Journal of Biomaterials and Nanobiotechnology
Vol.3 No.1(2012), Article ID:17008,7 pages DOI:10.4236/jbnb.2012.31016
Reducible Poly(2-dimethylaminoethyl) Methacrylate-Block-Polyvinylimidazole: Synthesis, Transfection Activity in Vitro
![]()
Shanghai Institute of Applied Physics, Chinese Academy of Sciences, Shanghai, China.
Email: yubozhang@sinap.ac.cn
Received October 16th, 2011; revised November 18th, 2011; accepted December 20th, 2011
Keywords: Reducible Polymer; Gene Expression; Transfection; Cytotoxicity
ABSTRACT
Reducible or imidazolyl polycations of block poly(imidazole/2-dimethylaminoethyl) are of promising in gene delivery. Dimeric poly(2-dimethylaminoethyl) methacrylate-block-polyvinylimidazole (rDPDMAEMAIM) and reducible poly (2-dimethylaminoethyl) methacrylate (rDPDMAEMA) with single disulfide bond in the backbone was synthesized by oxidizing their dithioester-terminated polymers. The polyplexes sizes, rDPDMAEMAIM/pDNA and rDPDMAEMA/ pDNA (plasmid DNA), are in the ranges of 100 nm - 150 nm at the weight ratio of 12:1, and the zeta potential of rDPDMAEMAIM/pDNA from 9.6 mV to 22.7 mV in phosphate buffered saline solutions increases with their weight ratios of 1:1 to 18:1. The results show that the rDPDMAEMAIM/pDNA polyplexes have higher transfection activity and lower cytotoxicity than that of rDPDMAEMAIM/pDNA against 293T cells in vitro in the presence of serum, indicating that the PDMAEMAIM present a promising nonviral gene vector.
1. Introduction
Reducible or imidazolyl non-viral vectors have enormous potential to treat vital human diseases because of safely and efficiently delivering therapeutic gene into the endosomes of target cell during endosomal processing and eventual lysosomal fusion [1-14]. Reducible polymers can improve the efficacy of the gene delivery and reduce intracellular toxicity due to their molecular backbone reversibility [1-7]. For examples, a reducible linear polyethylenimine (rPEI) has a low cytotoxicity and comparable transfection activity with non-reducible PEI [8,9]a reducible poly (amido-ethyl-enimines) (rPAEI) linked with cystamine bisacrylamide can deliver plasmid DNA (pDNA) and siRNA [10,11], a reducible poly(2-dimethylaminoethyl) methacrylate (rPDMAEMA) synthesized by reversible addition-fragmentation chain transfer (RAFT) polymerization has minimal cytotoxicity and better transfection activity than non-reducible PDMAEMA [7,12, 13], and linear reducible polycations polymerized by a 10-mer oligolysine can unpackage DNA due to cleaving disulfide bridges in a reducible medium like the cytosol [4]. Also, the histidine (His6 RPCs)-rich reducible polycations facilitate intracellular delivery of nucleic acids such as siRNA.
Polymers containing imidazole can induce membrane destabilization in acidic media endosomes and deliver nucleic acids into the cytosol, and their cell cytotoxicity are less than that of polyethylenimine (PEI) [14-29]. The polyplexes of poly(DMAEA/His (Boc)-OMe) phosphazene (PDHP)/DNA at the ratio of 10:1 (w:w) give a higher transfection efficiency and lower cytotoxicity against 293T cells than these of the histidine-free polymer and bPEI [18-23]. The poly (imidazole/2-dimethylaminoethylamino) phosphazenes/pDNA polyplexes enhance gene transfer activity and reduce cell cytotoxicity because of buffering capacity of imidazole groups in the endolysosomal pH range [24]. The pDNA and SiRNA polyplexes of β-cyclodextrin polycation modified with imidazole (CDPim) were almost 10 times more effective than these of CDP for tumor targeting and imaging [25,26,30]. After the chitosan amines (~37 mol%) are substituted with urocanic acid (UAC70) comprising an imidazole ring, the chitosan can improve its endosomal escape capacity [31]. When primary and secondary amines (39 mol%) were substituted with imidazolyl residues, the imidazolylated bPEI of 25 kDa and 750 kDa can increase the buffering effect, and the transfection efficiency against COS-1 and HEK293 cells was three times higher than bPEI [28]. The poly(1-vinylimidazole) partially alkylated with 1-bromobutane(PVIm-Bu) generates quaternary imidazole groups, and the transfection efficiency of PVImBu/DNA polyplexes against HepG2 cells was two orders of magnitude higher than with DNA/PVIm-NH2 polyplexes [29]. For above these reasons, the reducible or polymers with amino or imidazolyl residues should further enhance efficient gene delivery.
In this paper, dimeric poly (2-dimethylaminoethyl) methacrylate-block-polyvinylimidazole with single disulfide bond in the backbone (rDPDMAEMAIM) was synthesized by oxidizing their dithioester-terminated polymers. Compared with the dimeric reducible poly (2-dimethyl aminoethyl) methacrylate (rPDMAEMA)/pDNA polyplexes, the rPDMAEMAIM/DNA polyplexes have higher transfection activity and lower toxicity against 293T cells in vitro in the presence of serum. The rDPDMAEMAIM/pDNA and rDPDMAEMA/pDNA sizes at the weight ratio of 12:1 are in the ranges of 100 nm - 150 nm. The zeta potential of rDPDMAEMAIM/pDNA from 9.6 mV to 22.7 mV in PBS (phosphate buffered saline) solutions increases with their weight ratios of 1:1 to 18:1.
