American Journal of Plant Sciences
Vol.5 No.1(2014), Article ID:41947,6 pages DOI:10.4236/ajps.2014.51016
Karyotype Analysis of Ocimum basilicum in South-Eastern Nigeria
1Department of Crop Science, University of Calabar, Calabar, Nigeria; 2Department of Genetics & Biotechnology, University of Calabar, Calabar, Nigeria.
Email: paikpokpodion@yahoo.com
Copyright © 2014 Offiong Ukpong Edet, Peter Osobase Aikpokpodion. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Offiong Ukpong Edet, Peter Osobase Aikpokpodion. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received October 15th, 2013; revised December 18th, 2013; accepted January 3rd, 2014
KEYWORDS
Karyotype; Cytogenetics; Cytotype; Ideogram; Mitotic; Polyploidy
ABSTRACT
Ocimum basilicum is widely distributed in the tropical and subtropical regions of the world, with greatest variability in Africa and India. It is valued in many countries for its culinary, medicinal, industrial and religious importance. Although cytogenetic entries on the plant have been made in other geographical locations of the world, in Nigeria, such entries, prior to this report, have been limited if not completely unavailable. In this analysis, axillary buds, obtained from growing plants, were used to conduct mitotic study. Results from this study showed chromosome counts of 2n = 48 and 60, thus bringing to light the existence of chromosome number variation and the possibility of polyploidy at different levels in the plant species in this agro-ecological zone. This research has, therefore, established that at least there are two cytotypes in the population of Ocimum basilicum growing in the humid forest vegetation zone of Nigeria. Analysis of the two cytotypes revealed asymmetrical karyotypes, indicative of advancement in the evolutionary trend of the plant species.
1. Introduction
Ocimum basilicum, known commonly as sweet basil, is an important aromatic plant of the Lamiaceae family, which has about 252 genera and 6700 species, most of which are medicinal [1,2]. The genus, Ocimum, is cosmopolitan with its distribution spanning from the Mediterranean to Central Asia, with greatest variability in Africa and India. Among more than 150 species of Ocimum, sweet basil is the major essential oil crop commercially cultivated in many countries [3]. It is found in tropical Asia, Africa, America and subtropical regions of the world, from sea level to an altitude of about 1500 m [4,5].
Ocimum basilicum thrives in full sunshine, requiring 0.6 m - 4.2 m rainfall, soil pH of 4.3 to 8.2 and well drained sandy loam soils [6,7]. Clayey soils and waterlogged areas are unsuitable for the cultivation of the plant. It is susceptible to frost and the crop growth can be adversely affected in areas which experience heavy and continuous rainfall [7]. Sweet basil is an herbaceous plant with plant height ranging between 0.3 m and 1 m. The leaves are ovate, often puckered; flower pink or white, with fruits having four small nutlets which are mucilaginous when wet [4].
Elaborate scientific investigations, ranging from phytochemistry, pathogenic and insecticidal activities, utilization as a spice and flavouring ingredient etc. have been extensively catalogued for Ocimum basilicum. Reviews of the phytochemical and pharmacological studies on the plant indicate that it possesses analgesic, anti-inflammatory, antimicrobial, antioxidant, antiulcerogenic, cardiac stimulant, chemomodulatory, hepatoprotective, hypoglycemic, hypolipidemic, immunomodulatory and larvicidal activies [8-17]. Cardioprotective activity of ethanolic extract of sweet basil against isoproterenol-induced myocardial infarction has also been recently demonstrated [18]. Reports of cytogenetic studies have shown that polyploidy and chromosome number variations are common among Ocimum species. However, two basic chromosome numbers (x = 8 and x = 12) have been reported, on the basis of which the various species of the genus have been classified into two groups: basilcum group (x = 12) and sanctum group (x = 8) [19,20]. This cytogenetic variability presents a veritable resource to geneticists and plant breeders to develop plant varieties well adapted to specific agro-ecological zones in response to the ever increasing demand for this all-important plant species.
However, in Nigeria, where there exists a great genetic diversity of Ocimum species, recognized and utilized for various purposes by the locals, information regarding their cytogenetic features had, prior to this work, been limited or completely unavailable. This paper, therefore, reports on the cytogenetic characteristics (karyotype) of Ocimum basilicum accessions in the humid forest vegetation zone of Nigeria.
2. Materials and Methods
2.1. Plant Materials
Seeds of O. basilicum were obtained from different locations in Cross River State, South-eastern region of Nigeria (Akpabuyo-KTA, Calabar South-ANA, Calabar Municipality-MUN and Akamkpa-AKP) and raised in polybags in the experimental farm of the University of Calabar, Nigeria between the months of February and March, 2010. Table 1 provides geographic information of specific locations where plant materials were collected.
