Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2010, 3, 397-404 JBiSE doi:10.4236/jbise.2010.34055 Published Online April 2010 (http://www.SciRP.org/journal/jbise/). Published Online April 2010 in SciRes. http://www.scirp.org/journal/jbise Prokaryotic expression, purification of a novel candidate tumor suppressor gene FUS1 and characterization of its polyclonal antibodies Dong-Mei Zhang, Han-Shuo Yang, Xin-Yu Zhao, Wen Zhu, Zhi-Hua Feng, Yang Wan, Zhi-Wei Zhao, Ming-Hai Tang, Nong-Yu Huang, Yu-Quan Wei State Key Laboratory of Biotherapy, West China Hospital, West China Medical School and School of Life Sciences, Sichuan Univer- sity, Chengdu, China. Email: yuquawei@vip.sina.com; zwjulia@163.com Received 6 June 2009; revised 27 November 2009; accepted 4 December 2009. ABSTRACT FUS1 is a novel candidate tumor suppressor gene identified in human chromosome 3p21.3. Its expres- sion showed significantly reduction or even loss in lung cancer and other types of cancers. In order to further investigate the biological function of FUS1 protein, FUS1 cDNA from MRC-5 cells was amplified by RT-PCR and cloned into prokaryotic expression vector pQE-30. The recombinant expression plasmids were transformed into M15 strain and grown at 20℃ or 37℃. SDS–PAGE analysis revealed that the ac- cumulation of the recombinant protein FUS1 (rFUS1) in inclusion body forms reached maxium amount when induced with 0.5 mM IPTG for 5 h at 37℃. The inclusion bodies were solubilized in 2M urea and pu- rified by a 6 × His tagged affinity column under dena- turing condition. The purified rFUS1 was identified by electrospray ionization-mass spectr ometry (ESI-MS) and tested for purity by HPLC chromatography. The purified rFUS1 proteins were then used to immunize rabbits to obtain anti-human FUS1 polyclonal anti- bodies, which were suitable to detect both the recom- binant exogenous FUS1 and the endogenous FUS1 from tissues and cells by western blot and immuno- histochemistry, Available purified rFUS1 proteins and self-prepared polyclonal antibodies against FUS1 may provide effective tools for further studies on bio- logical function and application of FUS1. Keywords: FUS1; Polyclonal Antibody; Prokaryotic Expression; Recombinant Protein; Tumor Suppressor Gene 1. INTRODUCTION With a rapidly increasing incidence and a low cure rate, lung cancer has been the most common malignancy and leading cause of cancer deaths in the world [1]. There- fore, identification of new therapeutic targets and novel strategies are essential to improve the survival of pa- tients diagnosed with this disease. FUS1 is a novel tu- mor-suppressor gene located on the human chromosome 3p21.3 region [2]. Genetic alterations and allelic loss of 3p21.3 are among the most frequent and earliest cancer abnormalities detected in the pathogenesis of lung can- cers. This phenomenon occurs in almost 100% of small cell lung cancers (SCLCs) and more than 80% of non-small-cell lung cancers (NSCLCs) [3]. FUS1 func- tions as “gatekeepers” of human cancer and plays a very important role in lung cancer development [4,5]. A ma- jority of human lung cancers have been found loss of expression, haploinsufficiency or deficiency of post- translational of FUS1 [2,6,7]. FUS1 was recently dem- onstrated to mediate accumulation of p53 protein, and thus led to the apoptosis and cell cycle arrest in the early stages of lung cancer progression [8,9].However, many properties of FUS1 biological functions as well as its action mechanism are not well understood and little is known about its basic structure–function relationships. Therefore, the expression and the preparation of recom- binant protein FUS1 and its polyclonal antibody should provide effective experim ental tools for further identi fying its mechanism and biological functions against cancers. In the present study, th e full length FUS1 with six his- tidine (6 × His) residues was expressed in Escherichia coli M15. The recombinant protein was purified by urea and identified by electrospray ionization-mass spec- trometry analysis (ESI-MS). The polyclonal antibodies against FUS1 were prepared which were suitable to de- tect the presence of both the exogenous and endog enous FUS1 efficiently by western blot and immunohisto- chemistry. The self-prepared polyclonal antibodies may be applied as the effective tools for investigating the further molecular mechanisms of FUS1.  