, but most commonly MODY acts like a very mild version of type1 diabetes with continued and partial insulin production and normal insulin sensitivity [3].
The most common clinical presentation MODY is mild, asymptomatic hyperglycemia in nonobese children, adolescents and young adults having a prominent history of diabetes often in successive generations (a pattern consistent with autosomal mode of inheritance). Several clinical characteristics distinguish patients with MODY from those with Type 2 Diabetes including a prominent family history of diabetes in three or more generations, a young age at presentation and the absence of obesity [3,4].
Clinical studies of subjects with the forms of MODY indicate that each is associated with a different defect in the normal physiological pattern of stimulation of insulin secretion. The proteins encoded by these MODY loci have been identified and shown to be, hepatocyte nuclear factor, HNF-4 alpha (MODY 1), glucokinase (MODY 2), HNF1-alpha (MODY 3), Insulin Promoter Factor-1 (MODY 4), HNF-1 Beta (MODY 5) and NeuroD1 (MODY 6) respectively [5].
With the identification of genes responsible for MODY, it is possible to identify members of pedigrees who have inherited the specific mutations affecting their family, even before carbohydrate intolerance develops. Genetic screening for and identification of a specific MODY related mutation in children may have important prognostic and therapeutic implications.
Persons who are genetically susceptible to diabetes due to mutations in the genes for HNF1-alpha and HNF4-alpha should be monitored frequently so that appropriate therapy can be instituted early in the course of their hyperglycemia, because of the risk of progression to severe hyperglycemia and insulin requiring diabetes. Early treatments to achieve normoglycemia are necessary to prevent vascular nuropathic complications. Thus, MODY is one type of diabetes that warrants genetic counseling, because of the mode of inheritance and high penetrance [6,7].
This is the form of MODY, i.e. MODY 3 which can mostly resemble ordinary Type 1 Diabetes Mellitus, and one of the incentives for diagnosing it is that, insulin may be discontinued or deferred in favor of oral sulfonylureas. Some people treated with insulin for years due to a presumption of type1diabetes mellitus have been able to switch to oral hypoglycemic agents and discontinue injections. Long-term diabetic complications can occur if proper glycemic control is not achieved [7].
Genetic factors play a great role in this disorder. The genetic factors though appreciated are not been pointed out in sufficient details in our Indian Population. Hence we have taken up the said study in order to find out some of the known genetic factors or genes responsible for the development of this disease in our population.
Genetic diagnosis may also be advised for patients who have been classified as having Type 1 diabetes and who have a strong family history of diabetes. An appropriaciable fraction of these patients have been found to carry HNF1-alpha mutations. The diagnosis of HNF1-alpha related MODY rather than Type 1 diabetes has implications for prognosis in these patients.
It has been observed that the prevalence of MODY is probably higher in India than the Caucasian population. But the clinical types and the genetic profile of MODY in India and specially in our population is still unknown. Hence we have taken up the following study to investigate the mutation pattern in the different transcription factor genes with special reference to HNF1alpha which are highly penetrant with 63% mutation carriers having diabetes by the age of 25 years as study of mutation pattern in this gene is essential in our Eastern Indian poluation. Our study is focused on HNF1alpha related to MODY 3, which is the most commonly occurring one. Moreover the study is of immense importance as it would be easy to screen relatives of any subject in whom any mutation is observed, to identify other mutant subjects in the same family specially who are presymptomatic so that they can be monitored and rapid treatment can be initiated to minimize diabetic complications [8].
2. MATERIALS AND METHODS
Patient Selection Criteria
Inclusion Criteria
Study Subjects
Patients with diabetes with onset before the age of 25 years were selected in the study as probands. After receiving the detailed history (including family history) and duly signed informed consent forms, clinical examinations were done. The study protocol complied with the Declaration of Helsinki and was approved by the institution’s ethics committee. Informed consent was obtained from each subject.
Control: Non-diabetic subjects who did not have any history of Diabetes Mellitus were considered as the Control subjects. The family history of first and second degree relative of diabetes was reported for all the subjects.