2.2. Karyotype Study
Axillary buds were obtained from the growing plants and pretreated in 8-hydroxyquinoline (0.002M), fixed in carnoy’s solution and hydrolyzed in 1N hydrochloric acid at 60˚C. The hydrolyzed materials were rinsed in 70% alcohol and stained with FLP orcein before squashing. Prepared slides were viewed under a light microscope digitized with a Chinese made power shot A630 Canon camera (80 mega pixel, Canon PC 1201, NO 4126202101) and photographs of good cells were taken. Chromosomes were measured using R1370-19 ocular micrometer calibrated with OB2001 objective micrometer at 0.77 per unit. Ideograms of the species were accordingly constructed. Description of chromosomes was based on size, arm length, haploid set length (HSL), relative length, symmetry and form [21-23].
3. Results
The mitotic chromosome numbers, haploid set length, total long arm, total short arm, and karyotype formulae are presented in Table 2, while Table 3 shows the ka-
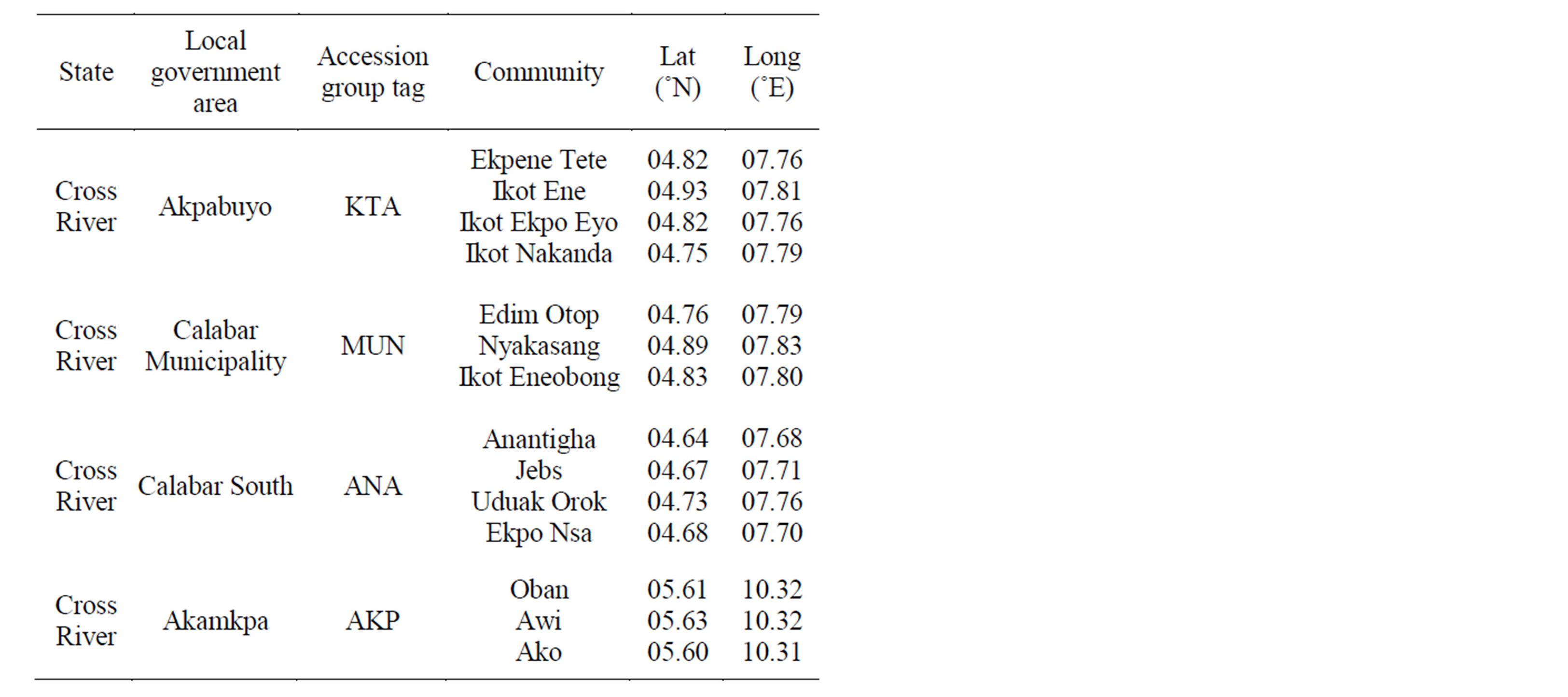
Table 1. Geographic information of locations of plant collection.