D. M. Zhang et al. / J. Biomedical Science and Engineering 3 (2010) 397-404 Copyright © 2010 SciRes. JBiSE 398 2. MATERIALS AND METHODS 2.1. Materials Escherichia coli strain JM109 an d M15 were maintained by our Lab. The human non-small-cell lung cancer (NSCLC) cell line A549 and normal human lung fibro- blast cell line MRC-5 were obtained from ATCC and were maintained in RPMI 1640 or DMEM supplemented with 10% FCS respectively. pQE-30 plasmid was pur- chased from Qiagen (Germany). Human paracancerous normal lung tissues and lung cancer tissues were ob- tained from Department of Thoracic and Cardiovascular Surgery, West China Hospital, Sichuan University. Re- striction endonucleases, T4 DNA ligase and prestained low range protein molecular weight marker were prod- ucts of MBI Fermentas (Lithuania). The High Fidelity PrimeScriptTM RT-PCR Kit was purchased from TaKaRa (Dalian, China). All PCR products used for cloning were confirmed by sequencing at Invitrogen Biotechnology Co., Ltd (Shanghai, China). pVITRO2-FUS1 plasmid was constructed by our Lab. 2.2. Total RNA Isolation Total RNA was isolated from MRC-5 cells using a stan- dard Trizol RNA isolation protocol. Prior to use, RNA concentration was spectrophotometrically determined, and RNA integrity was verified by electrophoresis. 2.3. RT-PCR Amplification of FUS1 cDNA The cDNA of full length FUS1 was prepared using a High Fidelity PrimeScriptTM RT-PCR Kit according to the manufacturer instructions. Based on the FUS1 cDNA sequence (GenBank accession No. AF055479), a pair of primers was designed as follows: 5'-CGCGGATCCATGGGCGCCAGCGGGTCCAAA G- 3'(sense) and 5'-CCGGTCGACTCACACCTCATAGAG GATCACAG-3'(anti-sense). The sense and anti-sense primers were introduced BamHI and SalI restriction sites (underlined) respectiv ely. 2.4. Plasmid Construction and Identification The amplified FUS1 cDNA and expression vector pQE-30 were digested with restriction enzymes BamHI/SalI re- spectively. The digested products were separated on a 1% agarose gel and the bands were extracted. The purified FUS1 cDNA and the linear vector were ligated overnight at 16℃ with T4 DNA ligase followed by transformed into E.coli JM109 competent cells. The transformation mixture was plated on to LB agar plates containing am- picillin (100 μg/ml). The plates were incubated for 16h at 37℃. The desired recombinant plasmid pQE-30-FUS1 was confirmed by PCR and restriction enzyme digestion with BamHI/SalI and DNA sequencing (Invitrogen). 2.5. Expression of Fusion Proteins E. coli M15 cells were transformed with recombinant plas- mid pQE-30-FUS1 extracted from JM 109. The bacteria cells were cultured at 37℃ in 5 ml LB liquid medium with 100 μg/ml ampicillin for 4-6 h at 220 rpm till to an absorb- ance of 0.6 at 600 nm. Expression o f the fusion protein was induced by isopropyl-β-D-thiogalactopyranoside (IPTG) with a final concentration of 0.5 mM for 5 h at 37℃. A time and temperature course o f expression and solubility to determine the optimal induction conditions for maxi- mum expression of protein was measured by taking ali- quots of cells at 1, 2, 3, 4 and 5 h after induction with IPTG at 37℃ and 20℃ These samples were harvested by centrifugation at 13,000 rpm for 1min. The pellets were resuspended in PBS (pH 8.0) and sonicated for 6 × 10 s with 10 s pauses at 20 W on ice. The total lysate induced with IPTG was divided into soluble and insolu- ble fractions by centrifugation at 15,000 rpm for 2 min at 4℃. The expression and solubility of FUS1 protein were then analyzed in parallel by 15% SDS–PAGE followed by staining with Coom assie Brill iant Bl ue R-250. The prot ein extracts of cells transformed with the uninduced bacteria were used as the control. 