2. Materials and Methods
2.1. Materials
All reagents (analytical grade) were purchased from Shanghai Chemical Reagents Corp., 2,2’-azobis (isobutyronitrile) (AIBN, 98%), butylamine (Sigma-Aldrich, 99%), anhydrous methanol (99.8%), ethyl bromide (99%), chloroform (99%), diethyl ether(99%), triton X-100 (SigmaAldrich), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich), potassium xanthogenate (99%), dithiothreitol (DTT, Fluka, >99%), and hexane(98.5%) were used as received unless otherwise noted. 2-(dimethylamino) ethyl methacrylate (Sigma-Aldrich, 99%), and vinylimidazole (Sigma-Aldrich, 99%) was passed through a column of activated basic alumina to remove the inhibitor and stored at 4˚C prior to use. Tetrahydrofuran (THF, 99%) was treated with KOH and distilled twice from Na in the presence of benzophenone.
2.2. Synthesis of Block Polymers
Ethyl xanthogenate (EX) was prepared by reaction of ethyl bromide with potassium xanthogenate. The DTPDMAEMA was synthesized by polymerization of DMAEMA using ethyl xanthogenate as chain transfer reagent. A stock solution of DMEMA (2 g), AIBN (8 mg), and EX (160 mg) in Schlenk tubes of 1.5 mL THF was thoroughly deoxygenated by four consecutive freeze-pump thaw cycles, placed in a water bath at 60˚C for 48 h, to precipitate and isolate the DTPDMAEMA by hexane. Under the identical conditions, the vinylimida-zole (2 g), AIBN (8 mg), and the DTPDMAEMA in THF was used to synthesize the DTPDMAEMAIM. Its 1H NMR was analyzed by Varian Mercury Plus-400 NMR spectrometer (Varian, USA) using d4-methanol d-chloroform as solvent, and molecular weight relative to the calibrated peptide standards were determined by gel permeation chromatograph (GPC, Waters 600) with N, N-dimethylformamide (DMF) as eluent solvent at flow rate of 0.5 mL/ min.
2.3. Synthesis of Dimeric Polymers
A reducible polymer was prepared according to the modified procedure as reported [4]. The DTPDMAEMA or DTPDMAEMAIM (0.5 g) was dissolved in THF (4 mL), then the butylamine (0.6 mL) and dimethyl sulfoxide (0.4 mL) was added dropwise under nitrogen atmosphere at room temperature for 3 h, and stirred for 14 days. After removing the solvent, the rPDMAEMA or rPDMAEMAIM was obtained by precipitation into excess hexane.
2.4. Preparation of DNA Polyplexes
The pDNA was amplified in DH5α strain of E.coli and prepared by End Free Plasmid Mega Kit (Qiagen GmbH, Hilden, Germany). The transfection activities of polyplexes against 293T (Human embronic kidney), HEK293 and HeLa cells were evaluated by pDNA encoding green fluorescent protein expression (EGFP) and microscopically observed [32-34].
The mixture of rPDMAEMAIM and pDNA at differrent weight ratios were stirred dropwise in 30 mmol/L sodium acetate (pH 5.0), and placed at 50˚C for 30 min. all experiments were conducted in triplicate unless otherwise specified. To visualize effect of the condensed pDNA, the polyplexes were examined by electrophoresis using ethidium bromide staining at 110 V/cm in a TrisacetateEDTA buffer system (pH 8.0) for 45 min. The polyplexes of rPDMAEMA/DNA were prepared by the same process. All polyplexes dropped on silicon substrates were sprayed by gold, and observed on scanning electron microscope (LEO, 1530VP). Diluted in PBS, the polyplexes solution was measured by zeta potential analyzer (ZetaSizer 3000HSA Malvern, UK).
2.5. Cell Culture and Cytotoxicity
The 293T, HEK293, and Hela cells were from the Cell Bank, Chinese Academy of Sciences, and grown in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), streptomycine (40 μg/mL) and ampenicillin (40 U/mL).
The cell cytotoxicity of polymers was determined by MTT assays. The 20,000 cells/well in 150 μL DMEM were seeded to 96 well plates, and incubated in a 5% CO2 incubator at 37˚C for 24 h. Next, rPDMAEMAIM or rPDMAEMA in the range of 10 μg/mL to 50 μg/mL were added, and incubated for additional 16 h. Replaced by fresh DMEM, the MTT solution of 5 mg/mL (20 μL per well) was added, and cells were incubated for 4 h. After removing the medium, the crystals on living cells were dissolved by adding 150 μL DMSO. The cell viability was expressed by 570 nm absorbance using a microplate reader (ELX800, BIO-TEX Instrument, Inc.), and calculated as mean percentage relative to untreated cells ± SD.
2.6. In Vitro Transfection Assays
To assay the amount of GFP expression, the 2.5 μg/mL of pDNA to each well was used for different ratio of polyplexes weights, and the 2 × 105 cells per well were seeded in 24 well plate and incubated by DMEM containing 10% FBS for 24 h. After removing DMEM, the cells were washed three times with DMEM without FBS and antibiotics, and incubated by adding polyplexes for 4 h, and further cultured by fresh DMEM medium with 10% FBS for additional 36 h. The transfected and controlled cells per well were washed by PBS, and lysed by 400 mL buffer (0.1 M KH2PO4/K2HPO4 (pH 7.5), 0.2% Triton X-100, and 1 mM DTT) using four times freezethaw cycles. The 100-μL lysate per well was transferred into a 96-well plates, and measured with a microplate fluorescence reader (Tecan GENios) at 488/510 nm (exication/emission) [23].