Table 2. Chromosome numbers, haploid set length, total long arm, total short arm and karyotype formulae of O. basilicum.
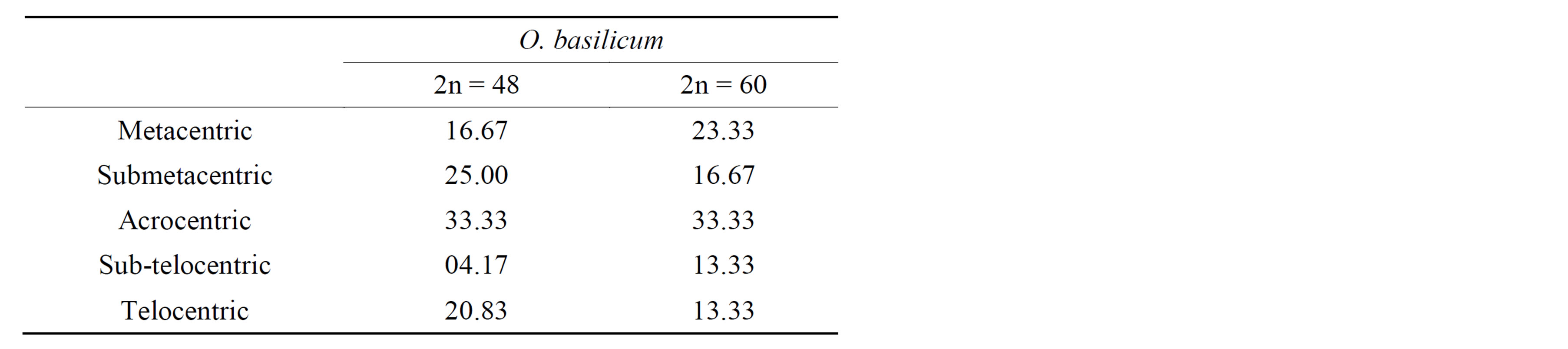
Table 3. Karyotype form (%).
ryotype form (%) of the species.
3.1. Karyotype Details of O. basilicum
Two chromosome numbers, 2n = 48 and 60 (Plates 1 and 2), were obtained in the four accession groups. The accessions with 24 pairs (2n = 48) of chromosomes (KTA, ANA and AKP) revealed a karyotype formula of 4m + 6sm + 8a + 1st + 5t. The mean total length of the chromosomes was 3.72 µm, while