2.6. Extraction of Fusion Proteins For large scale expression and purification, M15 strain was transformed with the plasmid and cultured in 1 L of LB medium at 37℃ until to an absorbance of 0.6 at 600 nm followed by induction of 0.5 mM IPTG at 37℃ for an additional 5 h before harvested. The cells were harvested by centrifugation at 4,000 rpm for 20 min at 4℃. The pellet was resuspended in PBS (20mM PB, 500 mM NaCl, pH 8.0). Extraction was performed using a French Pressur e Cell Press (APU Co., 04010008) at an internal pressure of 800 psi [10]. The remaining pellet was then harvested by centrifugation at 15,000 rpm for 30 min at 4℃. The supernatant (soluble fraction) was collected for analysis later. The inclusion bodies were weighted and solution buffer (20 mM PB, 500 mM NaCl, 2 M urea, Ph 8.0) were pulled to suspend the cells, then the supernatant were collected by centrifugation at 15000 rpm for 30 min at 4℃. The purified recombinant protein was confirmed by SDS-PAGE and Western blot- ting using anti-His monoclonal antibody conjugate to HRP. The concentration of the protein was determined according to Bradford [11]. 2.7. rFUS1 Identification by ESI-MS Analysis The presence of purified rFUS1 in the eluted fractions was separated on 15% SDS–PAGE and identified by electrospray ionization (ESI)-mass spectrometry (MS). The gel band stained with Coomassie brilliant blue R-250 was excised minced, reduced, alkylated with io-  D. M. Zhang et al. / J. Biomedical Science and Engineering 3 (2010) 397-404 Copyright © 2010 SciRes. JBiSE 399 doacetamide, In-gel digestion of proteins was carried out with 12.5 ng/μl mass spectrometry grade Trypsin Gold (Promega) for 12-16 h at 37℃ [12]. The tryptic peptides were extracted twice with the buffer containing 50% ACN /0.1% TFA for 15 min, and the solutions were com- bined together. Mass spectra were acquired using an ESI-Q-TOF mass spectrometer (Micromass, Manchester, UK) with 15 µl of tryptic peptides solution. The MS/MS data were acquired by the software of MassLynx (Mi- cromass) and converted to PKL files by the software of ProteinLynx 2.2.5 (Waters) were then analyzed using °MASCO T s ea rc h en gi ne (http://www.matrixscience. com). 2.8. Preparation, Purification of rFUS1 Poly- clonal Antibodies The purified rFUS1 was used to prepare antibodies in New Zealand white rabbit. The rabbit was first immu- nized subcutaneously with rFUS1 (200 μg) in complete Freund’s adjuvant. Two booster injections were given in incomplete Freund’s adjuvant every week. The serum was collected 7 days after the 3rd immunization to de- termine the antibody titer by enzymelinked immunosor- bent analysis (ELISA). The last immunization was per- formed one week later, and the antiserum was collected through heart after 7 days. The rabbit IgG fraction was precipitated from the immune seru m with 50% saturated (NH4)2SO4 and purified by DEAE-Sepharose column chromatography. 2.9. Specificity Analysis of the Polyclonal Antibodies by Western Blot The specificity of the antiserum was tested by western blot analysis using the to tal proteins of MRC-5 cells and those of A549 cells transfected with FUS1 constructs. rFUS1 and the total proteins of A549 cells untransfected were used as control. The cells were harvested and lysed in lysis buffer [10 mM Tris/HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100 and 1 mM DTT supplemented with protease and phosphatase inhibitors (2 mM sodium or- thovanadate, 100 nM okadaic acid, 1 mM NaF, 1 mM β-glycerophosphate and cocktail (Sigma)] according to the related methods [13]. The protein samples were separated on 15% SDS-PAGE and electrophoretically transferred onto PVDF (polyvinyllidenefluoride) mem- brane. After