3. Results and Discussion
3.1. Synthesis of rPDMAEMAIM
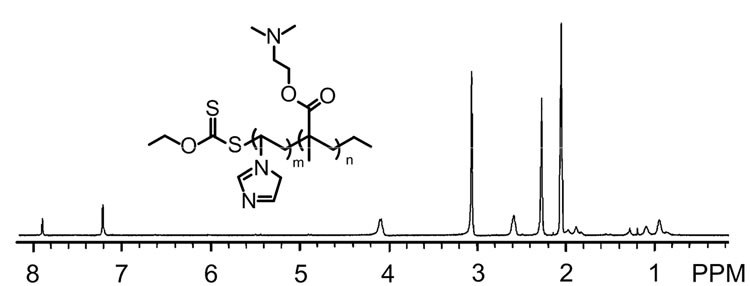
The biodegradable rPDMAEMA has significantly low cytotoxicity and the comparable transfection activity to non-reducible PDMAEMA [7,12], and the imidazolebased copolymers can improve transfection activity due to potent biocompatibility and protonation in the endo-lysosomal pH range [28,35,36]. In this study, the rPDMAEMAIM and rPDMAEMA were synthesized by RAFT polymerization using ethyl xanthogenate as chain transfer reagent (Figure 1). The dithioester-terminated DTPDMAEMAIM (δ = 1.1) was conducted by 1H NMR spectrum (Figure 1(a)). Further, the UV absorption of DTPDMAEMA and DTPDMAEMAIM at 270 nm confirmed the terminal dithioesters (Figure1 (b)). Coupled by disulfide bond, the rPDMAEMAIM and rPDMAEMA molecular weight were confirmed by GCP [7], the rPDMAEAM relative to peptide standards was 9.1 × 103; and rPDMAEMAIM, 1.12 × 104.
3.2. Polyplexes Characterization
To evaluate the ability of PDMAEMAIM to condense pDNA in 50 mM sodium acetate (pH 5.0), The PDMAEMAIM/pDNA at the weight ratios of 1:1 to 18:1 was conducted by gel retardation assays using ethidium bromide exclusion. Figure 2 shows that the PDMAEMAIM completely retarded pDNA at the ratio of 12:1 (w:w). Dulited by PBS solutions, the zeta potentials of 9.6 to 22.7 mV increases with weight ratios of 1:1 to 18:1, and is +16.2 mV at weight ratio of 12:1, indicating that the PDMAEMAIM/pDNA can induce membrane destabilization and promote cell uptake in endosome.
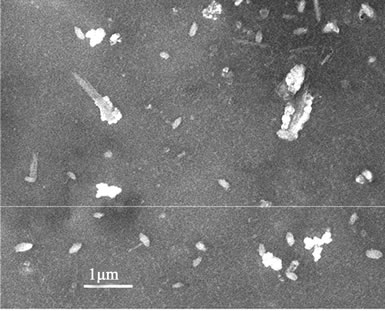
The polyplexes sizes affect the efficiency of gene delivery, and the nanoparticles of less than 150 nm are favorable to the endocytosis of many mammalian cells [24]. To observe the polyplexes sizes of rPDMAEMAIM/DNA, a mixture of the pDNA and rPDMAEMAIM at weight ratio of 12:1 were stirred by vortexing for 1 min, heated at 50˚C for 30 min, and dialyzed (12 kDa cutoff) extensively against the Milli-Q water for 24 h. The dialyzate was dropped on silicon substrate and air-dried. Also, polyplexes of rPDMAEA/pDNA was prepared under the same condition. SEM images show that the particles sizes of both polyplexes are in the range of 100 nm - 150 nm,
 (a)
(a)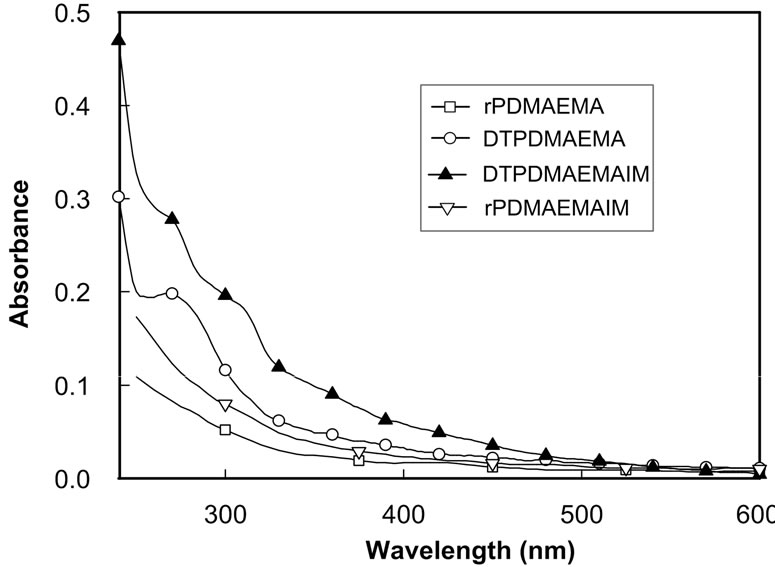 (b)
(b)
Figure 1. 1HNMR spectrum of DTPDMAEMAIM (a); UVVis absorption of DTPDMAEMAIM and DTPDMAEMA, rPDMAEMAIM, and rPDMAEMA (b).