Exclusion criteria: All patients were carefully examined and an investigation has been carried out to rule out Type 1 diabetes using GAD and IA2 Antibodies.
Sample processing and sample collection: Both patient and control samples were being collected from Department of Endocrinology and Metabolism, Central laboratory IPGME & R, and other Endocrinology clinics of the region.
Genomic DNA isolation: Genomic DNA from human whole blood was isolated using standard phenol-chloroform method of isolation and the quality of the DNA was checked by sphectrophotometric analysis [9].
Polymerase chain reaction (PCR): Amplification of the different exons of the HNF1-alpha gene was carried out with 120 ng of extracted DNA. The reaction Mixture in 50 µl consisted of 10 mM Tris-HCL (pH 8.3), 50 mM KCL, 2.5 mM MgCl2, 2.5 mM deoxyribonucleoside triphosphates, 1 U Taq DNA polymerase (MBI Fermentas) and 20 μmol primers specific for each exon. The reactions were performed at various annealing temperatures specific for each exon in a thermo cycler (Biometra) (Table 1).
Mutation detection: Molecular biological techniques of mutation detection has become more and more a focus of interest in clinical medicine because the responsible genes are now known for an increasing number of diseases and the detection is essential to enable reliable predictions and to design individual therapies. Therefore the SSCP (Single-Strand Conformation Polymorphism) analysis is one of the most commonly applied methods in detection of point mutations.
SSCP analysis: The use of SSCP for detection of the pathogenic mutations of MODY (Maturity Onset Diabetes in the Young) is described as an example in the study [10].
Statistical analysis: Statistical analysis for evaluation of SSCP variants among study subjects and control were evaluated by χ2 test. The Odds ratio (OR) and 95% confidence interval (95% CI) were evaluated by statistical value using Fishers exact probability test. The level of statistical significance was set at p < 0.05.
3. RESULT AND ANALYSIS
We have enrolled 98 subjects fulfilling stringent MODY criteria and 114 non diabetic subjects as healthy control (HC) out of 100 study subjects and 122 healthy controls that had positive amplification for all the exons.
3.1. Detection of Mutation by PCR-SSCP
Our PCR-SSCP based study revealed that there were a lot number of variations existing in the ten different exons of HNF1α gene among study subjects and controls. No statistically significant difference was observed regarding number of SSCP variants present in exon 1, 3, 4, 6, 8 and 10 respectively. [(8.16% vs 3.63%; p = 0.1437), (4.08% vs 1.78%; p = 0.3083), (7.14% vs 4.58%; p = 0.3865), (5.1% vs 1.78%; p = 0.1738), (4.08% vs 0.87%; p = 0.1253), (2.04% vs 0.87%; p = 0.4745) respectively] among study subjects and control. In exon 5 and 9 a statistically significant increased number of SSCP variants were found in the study subjects compared to the healthy controls [(18.36% vs 2.63%; p < 0.0001), (12.24% vs 2.63%; p = 0.0065) respectively]. Further increased trend of SSCP variants were also observed in exon 2 and 7 among study subjects compared to healthy controls [(7.14% vs 0.87%; p = 0.017), (6.12% vs 0.87%; p = 0.033) respectively] (Table 2, Figures 1-2).

Table 1. Primer sequences of the respective exons.