blocking overnight in 5% (w/v) non-fat milk, the PVDF membranes were incubated with serum at a dilution of 1:1000 for 2 h. The membranes were washed three times with TBST buffer and then incubated in goat anti-rabbit IgG conjugated with HRP at a dilution of 1:10000 for 1 h at 37℃.After washing two times with TBST buffer, one time with TBS then analyzed using the enhanced chemiluminescence detection system and ex- posed to Kodak BioMax X-ray film for 2-5 min. 2.10. Cell and Tissue Immunohistochemistry In order to further confirm that the polyclonal antibod ies against rFUS1 are suitable for application in recognizing the innate FUS1 proteins from cells or tissues, immuno- histochemistry was performed in A549 cells transfected with FUS1 constructs and normal lung tissue respec- tively. A549 cells were seeded onto coverslips and cul- tured in RPMI-1640 with 10% (v/v) FBS (fetal bovine serum) at 37℃ in 5% CO2. The cells were transfected with recombinant plasmid pVITRO2-FUS1 and vector pVITRO2 when cell confluence reached 60%. Hoechst 33258 staining was used to identify apoptosis induced by FUS1 expression in A549 cells. At 48 h post-transfection, cells were fixed with fresh Carnoy’s fixative, stained with Hoechst 33258 for 30 min at the concentration of 0.5 µg/ml. Stained nuclei were detected after washing twice with distilled water and observed under a fluores- cence microscope. At the same time, the coverslips transfected with pVITRO2-FUS1 was immersed in ice- cold acetone to fix for 20 min on ice. The cell was per- meabilized with 0.2% Triton X-100 for 10 min after washing two times with deionized water, the slips were blocked with goat serum albumin at 37℃ for 15 min, the anti-rFUS1 polyclonal serum was used as the pri- mary antibody (1:750). The second antibody was a biotinylated goat anti-rabbit IgG. The cells were then stained with HRP–streptavidin reagents (Dako) and DAB. Brown staining was considered positive. As for tissue immunohistochemical analysis, normal lung tissues were fixed in 10% buffered neutral formalin and embedded in paraffin. Then, indirect immunostain- ing for FUS1 was performed on paraffin-embedded tis- sues by using the LSAB (labelled streptavidin–biotin) method to visualize antibody response as described above. 3. RESULTS AND DISCUSSION 3.1. Construction and Identification of Expression Plasmid pQE-30-FUS1 For amplification of FUS1 cDNA, RT–PCR was per- formed with total RNA from MRC-5 cell as the template, using gene-specific primers containing a BamH I site or a Sal I site to facilitate cloning into expression plasmid pQE-30. A DNA fragment, approximate 350 bp in length, was obtained as shown (Figure 1(a)), which is consis- tent with the FUS1 cDNA 333 bp in length. The amplified FUS1 cDNA was inserted into the sites of BamH I and Sal I in the expression plasmid pQE-30. The recombinant plasmid pQE-30-FUS1 was verified by PCR using FUS1-specific primer and restriction en- donuclease digestion with BamH I/Sal I (Figure 1(b)) and DNA sequencing (data not shown). 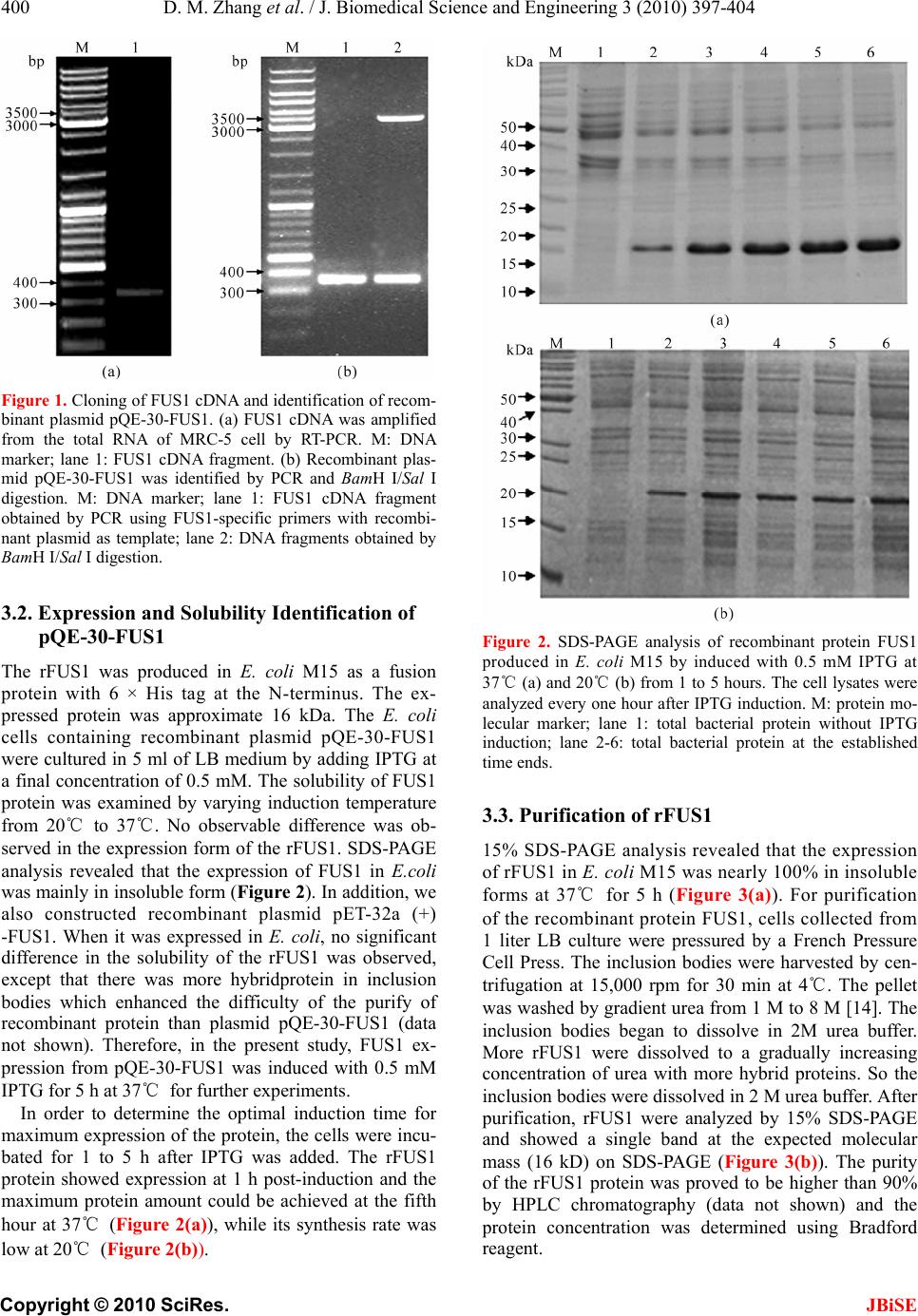 D. M. Zhang et al. / J. Biomedical Science and Engineering 3 (2010) 397-404 Copyright © 2010 SciRes. JBiSE 400 Figure 1. Cloning of FUS1 cDNA and identification of recom- binant plasmid pQE-30-FUS1. (a) FUS1 cDNA was amplified from the total RNA of MRC-5 cell by RT-PCR. M: DNA marker; lane 1: FUS1 cDNA fragment. (b) Recombinant plas- mid pQE-30-FUS1 was identified by PCR and BamH I/Sal I digestion. M: DNA marker; lane 1: FUS1 cDNA fragment obtained by PCR using FUS1-specific primers with recombi- nant plasmid as template; lane 2: DNA fragments obtained by BamH I/Sal I digestion. 