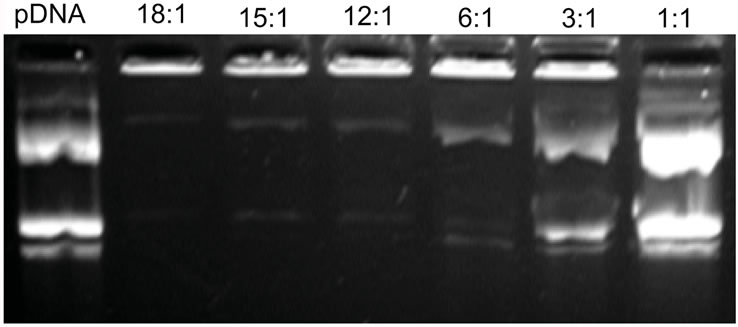
Figure 2. Gel retardation assay in a tris-acetate-EDTA buffer system (pH 8.0) at 110 V/cm for 45 min. rPDMAE MANIM/ pDNA at the different weight ratios of 1:1, 3:1, 6:1, 12:1, 15:1, and 18:1.
and the rPDMAEMA polyplexes had wire-like impurity. But non-dialyzate resulted in the strongly aggregation due to electrostatic attraction in buffer solution of 50 mM sodium acetate (Figure 3).
3.3. Cell Viability
The cell cytotoxicity of rPDMAEMAIM against 293T cells was measured by MTT assays. Figure 4 shows that the viability of 293T cells decreases with increasing polymer concentrations of 10 to 50 μg/mL, at 30 μg/mL, the viability was around 85% for rPDMAEMAIM; and 71%, for rPDMAEMA; but 30%, for PEI 25 kDa. This result shows that the single disulfide bond in the backbone was of intracellular degradation, and the imidazole moiety in rPDMAEMAIM can further enhance cell viability, but the PEI 25 kDa resulted in higher cytotoxicity against HEK-293 cells. Similarily, both polymers reached such viability against HEK293 cells, but over 85% against Hela cells (data not shown).
3.4. Transfection Activity in Virto
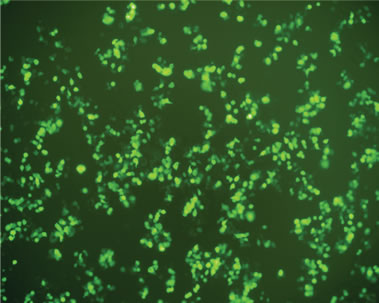
Transfection activity against 293T cells in vitro of rPDMAEMAIM polyplexes was performed by EGFP. At ratio of 12:1 (w:w), the fluorescent images show that the GFP expression of rPDMAEMAIM polyplexes against 293T cells was much higher than that of rPDMAEMA (Figures 5(a) and (b)). As a positive control, the inferior transfection activity of the PEI/pDNA could be due to caused by their lower cell viability (Figure 5(c)). As a control, naked pDNA in the cytoplasm of 293T cells showed only neglected fluorescence relative to background intensity, indicating that pDNA without any vector had very low transfection activity. (Figure 5(d)). The EGFP expressions of rPDMAEMAIM and rPDMAEMA polyplexes against Hela and HEK293 cells was lower than against 293T cells, indicating that the 293T cells were easy-to-transfect activity, and both transfection activities are related with cell types.
The transfection activity of rPDMAEMAIM/pDNA or rPDMAEMA/pDNA in vitro at different weight ratios was quantitatively analyzed by the total lysate of 293T cells. Because imidazole groups were further introduced by RAFT polymerization, the molecular weight of rPDMAEMAIM is more than rPDMAEMA. Here, the rPDMAEMAIM and rPDMAEMA polyplexes are prepared by different weight ratios of vector and pDNA. Figure 6 shows that GFP expression level of rPDMAEMAIM polyplexes was higher than that of rPDMAEMA at any given ratios, and reached maximum at weight ratio of 12:1, it further confirmed that the imidazole groups and redox-sensitive difulfide bonds in polymer can conduce to polyplexes transfection and improve cell viability. The lower GFP expression level of PEI 25k/pDNA was caused by cell cytotoxicity.
 (a)
(a)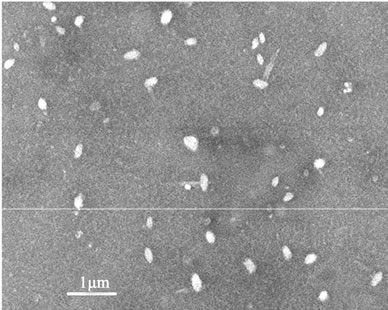 (b)
(b)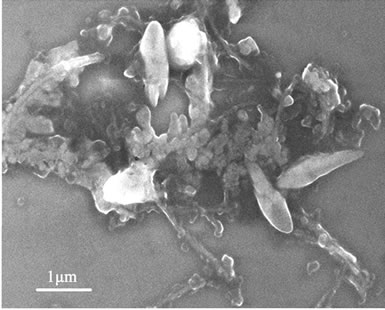 (c)
(c)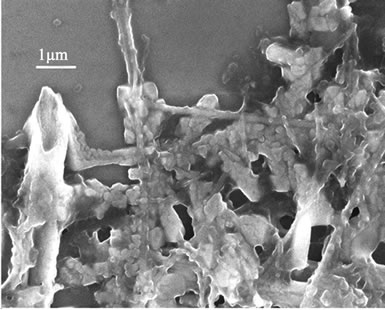 (d)
(d)
Figure 3. SEM images of dialyzate (a, b) and non-dialyzate (c, d); rPDMAEMA/DNA (a, c) and rPDMAEMAIM/DNA (b, d) at pH 7.0.