the means of the short and long arms were 1.23 µm and 2.49 µm, respectively (Table 4). The chromosomes ranged in size from 1.54 µm (chromosome 24) to 5.4 µm (chromosome 1), 16.67% of which were metacentric. The longest chromosome occupied 6.03% of the genome, while the shortest occupied 1.72% of the genome (Figure 1).
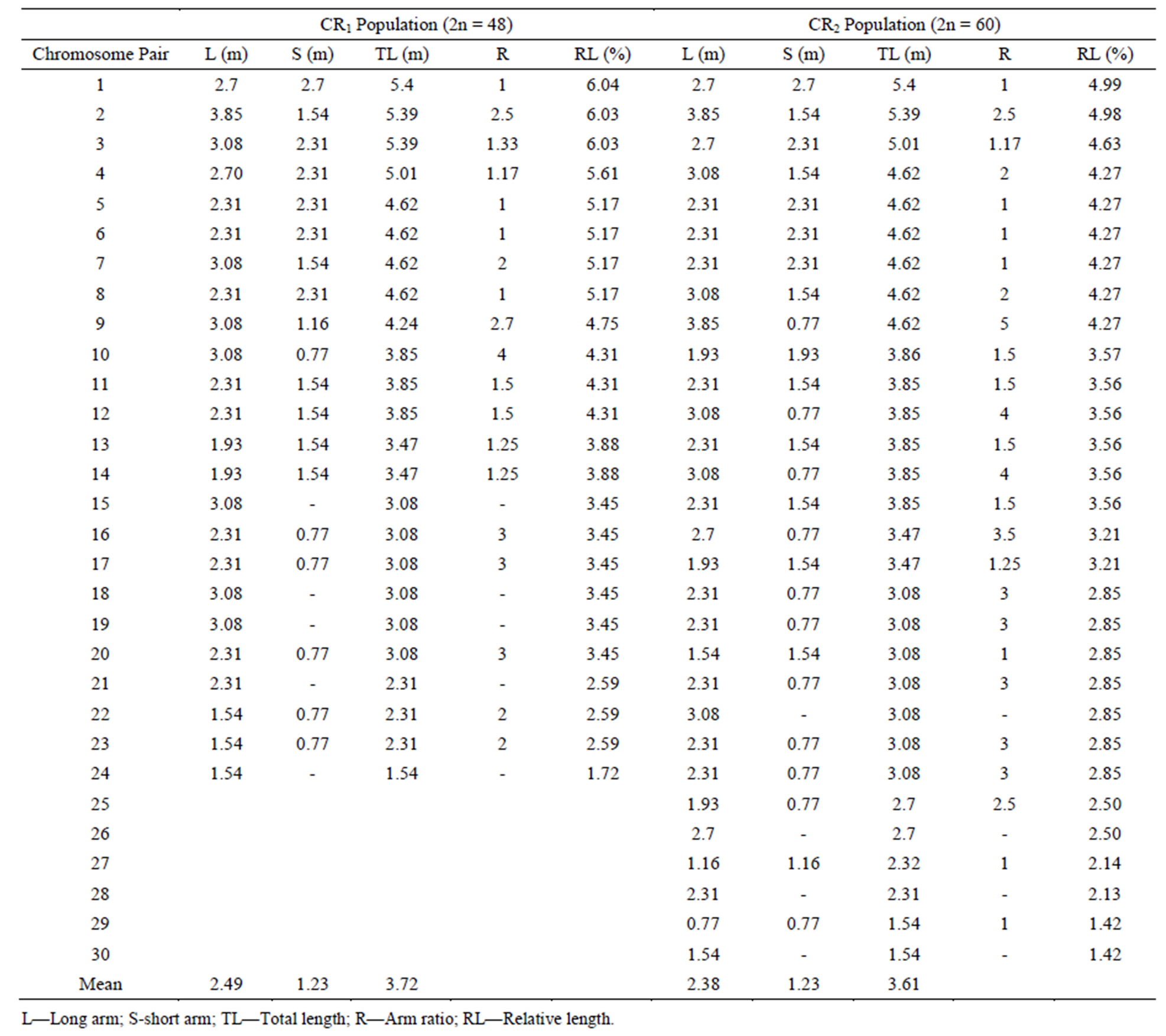
Table 4. Karyotype details of O. basilicum.
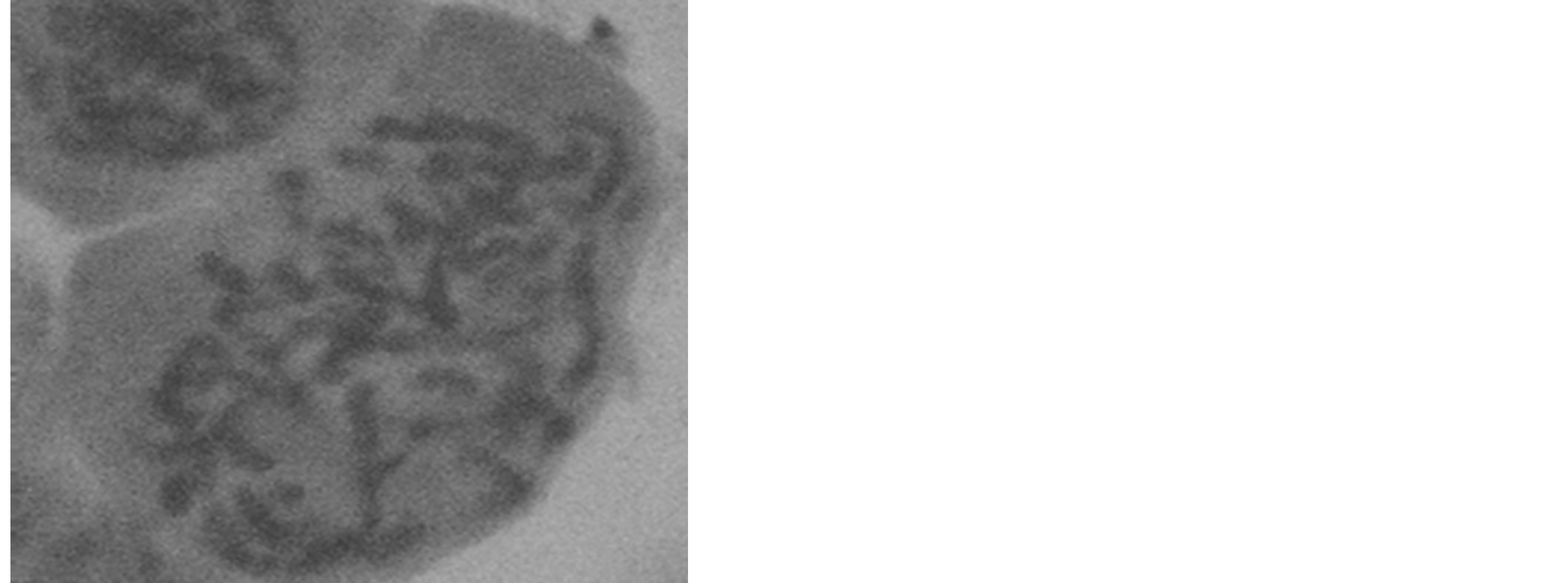
Plate 1. Somatic metaphase chromosomes of Ocimumbasilicum (CR1 population, 2n = 48, Magnification 7600×).