3.2. Expression and Solubility Identification of pQE-30-FUS1 The rFUS1 was produced in E. coli M15 as a fusion protein with 6 × His tag at the N-terminus. The ex- pressed protein was approximate 16 kDa. The E. coli cells containing recombinant plasmid pQE-30-FUS1 were cultured in 5 ml of LB medium by adding IPTG at a final concentration of 0.5 mM. The so lubility of FUS1 protein was examined by varying induction temperature from 20℃ to 37℃. No observable difference was ob- served in the expression form of the rFUS1. SDS-PAGE analysis revealed that the expression of FUS1 in E.coli was mainly in insoluble form (Figure 2). In addition, we also constructed recombinant plasmid pET-32a (+) -FUS1. When it was expressed in E. coli, no significant difference in the solubility of the rFUS1 was observed, except that there was more hybridprotein in inclusion bodies which enhanced the difficulty of the purify of recombinant protein than plasmid pQE-30-FUS1 (data not shown). Therefore, in the present study, FUS1 ex- pression from pQE-30-FUS1 was induced with 0.5 mM IPTG for 5 h at 37℃ for further experiments. In order to determine the optimal induction time for maximum expression of the protein, the cells were incu- bated for 1 to 5 h after IPTG was added. The rFUS1 protein showed expression at 1 h post-induction and the maximum protein amount could be achieved at the fifth hour at 37℃ (Figure 2(a)), while its synthesis rate was low at 20℃ (Figur e 2(b)). Figure 2. SDS-PAGE analysis of recombinant protein FUS1 produced in E. coli M15 by induced with 0.5 mM IPTG at 37℃ (a) and 20℃ (b) from 1 to 5 hours. The cell lysates were analyzed every one hour after IPTG induction. M: protein mo- lecular marker; lane 1: total bacterial protein without IPTG induction; lane 2-6: total bacterial protein at the established time ends. 3.3. Purification of rFUS1 15% SDS-PAGE analysis revealed that the expression of rFUS1 in E. coli M15 was nearly 100% in insoluble forms at 37℃ for 5 h (Figure 3(a)). For purification of the recombinant protein FUS1, cells collected from 1 liter LB culture were pressured by a French Pressure Cell Press. The inclusion bodies were harvested by cen- trifugation at 15,000 rpm for 30 min at 4℃. The pellet was washed by gradient urea from 1 M to 8 M [14]. The inclusion bodies began to dissolve in 2M urea buffer. More rFUS1 were dissolved to a gradually increasing concentration of urea with more hybrid proteins. So the inclusion bodies were dissolved in 2 M urea buffer. After purification, rFUS1 were analyzed by 15% SDS-PAGE and showed a single band at the expected molecular mass (16 kD) on SDS-PAGE (Figure 3(b)). The purity of the rFUS1 protein was proved to be higher than 90% by HPLC chromatography (data not shown) and the protein concentration was determined using Bradford reagent.  D. M. Zhang et al. / J. Biomedical Science and Engineering 3 (2010) 397-404 Copyright © 2010 SciRes. JBiSE 401 Figure 3. Solubility identification and purification of rFUS1 in E. coli M15. (a) the M15 cells were cultured at 37 , induced ℃ with 0. 5 mM IPT G for 5 h and assay e d by SDS- PAGE a naly si s. Lane 1: total bacterial protein without IPTG induction; lane 2: total bacterial protein with IPTG induction; lane 3: supernatant with IPTG induction; lane 4: precipitate with IPTG induction; (b) Purification of rFUS1.Lane 1: The precipitate of total bac- terial protein containing rFUS1 with IPTG induction. Lane 2: purified rFUS1; M: protein molecular marker. 3.4. Western Blot Anlysis of the Purified rFUS1 After SDS–PAGE, rFUS1 was transferred to a nitrocel- lulose membrane. The membrane was incubated with blocking buffer [0.1% Tween and 5% non-fat milk in Tris-buffered saline (TBS)] overnight at 4℃ and then with the anti-His-tag monoclonal antibody conjugate HRP diluted 1:1000. The membrane was washed 3 times with washing solution (Tween 0.1% in TBS) and incu- bated with an antimouse peroxidase-conjugated antibody (KPL) diluted 1:5000. After washed 3 times with wash- ing solution, the membrane was treated with SuperSignal West Pico Chemiluminescent Substrate (Pierce) for 5 min, then exposed to HyperWlm (Amersham Biosciences) for 5 min and visualized. The purified rFUS1 was verified successfully using Western-blot analysis through anti- His-tag mAb (Figure 4). 