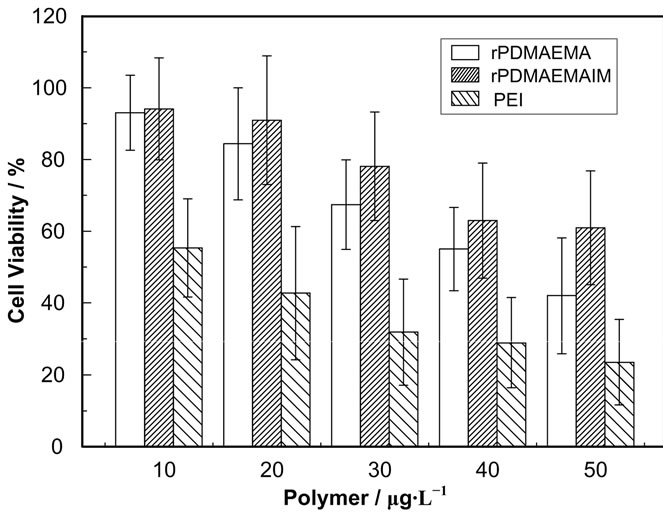
Figure 4. Cell cytotoxicity of rPDMAEMAIM, rPDMAEMA, and PEI 25 kDa in 293T cells, the cells density of 20,000 cells/well were seeded in 96-well plates and incubated for 24 h. The absorbance was conducted using a microplate reader at 570 nm (n = 4 ± SD).
 (a)
(a)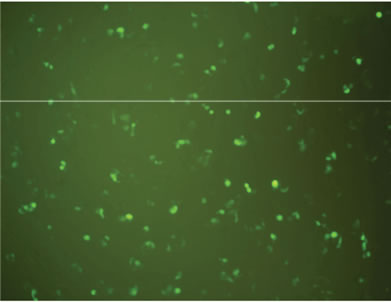 (b)
(b)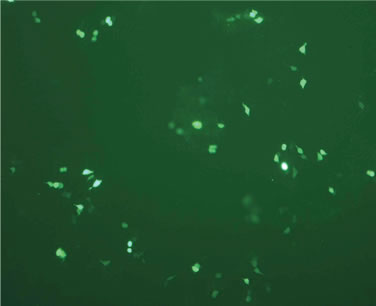 (c)
(c)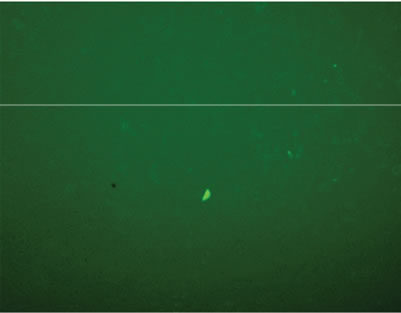 (d)
(d)
Figure 5. Fluorescent images of 293T cells transfected with polyplexes of polymer/DNA at weight ratio of 12:1, (a) rPDMAEMAIM/DNA; (b) rPDMAEMA/DNA; and (c) PEI 25 kDa/DNA; (d) Control. All transfection experiments were performed in DMEM supplemented with 10% FBS.
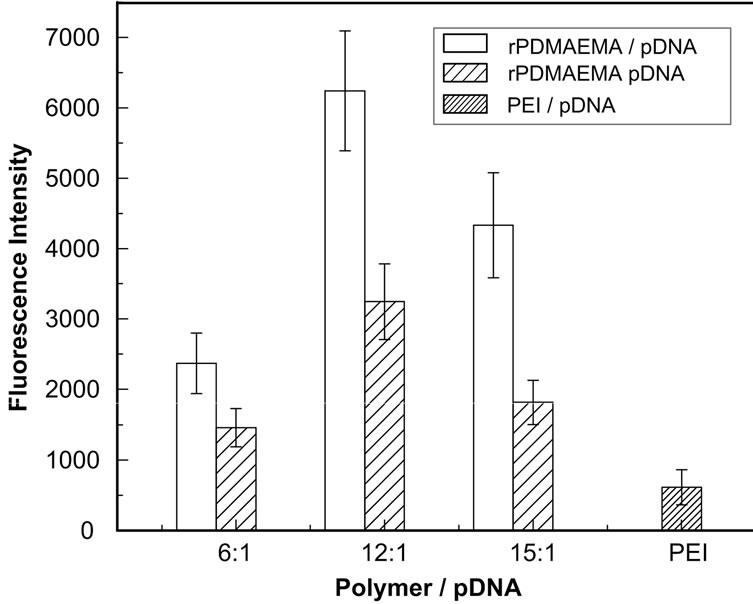
Figure 6. GFP expression of 293T cells transfected with polymer polyplexes at ratio of 12:1 (w/w). The cells were transfected by polyplexes at 37˚C C in 5% CO2 for 4 h, and incubated by fresh DMEM medium with 10% FBS for additional 36 h. GFP expression was quantified by fluorescent intensity of lysed solutions (n = 3 ± SD).