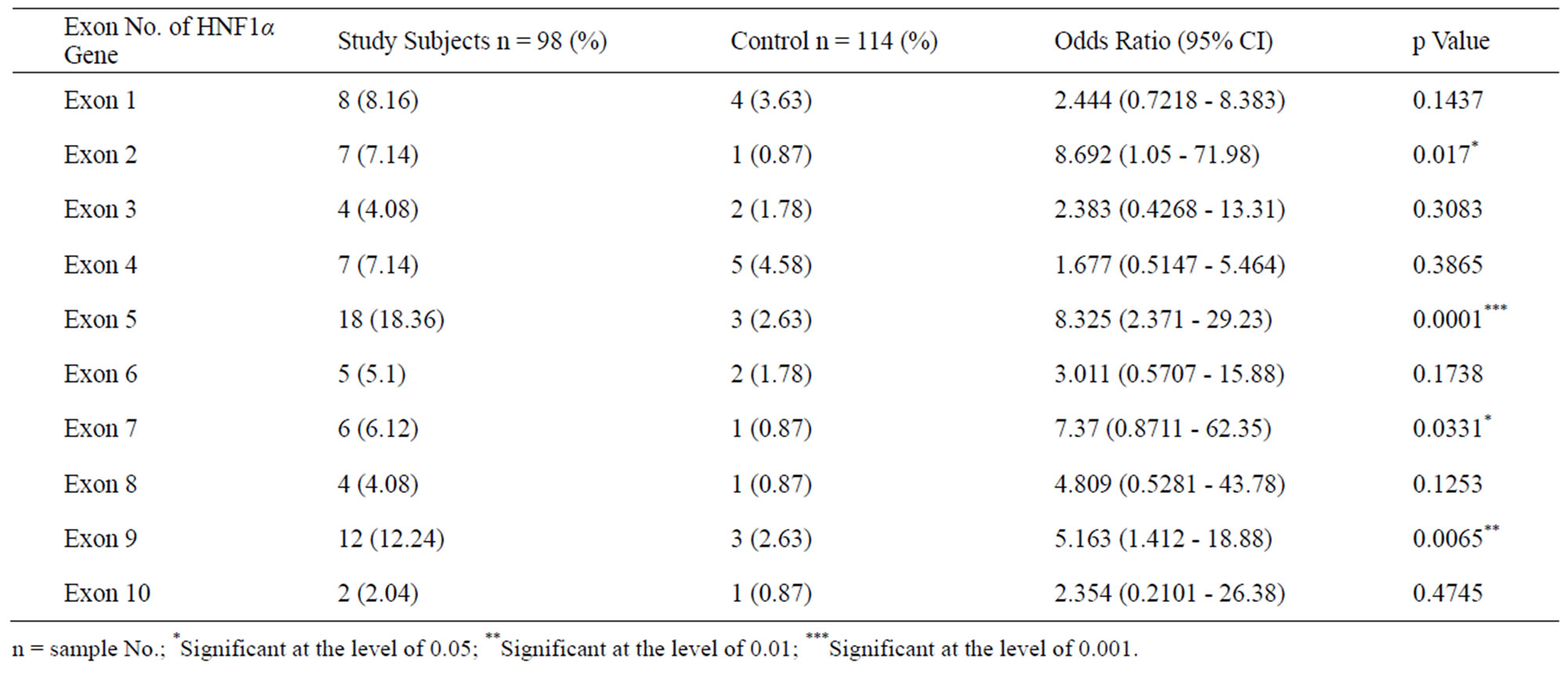
Table 2. Represents No. of SSCP variants present in following exons of HNF1α gene among study subjects and controls.
Figure 1. Representing PCR Amplicons of exon 5 of HNF1-alpha gene. Products amplified were run in a 1.5% agarose gel. L1 and L2 representing PCR amplicons. M representing 100-bp DNA ladder molecular marker.
3.2. Identification Novel Variants in the Study Subjects
Mutations are located throughout the HNF1-alpha gene, here in our study mostly affecting exon 5. Screening 98 subjects fulfilling stringent MODY criteria for 10 exons of the HNF1-alpha gene we identification of 3 different polymorphisms all of which are novel in nature. These three single nucleotide polymorphisms were observed in exon 5 of the HNF1-alpha gene. The single nucleotide polymorphisms identified are, c.1104A > C, c.1155C > G c.1153C > G. In case of exon 5, PCR-SSCP study revealed about 18.36% variations and 3 novel polymorphisms were identified after sequencing (3.06%) in this case (Table 3, Figure 3).