3.5. rFUS1 Identification by ESI-MS Analysis The purified recombinant FUS1 proteins were further validated by ESI-MS analysis. The result from MS data suggested that the identified protein exactly matched with human FUS1 protein, which had a Mascot score 1018. MS/MS analysis revealed that seven unique pep- tide unambiguously matched to the target FUS1 protein (Figure 5(a)). For example, MS/MS spectrum of parent ion 861.7737 and the result of peptide sequence query were shown in Figure 5(b). All matched peptides were shown in Figure 5(c) (underlined), which indicated that the purified recombinant protein FUS1 were completely correct. A sufficient amount of purified FUS1 protein would make it possible to prepare polyclonal antibodies against FUS1 and to further analyze its interacting pro- teins or structure by X-r a y crystallography. Figure 4. Analysis of purified rFUS1 by Western blot. The purified recombinant protein FUS1 was separated on 15% SDS- PAGE and probed by with anti-His tag mAb conjugated with HRP (1:10000). Che- miluminescence immunoassay was used for color development. Figure 5. ESI–MS/MS identification of tryptic peptides from the purified recombinant protein. (a) mass spectrogram of tryptic peptides from purified FUS1 protein. Totally 7 unique peptides were matched to target protein; (b) MS/MS spectrum of parent ion 861.7737 as an example. Data base search indicated its pep- tide sequence was GLWPFASAAGGGGSEAAGAEQALVRPR, which was a part of the sequence of FUS1; (c) matched pep- tides (underlined). 3.6. Characteristics of Antiserum Against FUS1 The specificity of the an tiserum from rabbit again st puri- fied rFUS1 and total proteins extracted from MRC-5 was checked by Western blot analysi s. FU S1 h as been proved no protein level expression in A549 cells though FUS1  D. M. Zhang et al. / J. Biomedical Science and Engineering 3 (2010) 397-404 Copyright © 2010 SciRes. JBiSE 402 mRNA was detectable [3,6]. Therefore, total proteins f ro m A549 cells transfected with expression plasmid pVI- TRO2-FUS1 was also used in Western blot. The result indicated that the antiserum from rabbit can recognize both exogenous recombinant FUS1 and endogenous FUS1 effectively. There were positive bands at the po si- tion of approximately 16 kDa (Figure 6). However, non- immunized serum was negative (data not shown). The FUS1 protein is proved to be indeed present in the cytoplasm and may have a role in signal transduction [8]. In the present study, FUS1-negative A549 cells were stained with Hoechst 33258 af ter 24 h transient transfec- tion with pVI TRO2-FUS1. More hyp ercondensed nuclei and apoptotic bodies appeared in the transfected A549 cells (brightly stained; Figure 7(c)) comparing with that in the control cells (Figure 7(a) and (b)). The result of FUS1 inducing apoptosis consisted with the findings reported previously by Ji et al. and Ito et al. [8,15] FUS1-mediated apoptosis is proved to be associated with the accumulation of p53 protein, the down-regulating expression of MDM2 and the activation of Apaf1-drivened mitochondrial apopto tic pathway [9]. However, the exact mechanism of inactivation of FUS1 in human tumori- genesis and its role in FUS1-mediated tumor suppression are still unclear and need to be investigated in detail. Figure 6. Western-blot analysis of FUS1 expression using the rabbit antiserum (1:1000). (a) the total protein from A549 (lane 1), purified rFUS1 (lane 2), the total protein from MRC-5 cells (lane 3) and the total protein from A549 cells after transient transfection with FUS1 constructs (line 4) were loaded. Horse radish peroxidase-conjugated goat anti-rabbit IgG (1:10000) and enchanced chemiluminescence were used for color devel- opment; (b) the same blots were probed by β-actin as contr ol. Figure 7. Induction of apoptosis in A549 cells by the expres- sion