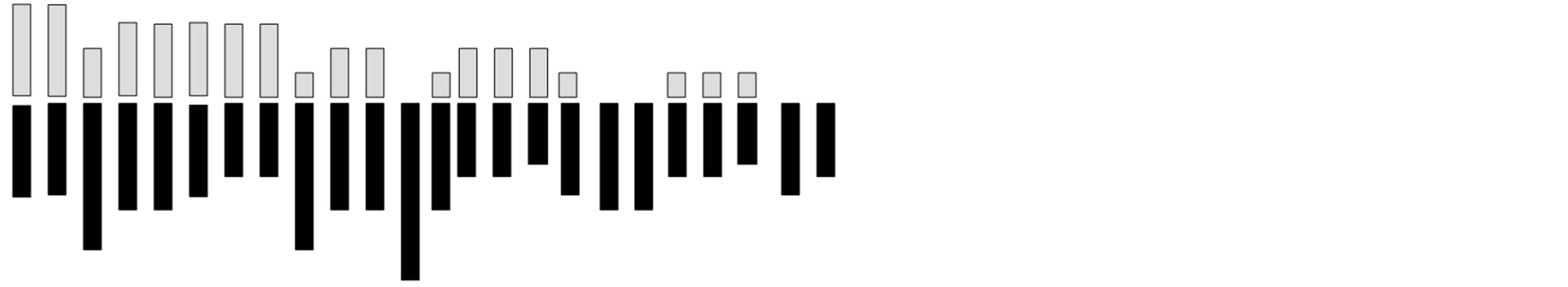
Figure 1. Idiogram of Ocimumbasilicum (CR1 population, 2n = 48).
On the other hand, 30 pairs (2n = 60) of chromosomes were recorded in the MUN accession group, with 23.33% of metacentric chromosomes. Chromosomes of this group ranged from 1.54 µm (chromosome 30) to 5.4 µm (chromosome 1) in size, with a total mean length of 3.61 µm, mean short arm of 1.23 µm and mean long arm of 2.38 µm (Table 4). The population revealed a karyotype formula of 7m + 5sm + 10a + 4st + 4t, with 1.42% of the genome being occupied by the shortest chromosome, whereas the longest chromosome occupied 4.98% of the genome (Table 4, Figure 2).
4. Discussion
4.1. Variable Chromosome Numbers and Polyploidization in Ocimum basilicum
The chromosome counts of 2n = 48 and 60 recorded in this research groups the evaluated accessions into two cytotypes: group I—KTA, ANA and AKP (2n = 48) and group II—MUN (2n = 60). Chromosome number, 2n = 48 obtained in this work for this plant species agrees with earlier reports which posited that O. basilicum occurs in nature as a tetraploid species with chromosome count of 2n = 4x = 48 [7,24]. However, 2n = 60 in this plant species is a new number obtained in this study for the species. Other research results had shown variable chromosome numbers for this species: 2n = 48, 52 and 72 [7,25]. Based on the basic number of 12 for O. basilicum [20,24], it can be suggested that polyploidization, occurring at different levels, is a feature of this plant species. Hence, 2n = 48 and 60 indicate tetraploid and pentaploid, respectively.
The importance of this study, therefore, is that in the O. basilicum population in South-eastern Nigeria, at least one new cytotype (2n = 60) has evolved. This is an indication of the occurrence of polyploidy and new cytotypes in this species.