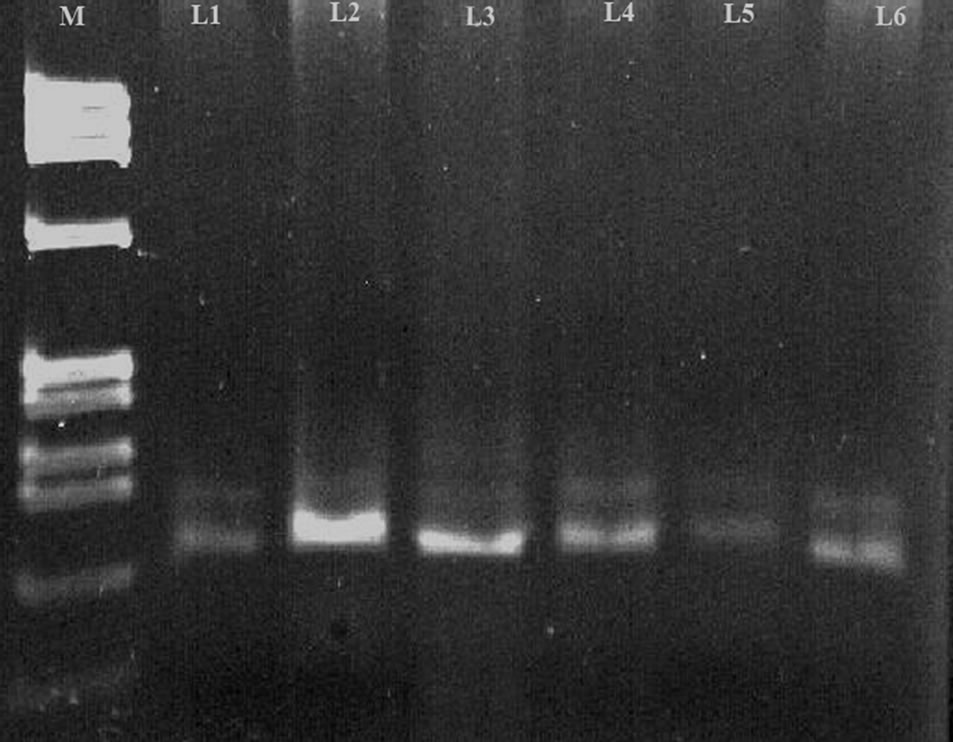
Figure 2. Representing single strand conformational analysis of exon 5 HNF1-alpha gene. The denatured amplicons were run in a 6% polyacrylamide gel. L2 and L5 representing the abnormal SSCP conformers with respect to the other subjects in L1, L3, L4 and L6 M shows BsuRI (HaeIII) marker.
4. DISCUSSION
A study was initiated in our department to evaluate the contribution of the MODY genes to the etiology of early onset Diabetes. A better understanding of the causes and pathophysiology of MODY is emerging as an important entity of genetic and molecular biology. HNFs are transcription factors first identified because of their role in the tissue-specific regulation of gene expression in liver. It is seen that mutations in the gene encoding the transcription factor hepatocyte nuclear factor (HNF1-alpha) account for most of the mutations associated with MODY and are the cause of one of the clinical entity of diabetes, maturityonset of the young (MODY 3). HNF1-alpha, HNF1-beta and HNF4-alpha constitute a part of transcription factors
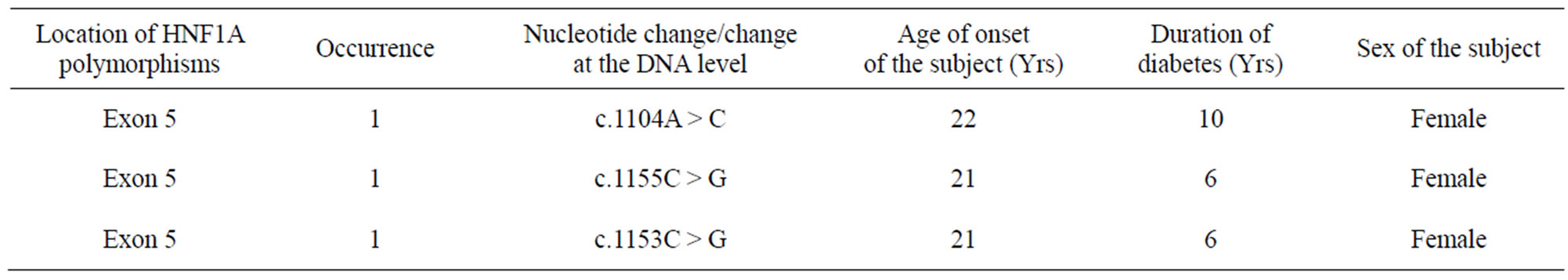
Table 3. showing novel polymorphisms identified in study subjects of HNF1-alpha.