of FUS1. (a) the untreated A549 cells as negative control; (b) A549 cells transfected with pVITRO2; (c) A549 cells transfected with pVITRO2-FUS1. At 24 h post-transfection, the cells were stained with Hoechst 33258. The arrows indi- cated the representatives of apoptotic cell. Original magnifica- tion, × 200. To further elucidate the relationship between FUS1 expression and apoptosis, the self-prepared anti-rFUS1 polyclonal antibodies were used for immunostaining in A549 cells transfected with pVITRO2-FUS1 and un- treated cells. At the same time, immunohistochemistry was also performed in paracancerous normal lung tissues and lung cancer tissues. There were strong brown stain- ing in the cytoplasm of A549 cells transfected with FUS1 constructs and that of paracancerous normal lung tissues (Figure 8(a) and (c)). However, no immunostaining was observed in the cytoplasm of non-transfected A549 cells and that of lung cancer tissues (Figure 8(b) and (d)). These results are consistent with previous findings and provide further evidence that FUS1 plays imp ortant roles in tumor-suppression function and lung cancer develop- ment. The similar results were obtained in liver, stomach, Figure 8. Analysis of FUS1 expression using the self-prepared rabbit anti-human rFUS1 polyclonal antibodies (1:750). Im- munostaining was performed in A549 cells transfected with pVITRO2-FUS1 or not (a and b) and paracancerous normal lung tissues and lung cancer tissues (c and d). The anti-FUS1 polyclonal antibodies (1:750) can recognize the FUS1 protein in the cytoplasm of A549 cells transfected with FUS1 con- structs and that of paracancerous normal lung tissues (a and c) comparing with the corresponding controls (b and d). Brown staining showed the positive results. Haematoxylin staining showed the cell nuclei. Original magnification, × 200.  D. M. Zhang et al. / J. Biomedical Science and Engineering 3 (2010) 397-404 Copyright © 2010 SciRes. JBiSE 403 cervix, endometrial and ovarian carcinomas when using the self-prepared anti-FUS1 polyclonal antibodies (data not shown). 4. CONCLUSIONS FUS1 is a novel tumor-suppressor gene located on hu- man chromosome 3p21.3 that is frequently deleted in human lung and breast cancers. But there is no commer- cial antibody against full-length FUS1 until now. In the present work, human full-length FUS1 cDNA was cloned and expressed as a fusion protein with six his- tidine (6 × His) tag in E. coli. The purified recombinant protein was identified by ESI–MS (electrospray ioniza- tion MS) analysis. Furthermore, the specific and sensi- tive polyclonal antibod ies against full-length FUS1 were raised, which were suitable to detect both the recombi- nant exogenous FUS1 and the endogenous FUS1 from tissues and cells by western blot and immunohistochem- istry. To our knowledge, this is the first report of soluble expression and purification of rFUS1 protein and gen- eration of antifull-length FUS1 polyclonal antibodies so far. The purified rFUS1 proteins and self-prepared poly- clonal antibodies against FUS1 may provide effective tools for further studies on biological function and me- chanism of FUS1 in pathogenesis of lung and other car- cinomas in the future. FUS1 protein expression rarely existed in human pri- mary lung cancer tissue using the self-prepared poly- clonal antibodies, while it can be detected in cytoplasm of normal lung tissues. The similar result was also testi- fied in A549 cells transfected with or without FUS1 con- structs. These results are consistent with previous find- ings and provide further evidence that FUS1 plays im- portant roles in tumor-suppression function and lung cancer development. 