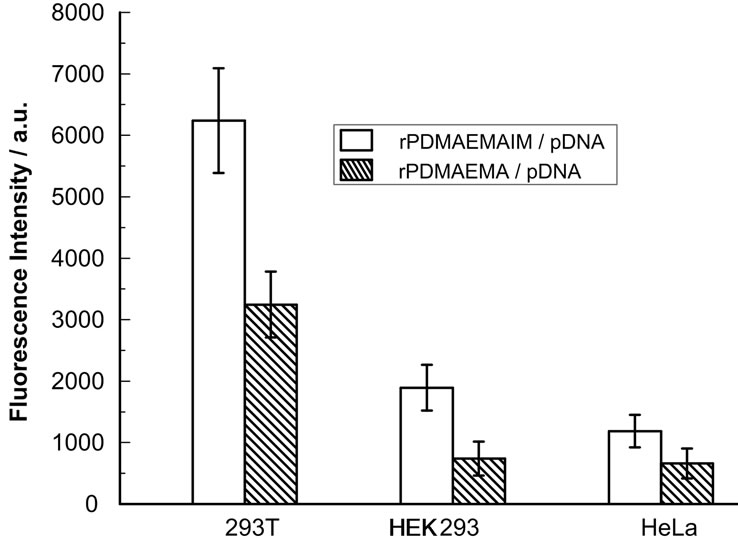
Figure 7. GFP expression of 293 T, HEK293 and HeLa cells transfected with polymer polyplexes at ratio of 12:1 (w/w). The cells were transfected by polyplexes at 37˚C in 5% CO2 for 4 h, and incubated by fresh DMEM medium with 10% FBS for additional 36 h. GFP expression was quantified by fluorescent intensity of lysed solutions (n = 3 ± SD).
Although 293T and HEK293 cells are isogenous, the transfection efficiency of rPDMAEMAIM and rPMAEMA polyplexes against 293T cells was higher than against HEK293 cells (Figure 7), and against HeLa cells was further lower. It is concluded that the transfection activity of rPDMAEMAIM and rPDMAEMAIM could be relative to following reasons: 1) Cell types, both HEK- 293 and 293T cells were from human embryonic kidney epithelial cell lines, but 293T cells were easily transfected by rPDMAEMA or rPDMAEMAIM; 2) The protonated effect. The imidazole in rPDMAEMAIM could induce destabilization of cell membranes to deliver pDNA into the cytosol, and make their polyplexes escape from endosome timely [24]; 3) The difference of cytotoxicity. The free rPDMAEMAIM display much higher cell viability on 293T, HEK293 and Hela cells than rPDMAEMA; 4) The ability of polyplexes to condense DNA. A feasible molecular weight would be an important in gene delivery, for example, the transfection activity of thiolated chitosan was better than that of crossed-chitosan by disulfide bonds [29], the transfection efficiency of rPDMAEMA did not increase with amounts of oligomers [7]; 5) Effect of molecular weights, the rPDMAEMAIM polyplexes have higher transfection than that of rPDMAEMA, and TPDMAEMAIM and TPDMEAM polyplexes are of lower transfection than their reducible dimmers (not shown data), resulting in that amounts of pDNA per polyplex increase with molecular weights.
4. Conclusion
The rPDMAEMAIM was successfully prepared by RAFT polymerization and oxidation. Both rPDMAEMAIM and rPDMAEMA can condense pDNA into polyplexes with lower 150 nm sizes. The rPDMAEMAIM polyplexes reach better transfection efficiency and lower cytotoxicity against 293T than the rPDMAEMA. The rPDMAEMAIM should be a promising cationic polymer in gene delivery.
REFERENCES
- D. S. Manickam and D. Oupický, “Polyplex gene delivery modulated by redox potential gradients,” Journal of Drug Targeting, Vol. 14, No. 8, 2006, pp. 519-526. doi:10.1080/10611860600834409
- K. Miyata, Y. Kakizawa, N. Nishiyama, A. Harada, Y. Yamasaki, H. Koyama and K. Kataoka, “Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression,” Journal of the American Chemical Society, Vol. 126, No. 8, 2004, pp. 2355-2361. doi:10.1021/ja0379666
- D. L. McKenzie, K.Y. Kwok and K. G. Rice, “A potent new class of reductively activated peptide gene delivery agents,” Journal of Biological Chemistry, Vol. 275, No. 14, 2000, 9970-9977. doi:10.1074/jbc.275.14.9970
- M. L. Read, K. H. Bremner, D. Oupicky, N. K. Green, P. F. Searle and L. W. Seymour, “Vectors based on reducible polycations facilitate intracellular release of nucleic acids,” The Journal of Gene Medicine, Vol. 5, No. 3, 2003, pp. 232-245. doi:10.1002/jgm.331
- M. Oishi, T. Hayama, Y. Akiyama, S. Takae, A. Harada, Y. Yamasaki, F. Nagatsugi, S. Sasaki, Y. Nagasaki and K. Kataoka, “Supramolecular assemblies for the cytoplasmic delivery of antisense oligodeoxynucleotide: polyion complex (PIC) micelles based on poly(ethylene glycol)- SS-oligodeoxynucleotide conjugate,” Biomacromolecules, Vol. 6, No. 5, 2005, pp. 2449-2454. doi:10.1021/bm050370l