Figure 3. Representing dendogram showing sequencing analysis of HNF1-α exon 5, sequenced with primers specific to the exon.
that function together to control gene expression during embryonic development and during adulthood in tissues in which they are co expressed [11].
Recent evidences have gathered in favour of HNF1-alpha gene which can be implicated for MODY 3 due to mutations in said gene. It is a homeobox gene on chromosome 12. This is the most common type of MODY in populations with European ancestry, accounting for about 70% of all cases in Europe [12]. HNF1-alpha is a transcription factor (also known as transcription factor or TCF1) which is thought to control a regulatory network (including, among other genes, HNF1α) important for differentiation of beta cells in islets of Langerhans of pancreas. Mutations of this gene lead to reduced beta cell mass or impaired function. HNF1A consists of ten exons, coding 631 amino acids and has three different isomers namely, isoform A (exons 8 - 10), isoform AB (exon 7) and isoform ABC (exons 1 - 6) formed by alternative splicing and polyadenylation. The isoform A has a lower transactivation activity compared with the isoforms AB and ABC. Mutations affecting different isoforms have been associated with age at disease onset in certain studies; thus, isoforms may influence the phenotype of disease differently. The HNF1A protein consists of a dimerization domain (amino acids 1 - 33), a bipartite DNA-binding domain (POU domain: 100 - 184 and homeodomain: 198 - 281) and a transactivation domain (amino acids 282 - 631). Mutations occurring within different domains may also influence age at manifestation differently, depending on type and location of mutations occurrence [12]. Clinical manifestations of MODY 1 and MODY 3 diabetes are more or less identical. About 70% of people develop this clinical condition by age 25 years, but it occurs at much later stage of life in a few. This type of diabetes can often be treated with sulfonylurea with excellent results for decades. However, the loss of insulin secretary capacity is slowly progressive and most of the cases eventually need insulin therapy. Clinical expression of MODY 3 is highly variable from one family to another or even within the same family, which might be due to genetic or environmental factors including molecular characteristics of HNF1-α gene mutation. The symptoms at diagnosis may be variable. Severity and course of insulin secretion defect also may vary since approximately one third patients are treated with insulin after 15 years of diabetes duration, while others control their diabetes by diet or oral hypoglycemic agents [13].
In this study we screened 98 subjects, clinically fulfilling stringent MODY criteria and 114 non-diabetic control subjects for mutational analysis by the standard PCRSSCP method. The subjects selected in the study, were all patients of Diabetes before the age of 25 years with detailed family history for at least unto 2 generations. The control subjects were those who did not have any diabetic history i.e. they are non-diabetic (up to 2 generations). The study done in the Eastern Indian population as published Indian studies on MODY is quite sparse [14].
Moreover, patients with MODY may be erroneously be classified frequently as Type 1 Diabetes Mellitus (T1- DM) as well as Type 2 DM (T2DM). Diffferential diagnosis between MODY and early onset Type 2 DM is difficult because Type 2 DM tend to occur at an early age [3,15]. Hence, our study in its own way, may help to diagnose MODY by molecular genetic screening methods, utilizing molecular tools, which would help the clinicists of our region to throw some light in the proper and strict diagnosis of MODY, which definitely requires and warrants extra attention in the field of juvenile and adolescent diabetes [13,16,17].