5. ACKNOWLEDGEMENTS This work was supported by the grant from the National Key Research Program for New Drug Development (2009Z X09301-004). REFERENCES [1] Jemal, A., Siegel, R., Ward, E., Hao, Y., Xu, J., Murray, T. and Thun, M.J. (2008) Cancer statistics. A Cancer Jour- nal for Clinicians, 58, 71-96. [2] Kondo, M., Ji, L., Kamibayashi, C., Tomizawa, Y., Randle, D., Sekido, Y., Yokota, J., Kashuba, V., Zabarovsky, E., Kuzmin, I., Lerman, M., Roth, J. and Minna, J.D. (2001) Overexpression of candidate tumor suppressor gene FUS1 isolated from the 3p21 .3 homozygous deletion region leads to G1 arrest and growth inhibition of lung cancer cells. Oncogene, 20, 6258-6262. [3] Lerman, M.I., Glenn, G.M., Daniel, L., Latif, F., Hosoe, S., Brauch, H., Hampsch, K., Delisio, J., Orcutt, M. and Zbar, B. (1990) A new polymorphic probe on chromo- some 3p: Lambda LIB28-77 (D3S169E). Nucleic Acids Research, 18, 205. [4] Lerman, M.I. and Minna, J.D. (2000) The 630-kb lung c a n- cer homozygous deletion region on human chromosome 3p21.3: Identification and evaluation of the resident can- didate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Co n- sortium. Cancer Research, 60, 6116-6133. [5] Zabarovsky, E.R., Lerman, M.I. and Minna, J.D. (2002) Tu- mor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene, 21, 6915- 6935. [6] Uno, F., Sasaki, J., Nishizaki, M., Carboni, G., Xu, K., Atkinson, E.N., Kondo, M., Minna, J.D., Roth, J.A. and Ji, L. (2004) Myristoylation of the FUS1 protein is re- quired for tumor suppression in human lung cancer cells. Cancer Research, 64, 2969-2976. [7] Prudkin, L., Behrens, C., Liu, D.D., Zhou, X., Ozburn, N.C., Bekele, B.N., Minna, J.D., Moran, C., Roth, J.A., Ji, L. and Wistuba, L.L. (2008) Loss and reduction of FUS1 protein expression is a frequent phenomenon in the patho- genesis of lung cancer. Clinical Cancer Research, 14, 41-47. [8] Ito, I., Ji, L., Tanaka, F., Saito, Y., Gopalan, B., Branch, C.D., Xu, K., Atkinson, E.N., Bekele, B.N., Stephens, L.C., Minna, J.D., Roth, J.A. and Ramesh, R. (2004) Li- posomal vector mediated delivery of the 3p FUS1 gene demonstrates potent antitumor activity against human lung cancer in vivo. Cancer Gene Therapy, 11, 733-739. [9] Deng, W.G., Kawashima, H., Wu, G., Jayachandran, G., Xu, K., Minna, J.D., Roth, J.A. and Ji, L. (2007) Syner- gistic tumor suppression by coexpression of FUS1 and p53 is associated with down-regulation of murine double minute-2 and a ctivation of the apoptotic prote ase-act iva t ing factor 1-dependent apoptotic pathway in human non- small cell lung cancer cells. Cancer Research, 67, 709-717. [10] Tsukamoto, H., Fukudome, K., Kohara, J., Nakatake, H. and Kimoto, M. (2007) Preparation of recombinant mur- ine tumor necrosis factor-alpha in Escherichia coli: A rapid method to remove tags from fusion proteins by thrombin-cleavage and ion-exchange chromatography. Protein Expression and Purification, 56, 138-144. [11] Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein util- izing the principle of protein-dye binding. Analytical Bio- chemistry, 72, 248-254. [12] Zhao, X.Y., Li, H.X., Liang, S .F., Yuan, Z., Yan , F., Ruan, X.Z., You, J., Xiong, S.Q., Tang, M.H. and Wei, Y.Q. (2008) Soluble expression of human DRR1 (down-regulated in renal cell carcinoma 1) in Escherichia coli and preparation of its polyclonal antibodies. Biotechnology and Appied Biochemis- try, 49, 17-23. [13] Benzinger, A., Muster, N., Koch, H.B., Yates, J.R. and Hermeking, H. (2005) Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in can- cer. Molecular & Cellular Proteomics, 4, 785-795. [14] Wilkinson, R.J., Elliott, P., Carragher, J.F. and Francis, G. (2004) Expression, purification, and in vitro characteri- zation of recombinant salmon insulin-like growth fac- tor-II. Protein Expression and Purification, 35, 334-343. [15] Ji, L., Nishizaki, M., Gao, B., Burbee, D., Kondo, M., Kamibayashi, C., Xu, K., Yen, N., Atkinson, E.N., Fang, B., Lerman, M.I., Roth, J.A. and Minna, J.D. (2002) Ex-  D. M. Zhang et al. / J. Biomedical Science and Engineering 3 (2010) 397-404 Copyright © 2010 SciRes. JBiSE 404 pression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Research, 62, 2715-2720. |