- D. S. Manickam and D. Oupicky, “Multiblock reducible copolypeptides containing histidine-rich and nuclear localization sequences for gene delivery,” Bioconjugate Chemistry, Vol. 17, No. 6, 2006, 1395-1403. doi:10.1021/bc060104k
- Y. Z. You, D. S. Manickam, Q. H. Zhou and D. Oupický, “Reducible poly(2-dimethylaminoethyl methacrylate): Synthesis, cytotoxicity, and gene delivery activity,” Journal of Controlled Release, Vol. 122, No. 3, 2007, pp. 217-225. doi:10.1016/j.jconrel.2007.04.020
- Y. Lee, H. Mo, H. Koo, J. Y. Park, M. Y. Cho, G. W. Jin and J. S. Park, “Visualization of the degradation of a disulfide polymer, linear poly(ethylenimine sulfide), for gene delivery,” Bioconjugate Chemistry, Vol. 18, No. 1, 2007, pp. 13-18. doi:10.1021/bc060113t
- J. Kloeckner, E. Wagner and M. Ogris, “Degradable gene carriers based on oligomerized polyamines,” European Journal of Pharmaceutical Sciences, Vol. 29, No. 5, 2006, pp. 414-425. doi:10.1016/j.ejps.2006.08.002
- C. Lin, Z. Zhong, M. C. Lok, X. Jiang, W. E. Hennink, J. Feijen and J. F. J. Engbersen, “Novel bioreducible poly (Amido amine)s for highly efficient gene delivery,” Bioconjugate Chemistry, Vol. 18, No. 1, 2007, pp. 138- 145. doi:10.1021/bc060200l
- L. V. Christensen, C. W. Chang, W. J. Kim, S. W. Kim, Z. Zhong, C. Lin, J. F. J. Engbersen and J. Feijen, “Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery,” Bioconjugate Chemistry, Vol. 17, No. 5, 2006, pp. 1233-1240. doi:10.1021/bc0602026
- A. M. Funhoff, C. F. van Nostrum, M. C. Lok, J. A. Kruijtzer, D. J. Crommelin and W. E. Hennink, “Cationic polymethacrylates with covalently linked membrane destabilizing peptides as gene delivery vectors,” Journal of Controlled Release, Vol. 101, No. 1-3, 2005, pp. 233-246. doi:10.1016/j.jconrel.2004.06.023
- R. A. Jones, M. H. Poniris and M. R. Wilson, “pDMAEMA is ransferring by endocytosis but does not physically disrupt endosomes,” Journal of Controlled Release, Vol. 96, No. 3, 2004, pp. 379-391. doi:10.1016/j.jconrel.2004.02.011
- M. Zenke, P. Steinlein, E. Wagner, M. Cotton, H. Beug and M. L. Birnstiel, “Receptor-mediated endocytosis of ransferring-polycation conjugates: an efficient way to introduce DNA into hematopoietic cells,” Proceedings of the National Academy of Sciences, Vol. 87, No. 10, 1990, pp. 3655-3659. doi:10.1073/pnas.87.10.3655
- P. Midoux, C. Mendes, A. Legrand, J. Raimond, R. Mayer, M. Monsigny and A. C. Roche, “Specific gene transfer mediated by lactosylated poly-L-lysine into hepatoma Cells,” Nucleic Acids Research, Vol. 21, No. 4, 1993, pp. 871-878. doi:10.1093/nar/21.4.871
- O. Boussif, F. Lezoualc’h, M. A. Zanta, M. D.Mergny, D. Scherman, B. Demeneix and J. P. Behr, “A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine,” Proceedings of the National Academy of Sciences, Vol. 92, No. 16, 1995, pp. 7297-7301. doi:10.1073/pnas.92.16.7297
- B. Demeneix and J. P. Behr, “Polyethylenimine (PEI),” Advances in Genetics, Vol. 53, 2005, pp. 215-230. doi:10.1016/S0065-2660(05)53008-6
- Y. Yang, Z. Xu, S. Chen, Y. Gao, W. Gu, L. Chen, Y. Pei and Y. Li, “Histidylated cationic polyorganophosphazene/ DNA self-assembled nanoparticles for gene delivery,” International Journal of Pharmaceutics, Vol. 353, No. 1- 2, 2008, pp. 277-282.