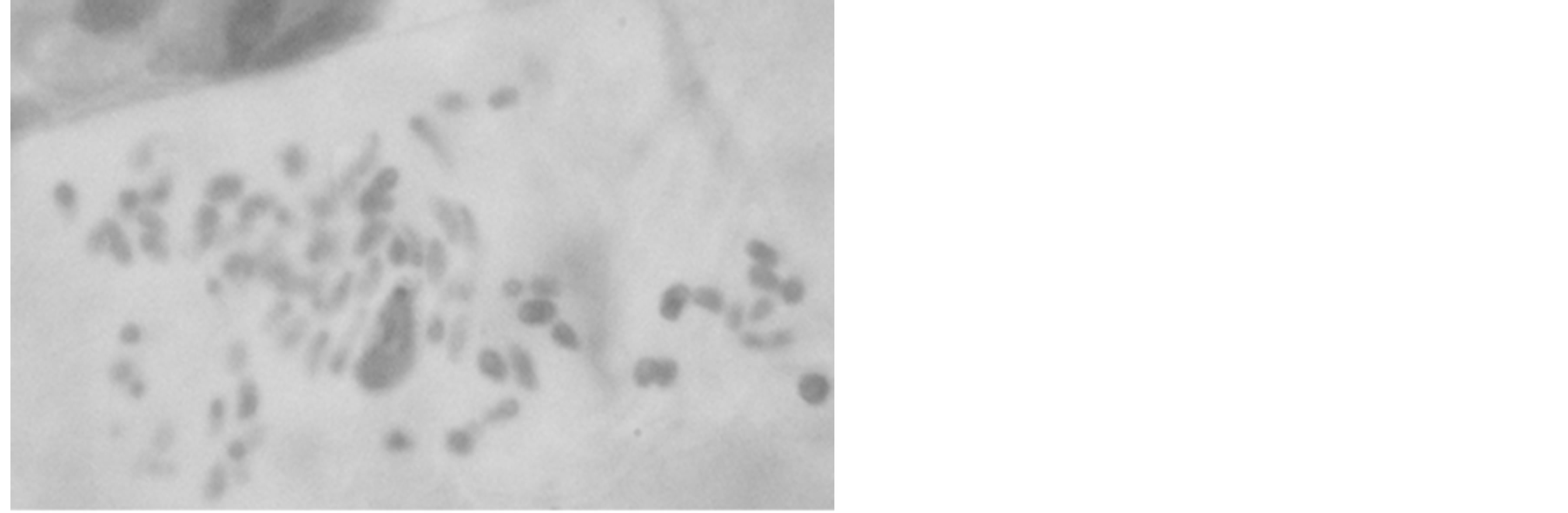
Plate 2. Somatic metaphase chromosomes of Ocimumbasilicum (CR2 population, 2n = 60, Magnification 7300×).
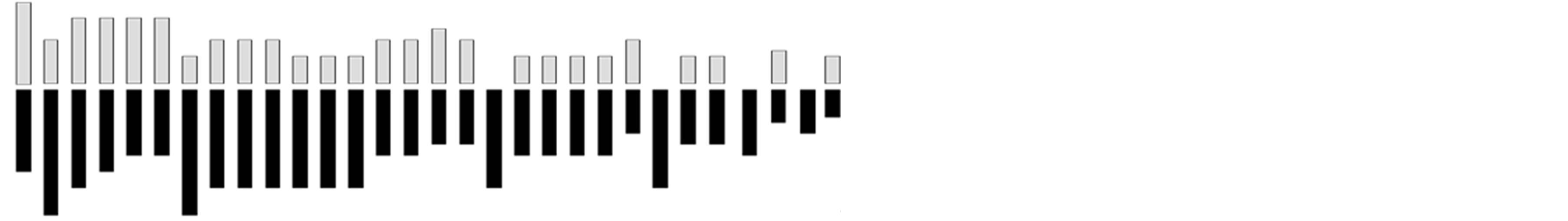
Figure 2. Idiogram of Ocimumbasilicum (CR2 Population, 2n = 60).
The absence of diploid species in this species may be attributed to the observable fluctuations in the climatic conditions of this humid ecological zone, since polyploids are at a selective advantage in habitats which have been subjected to frequent changes in climatic and edaphic factors [26].
4.2. Karyotype Symmetry and Evolution
From the karyotype details obtained from this study, the plant species has asymmetrical karyotype. Asymmetrical karyotypes as those possessing many chromosomes with subterminal centromeres or greater differences in size between the largest and smallest chromosomes [26]. The karyotypes showed higher proportions of acrocentric chromosomes as compared to metacentric chromosomes and by implication cannot be said to be primitive. Chromosomes become more asymmetrical as evolution progresses, moving from submetacentric state towards acrocentric state in extreme cases [27]. However, a reversal of this trend occurs periodically, giving rise to a reduction in chromosome number [26]. This is possible because at the peak of asymmetry, all the chromosomes can be acrocentric and/or telocentric. Hence, two telocentric chromosomes can fuse (centric fusion) to form large metacentric or submetacentric chromosomes, resulting in a reduced number of chromosomes in a species. Although variable chromosome numbers have been earlier on reported for this species (2n = 48, 52 and 72), it is not certain that the difference in number of chromosomes observed in this study is attributable to centric fusion since there is no pronounced difference in the size of the chromosomes of the two karyotypes. However, polyploidy may have been operative at different levels.