There are two important findings from our work .The primary finding is that in the 10 different exons of the gene HNF1-alpha which is the responsible gene for causing MODY 3, there exists variations in clinically diagnosed MODY subjects of our region and the variations vary quite fairly from exon to exon. As revealed from our study, maximum number of variations exists in exon 5 of the HNF1- alpha gene.
Since sequencing is cost effective, hence one can use these methods to screen MODY subjects in our region, initially for exon 5 then followed by other exons of the said gene. Thus exon 5 of the HNF1-alpha gene may expected to be the mutational hotspot in our region and may serve as a molecular marker while screening the MODY subjects by molecular genetic screening method.
Mutation detection by molecular genetic screening method is now a focus of interest in clinical medicine. Identification of sequence variants by PCR-SSCP analysis is one of the most commonly applied methods used for detection of mutations. Here the efficacy of PCR-SSCP for detection of pathogenic mutation of MODY 3 is described, in the study which was confirmed by direct sequencing method [18].
The second finding is derived from the results of the sequencing study which revealed three novel variants all of which were identified in exon 5. It is mostly observed that the age at which the disease was diagnosed is around 20 - 22 years in subjects who had these polymorphisms in exon 5. Therefore the disease onset is likely to be at this age in subjects with polymorphisms in exon 5.
Thus the result of our PCR-SSCP study goes in agreement with the results of the sequencing study, which revealed that existence of novel polymorphisms in exon 5 in our Eastern Indian population which may be referred to as the “Mutational Hotspot” in our population.
Our study expressed relatively fewer numbers of HNF1- alpha gene mutations in comparison with studies done on China [19], Japan [20], Mexico [21], UK [12], which may suggest that further additional MODY gene screening is essential in our population.
5. CONCLUSION
Our study thus concludes that there is a high probable chance of mutations in exon 5, of HNF1-alpha gene responsible for MODY 3 in our population as evident by sequencing profiles study. Since these novel variants were all observed in exon 5, it appears to be the most signifycant exon hence screening studies should begin with this exon in our population. Therefore exon 5 is likely to be rich in MODY 3 mutations and may be referred to as THE MUTATIONAL HOTSPOT and to the best of our knowledge, our study is the pilot study from this region.
6. ACKNOWLEDGEMENTS
The authors sincerely thank M.P Birla Medical Research Centre, Kolkata, for their financial support in our study. The authors express sincere gratitude to the medical institute for their cooperation in allowing us to contact the patients and to obtain information from their clinical records.
REFERENCES
- Mclntyre, E.A. and Walker, M. (2002) Genetics of type 2 diabetes and insulin resistance: Knowledge from human studies. Clinical Endocrinology, 57, 303-311.
- Winter, W.E. (2000) Molecular and biochemical analysis of the MODY Syndromes. Pediatric Diabetes, 1, 88-117. doi:10.1034/j.1399-5448.2000.010206.x
- Fajans, S.S. and Bell, G.I. (2001) Molecular mechanisms and clinical pathophysiology of Maturity Onset Diabetes of the Young. New England Journal of Medicines, 345, 971-980. doi:10.1056/NEJMra002168
- Elbein, S.C., Teng, K., Yount, P., et al. (1998) Linkage and molecular scaning analysis of MODY 3/hepatocyte nuclear factor 1-alpha gene in typical familial type 2 diabetes: Evidence for novel mutation in exon 8 and 10. Journal of Clinical Eendocrinology and Metabolism, 83, 2059-2065. doi:10.1210/jc.83.6.2059
- Froguel, P. and Velho, G. (1999) Molecular genetics of Maturity-Onset Diabetes of the Young. Trends Endocrinol Metab, 10, 142-146. doi:10.1016/S1043-2760(98)00134-9
- Hattersley, A.T. (1998) Maturity-onset diabetes of the young: Clinical heterogeneity explained by genetic heterogeneity. Diabetes Care, 15, 15-24.