- Q. R. Chen, L. Zhang, S. A. Stass and A. J. Mixson, “Copolymer of histidine and lysine markedly enhances transfection efficiency of liposomes,” Gene Therapy, Vol. 7, No. 19, 2000, pp. 1698-1705. doi:10.1038/sj.gt.3301294
- Q. R. Chen, L. Zhang, S. A. Stass and A. J. Mixson, “Branched co-polymers of histidine and lysine are efficient carriers of plasmids,” Nucleic Acids Research, Vol. 29, No. 6, 2001, pp. 1334-1340. doi:10.1093/nar/29.6.1334
- Q. R. Chen, L. Zhang, P. W. Luther and A. J. Mixson, “Optimal transfection with the HK polymer depends on its degree of branching and the pH of endocytic vesicles,” Nucleic Acids Research, Vol. 30, No. 6, 2002, pp. 1338- 1345. doi:10.1093/nar/30.6.1338
- Q. Leng and A. J. Mixson, “Modified branched peptides with a histidine-rich tail enhance in vitro gene transfection,” Nucleic Acids Research, Vol. 33, No. 4, 2005, p.e40. doi:10.1093/nar/gni040
- Q. Leng, P. Scaria, P. Lu, M. C. Woodle and A. J. Mixson, “Systemic delivery of HK Raf-1 siRNA polyplexes inhibits MDA-MB-435 xenografts Tumor xenograft reduction by HK: siRNA polyplexes,” Cancer Gene Therapy, Vol. 15, 2008, pp. 485-495. doi:10.1038/cgt.2008.29
- Y. Yang, Z. Xu, J. Jiang, Y. Gao, W. Gu, L. Chen, X. Tang and Y. Li, “Poly(imidazole/DMAEA) phosphazene/DNA self-assembled nanoparticles for gene delivery: Synthesis and in vitro transfection,” Journal of Controlled Release, Vol. 127, No. 3, 2008, pp. 273-279. doi:10.1016/j.jconrel.2008.01.012
- D. W. Bartlett, H. Su, I. J. Hildebrandt, W. A. Weber and M. E. Davis, “Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging,” Proceedings of the National Academy of Sciences, Vol. 104, No. 39, 2007, pp. 15549-15554. doi:10.1073/pnas.0707461104
- J. D. Heidel, Z. Yu, J. Y. Liu, S. M. Rele, Y. Liang, R. K. Zeidan, D. J. Kornbrust and M. E. Davis, “Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA,” Proceedings of the National Academy of Sciences, Vol. 104, No. 14, 2007, pp. 5715- 5721. doi:10.1073/pnas.0701458104
- T. H. Kim, J. E. Ihm, Y. J. Choi, J. W. Nah and C. S. Cho, “Efficient gene delivery by urocanic acid-modified chitosan,” Journal of Controlled Release, Vol. 93, No. 3, 2003, pp. 389-402. doi:10.1016/j.jconrel.2003.08.017
- A. Swami, A. Aggarwal, A. Pathak, S. Patnaik, P. Kumar, Y. Singh and K. C. Gupta, “Imidazolyl-PEI modified nanoparticles for enhanced gene delivery,” International Journal of Pharmaceutics, Vol. 335, No. 1-2, 2007, pp. 180-192.
- T. Hakamatani, S. Asayama and H. Kawakami, “Synthesis of alkylated Poly(1-vinylimidazole) for a new pHsensitive DNA carrier,” Nucleic Acids Symposium Series, Vol. 52, No. 1, 2008, pp. 677-678. doi:10.1093/nass/nrn342
- D. W. Bartlett and M. E. Davis, “Physicochemical and biological characterization of targeted, nucleic acidcontaining nanoparticles,” Bioconjugate Chemistry, Vol. 18, No. 2, 2007, pp. 456-468. doi:10.1021/bc0603539
- H. Jin, C. X. Xu, H. W. Kim, Y. S. Chung, J. Y. Shin, S. H. Chang, S. J. Park, E. S. Lee, S. K. Hwang, J. T. Kwon, A. Minai-Tehrani, M. Woo, M. S. Noh, H. J. Youn, D. Y. Kim, B. I. Yoon, K. H. Lee, T. H. Kim, C. S. Cho and M. H. Cho, “Urocanic acid-modified chitosan-mediated PTEN delivery via aerosol suppressed lung tumorigenesis in K-rasLA1 miceAerosol delivery of PTEN for lung Cancer Treatment,” Cancer Gene Therapy, Vol. 15, 2008, pp. 275-283. doi:10.1038/sj.cgt.7701116
- H. T. Wu, C. S. Lin and M. C. Huang, “In vitro and ex vivo green fluorescent protein expression in alveolar mammary epithelial cells and mammary glands driven by the distal 5'-regulative sequence and intron 1 of the goat β-casein gene,” Reproduction, Fertility and Development, Vol. 15, No. 4, 2003, pp. 231-239. doi:10.1071/RD01050
- Y. Gao, W. W. Gu, L. L. Chen, Z. H. Xu and Y. P. Li, “A multifunctional nanodevice as non-viral vector for gene delivery: in vitro characteristics and transfection,” Journal of Controlled Release, Vol. 118, No. 3, 2007, pp. 381- 388. doi:10.1016/j.jconrel.2007.01.006
- J. Hoon Jeong, L. V. Christensen, J. W. Yockman, Z. Zhong, J. F. Engbersen, W. J. Kim, J. Feijen and S. WanKim, “Reducible poly(amido ethylenimine) directed to enhance RNA interference,” Biomaterials, Vol. 28, No. 10, 2007, pp. 1912-1917. doi:10.1016/j.biomaterials.2006.12.019
- S. Mishra, J. D. Heidel, P. Webster and M. E. Davis, “Imidazole groups on a linear, cyclodextrin-containing polycation produce enhanced gene delivery via multiple processes,” Journal of Controlled Release, Vol. 116, No. 2, 2006, pp. 179-191. doi:10.1016/j.jconrel.2006.06.018
- J. S. Park, T. H. Han, K. Y. Lee, S. S. Han, J. J. Hwang, D. H. Moon, S. Y. Kim and Y. W. Cho, “N-acetyl histidine-conjugated glycol chitosan self-assembled nanoparticles for intracytoplasmic delivery of drugs: endocytosis, exocytosis and drug release,” Journal of Controlled Release, Vol. 115, No. 1, 2006, pp. 37-45. doi:10.1016/j.jconrel.2006.07.011