5. Conclusion
From this work, it has been established that the population of Ocimum basilicum growing in the South-eastern region of Nigeria are characterized by polyploidy and chromosome number variation, with at least two cytotypes. The chromosome count of 2n = 60 is a new number discovered in this study for this species. The plant species has asymmetrical karyotype with a higher proportion of acrocentric chromosomes, as compared to other forms of chromosomes, indicative of advancement in its evolutionary trend. However, further investigation utilizing flow cytometric analysis and molecular markers is strongly recommended.
REFERENCES
- D. J. Mabberley, “Mabberly’s Plant-Book: A Potable Dictionary of Plants, Their Classification and Uses,” 3rd Edition, Cambridge University Press, Cambridge, 2008.
- R. C. Wren, “Potter’s New Cyclopaedia for Botanical Drugs and Preparations,” Health Science Press, Rustington, Sussex, 1968.
- X. Chang, P. G. Anderson and C. J. Wright, “Variation in the Essential Oils in Different Leaves of Basil (O. basilicum) at Day Time,” The Open Horticulture Journal, Vol. 2, 2009, pp. 13-16. www.researchgate.net/...Variation...Essential_Oils_in_Different_Leaves_
- A. Paton and E. Putievsky, “Taxonomic Problems and Cytotaxonomic Relationships between and within Varieties of Ocimum basilicum and Related Species,” Kew Bulletin, Vol. 51, No. 3, 1996, pp. 1-16. http://dx.doi.org/10.2307/4117026
- D. L. Sulistiarini, “Ocimum gratissimum L.,” In: A. P. L. Oyen and D. X. Nkuyen, Eds., Essential Oil Plants, Plant Resources of South East Asia, Vol. 19, 1999, pp. 140-142. www.worldforestrycentre.org/sea/Products/.../af/.../SpeciesInfo.asp
- H. R. Juliani and J. E. Simon, “Antioxidant Activity of Basil,” In: J. Janick and A. Whipkey, Eds., Trends in New Crops and New Uses, ASHS Press, Alexandria, 2002, pp. 575-579. www.hort.purdue.edu/newcrop/ncnu02/v5-575.html
- H. Panda, “Aromatic Plants Cultivation, Processing and Uses,” Asia Pacific Business Press Inc., 2005.
- A. Bilal, N. Jahan, A. Ahmet, S. N. Bilal, S. Habib and S. Hajra, “Phytochemical and Pharmacological Studies on Ocimum basilicum L: A Review,” International Journal of Current Research and Review, Vol. 4, No. 23, 2012, pp. 73-83. www.scopmed.org/?mno=30017
- G. B. Choudhury, K. J. Prabhat, B. S. Nayak, S. K. Panda and S. K. Tripathy, “Phytochemical Investigation and Evaluation of Analgesic Activity of Leafy Extracts of Various Ocimum Species,” The Indian Pharmacist, Vol. 8, No. 12, 2010, pp. 67-70. http://cat.inist.fr/?amodele=afficheN&cpsidt=23062187
- D. Benedec, A. E. Parvu, I. Oniga, A. Toiu and B. Tiperciuc, “Effects of Ocimum basilicum L. Extract on Experimental Acute Inflammation,” Revista Medico-Chirurgicala, Vol. 111, No. 4, 2007, pp. 1065-1069. www.ncbi.nlm.nih.gov/pubmed/18389806
- S. Chinnasamy, G. Balakrishnan, S. V. Kontham, S. L. Baddireddi and A. Balakrishnan, “Potential Anti-Inflammatory Properties of Crude Alcoholic Extract of Ocimum basilicum L. in Human Peripheral Blood Mononuclear Cells,” Journal of Health Science, Vol. 53, No. 4, 2007, pp. 500-505. http://jhs.pharm.or.jp/data/53_500.pdf http://dx.doi.org/10.1248/jhs.53.500
- I. Kaya, N. Yigit and M. Benli, “Antimicrobial Activity of Various Extracts of Ocimum basilicum L. and Observation of the Inhibition Effect on Bacterial Cells by Use of Scanning Electron Microscopy,” African Journal of Traditional, Complementary and Alternative Medicines, Vol. 5, No. 4, 2008, pp. 363-369. www.ncbi.nlm.nih.gov/pubmed/20161958