- Lehto, M., Wipemo, C., Ivarsson, S.A., et al. (1999) High frequency of mutations in MODY and mitochondrial genes in Scandinavian patients with familial early-onset diabetes. Diabetologia, 42, 1131-1137. doi:10.1007/s001250051281
- Mohan, V., Ramachandran, A., Snehalatha, C., et al. (1985) High prevalence of Maturity Onset Diabetes of the Young (MODY) among Indians. Diabetes Care, 8, 371- 374. doi:10.2337/diacare.8.4.371
- Sambroock, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular cloning: A laboratory manual. Cold Spring Harbour Publishing, New York.
- Baroni, M.G., Sentinelli, F., Massa, O., et al. (2001) Single-strand conformation polymorphism analysis of the glucose transporter gene GLUT1 in Maturity-Onset Diabetes of the Young. Journal of Molecular Medicine, 79, 270-274. doi:10.1007/s001090100220
- Ellard, S. (2000) Hepatocyte nuclear factor 1 alpha (HNF-1 alpha) mutations in Maturity-Onset Diabetes of the Young. Human Mutation, 16, 377-385. doi:10.1002/1098-1004(200011)16:5<377::AID-HUMU1>3.0.CO;2-2
- Christine, B., Claire, C., Jean, R., et al. (2008) The type and the position of HNF1A mutation modulate age at diagnosis of diabetes in patients with Maturity-Onset Diabetes of the Young (MODY)-3. Diabetes, 57, 503-508.
- Shih, D.Q. and Stoffel, M. (2002) Molecular etiologies of MODY and other early-onset forms of diabetes. Current Diabetes Reports, 2, 125-134. doi:10.1007/s11892-002-0071-9
- Radha, V., Ek, J., Anuradha, S., et al. (2009) Identification of novel variants in the hepatocyte nuclear factor-1α gene in South Indian patients with Maturity Onset Diabetes of Young. The Journal of Clinical Endocrinology & Metabolism, 94, 1959-1965. doi:10.1210/jc.2008-2371
- Byrne, M.M., Sturis, J., Menzel, S., et al. (1996) Altered insulin secretory responses to glucose in diabetic and non-diabetic subjects with mutations in the diabetes susceptibility gene MODY 3 on chromosome 12. Diabetes, 45, 1503-1510. doi:10.2337/diabetes.45.11.1503
- Vaxillaire, M., Rouard, M., Yamagata, K., et al. (1997) Identification of nine novel mutations in the hepatocyte nuclear factor-1 gene associated with Maturity-Onset Diabetes of the Young (MODY 3). Human Molecular Genetics, 6, 583-586. doi:10.1093/hmg/6.4.583
- Yamagata, K., Oda, N., Kaisaki, P.J., Menzel, S., et al. (1996) Mutations in the hepatocyte nuclear factor-1 alpha gene in Maturity-Onset Diabetes of the Young (MODY 3). Nature, 384, 455-458. doi:10.1038/384455a0
- Kaisaki, P.J., Menzel, S., Lindner, T., et al. (1997) Mutations in the hepatocyte nuclear factor-1-α gene in MODY and early-onset NIDDM. Evidence for a mutational hotspot in exon 4. Diabetes, 46, 528-535. doi:10.2337/diabetes.46.3.528
- Zhen, Y., Song-Hua, W.U., Tai-Shan, Z., et al. (2006) Identification of four novel mutations in the HNF-1A gene in Chinese early-onset and/or multiplex diabetes pedigrees. The Journal of Chinese Medicine, 119, 1072- 1078.
- Tonooka, N., Tomura, H., Takahashi, Y., et al. (2002), High frequency of mutations in the HNF-1alpha gene in non-obese patients with diabetes of youth in Japanese and identification of a case of digenic inheritance. Diabetologia, 45, 1709-1712. doi:10.1007/s00125-002-0978-3
- Domínguez-López, A., Miliar-García, A., Segura-Kato, Y.X., et al. (2005) Mutations in MODY genes are not common cause of early-onset type 2 diabetes in Mexican families. JOP, 10, 238-245.