- H. P. Bais, T. S. Walker, H. P. Schweizer and J. M. Vivanco, “Root Specific Elicitation and Antimicrobial Activity of Rosmarinic Acid in Hairy Root Cultures of Ocimum basilicum,” Plant Physiology and Biochemistry, Vol. 40, No. 11, 2002, pp. 983-995. http://dx.doi.org/10.1016/S0981-9428(02)01460-2
- R. Meera, P. Devi, B. Kameshwari, B. Madhumita and N. J. Merlin, “Antioxidant and Hepatoprotective Activities of Ocimum basilicum L. and Trigonella foenumgraecum L. against H2O2 and CCl4 Induced Hepatotoxicity in Goat Liver,” Indian Journal of Experimental Biology, Vol. 47, 2009, pp. 584-590. http://nopr.niscair.res.in/bitstream/.../5044/1/IJEB%2047(7)%20584-590.pdf
- L. Seung-Joo, U. Katumi, S. Takayuki and L. KwangGeun, “Identification of Volatile Components in Basil (Ocimum basilicum L.) and Thyme Leaves (Thymus vulgaris L.) and Their Antioxidant Properties,” Food Chemistry, Vol. 91, 2005, pp. 131-137. www.kmitl.ac.th/sisc/GC-MS/paper/natural%20nol.pdf http://dx.doi.org/10.1016/j.foodchem.2004.05.056
- M. S. Akhtar and M. Munir, “Evaluation of the Gastric Antiulcerogenic Effects of Solanum nigrum, Brassica oleracea and Ocimum basilicum in Rats,” Journal of Enthnopharmacology, Vol. 27, No. 1-2, 1989, pp. 163-176. www.ncbi.nlm.nih.gov/pubmed/2515396 http://dx.doi.org/10.1016/0378-8741(89)90088-3
- T. Dasgupta, A. R. Rao and P. K. Yadava, “Chemomodulatory Efficacy of Basil Leaf (Ocimum basilicum) and Drug Metabolizing and Antioxidant Enzymes and on Carcinogen-Induced Skin and Forestomach Papillomagenesis,” Phytomedicine, Vol. 11, No. 2-3, 2004, pp. 139- 151. www.ncbi.nlm.nih.gov/pubmed/15070164 http://dx.doi.org/10.1078/0944-7113-00289
- F. Fathiazad, A. Matlobi, A. Khorrami, S. Hamedeyazdan, H. Soraya, M. Hammami, N. Maleki-Dizaji and A. Garjani, “Phytochemical Screening and Evaluation of Cardioprotective Activity of Ethanolic Extract of Ocimum basilicum L. (basil) against Isoproterenol-Induced Myocardial Infarction in Rats,” DARU Journal of Pharmaceutical Sciences, Vol. 20, No. 87, 2012, pp. 1-10. http://www.darujps.com/content/20/1/87
- C. D. Darlington, “Chromosome Botany and the Origins of Cultivated Plants,” George Allen and Unwin Hyman, London, 1964.
- B. P. Skaria, “Aromatic Plants,” Horticultural Scientific Series, New India Publishing, India, 2007.
- A. F. Levan, K. Fredga and A. A. Sandberg, “Nomenclature for Centromere Position of Chromosomes,” Hereditas, Vol. 52, No. 2, 1964, pp. 201-220. http://onlinelibrary.wiley.com>Genetics>Genetics>Hereditas>Vol52Issue2
- S. M. Vargas, G. A. Torres, F. S. Pereira and L. C. Davide, “Karyotype Studies of Cratylia argentea (Desv.), O. kuntze and C. Mollis.,” Journal of Genetics and Molecular Research, Vol. 6, No. 3, 2007, pp. 707-712. www.ncbi.nlm.nih.gov/pubmed/18050091
- M. Pedro and A. Delgado-Salinas, “Karyotype Analysis in Six Species of Phaseolus,” Caryologia, Vol. 62, No. 3, 2009, pp. 1167-1170. www1.unifi.it/caryologia/past_volumes/62.../01-1490-(167-170)-VF-pdf
- C. Klaudija, L. Zlatko, B. Visnja, J. Branka, B. Borut, K. Ivan and S. Zlatko, “Genetic Relations among Basil Taxa Based on Molecular Markers, Nuclear DNA Content and Chromosome Number,” Plant Systematics and Evolution, Vol. 285, No. 1-2, 2010, pp. 13-22. http://dx.doi.org/10.1007/s00606-009-0251-z
- M. J. Moumita, A. K. Dalta and M. G. Gopal, “Chromosomes Number Variation in Ocimum basilicum L.,” Cytologia, Vol. 70, No. 4, 2005, pp. 455-458. http://dx.doi.org/10.1508/cytologia.70.455
- G. L. Stebbins, “Chromosomal Evolution in Higher Plants,” Edward Arnold Pub. Ltd., London, 1971.
- G. A. Levitzky, “The Morphology of Chromosomes,” Bulletin of Applied Botany, Genetics and Plant Breeding, Vol. 27, 1931, p. 19174. www.antiqbook.com/books/bookinfo.phtml?0=aquila&bnr=5781

