American Journal of Plant Sciences
Vol.3 No.10(2012), Article ID:24157,4 pages DOI:10.4236/ajps.2012.310177
System Position and Divergent Time of Based on ITS Sequence Humulus scandens
![]()
College of Biotechnology, Southwest University/Key Sericultural Laboratory of Ministry of Agriculture, Chongqing, China.
Email: crf55@163.com
Received May 29th, 2012; revised July 10th, 2012; accepted July 26th, 2012
Keywords: Moraceae; Humulus scandens; ITS; Systematic Position; Divergent Time
ABSTRACT
This article mainly studied Humulus scandens’ ITS sequence, its system position and its divergent time. ITS is 624 bp with a GC percentage content of 57.21%. It only has the variation in 585 (C changes T), systematic position in Humulus lupulus (GenBank: EF136401) and Humulus scandens (GenBank: FJ980285), Humulus lupulus (GenBank: DQ005990), with Humulus scandens (GenBank: FJ980285), Humulus lupulus (GenBank: DQ005990) has the near sibship. Humulus in Moraceae family system position between Cudrania tricuspidata and Artocarpeae, support the Cannabiodeae promotion for Cannabinaceae. The divergent time of Humulus is 70.88 mya in Moraceae, Humulus scandens and Humulus lupulus (GenBank: EF136401) is 12.78 mya, with Humulus scandens (GenBank: FJ980285), Humulus lupulus (GenBank: DQ005990) is 1.10 mya. In the Moraceae branch’s molecular systematics research, the suitable choice is to choose Humulus scandens as Moraceae’s outgroup.
1. Introduction
Humulus scandens is a twining or prostrate vine that grows as an annual. The opposite leaves are 7 - 10 cm in length and deeply divided into five distinct palmate lobes with a serrate margin and rough surface. The underside of the leaf is pubescent, bearing yellow glands. The stems and petiole are covered with sharp, downward-curving hairs. Male flowers are yellowish green panicles 15 - 25 cm long. Female flowers are catkin-like drooping spikes about 5 mm in diameter. The ovary, covered in a white tomentum, is triangular with an acuminate apex, and enclose in a papery bract with two external stigmata. Humulus scandens is distributed widely in most provinces in China, with the exception of Hainan, Tibet, and Qinghai. It occurs in ditch, wastelands, debris, and forest margins, It also occurs in Vietnam and Japan [1]. Humulus scandens is a useful traditional herb which contains various nutriaents. Young Humulus scanders can be used as fodders. The stem fibers can be used for papermaking, and the seed oils are used in soap production. The flowers can be substituted for Humulus lupulus in brewing. It has strong rresistance, and can be used to conserve soil and water. As a climbing twining vine, Humulus scandens may cause damage to fruit trees and grain crops or decrease production because of its climbing and twining tendency. Meanwhile, due to the spinibarbus, it also causes damage on the human skin and hinders the production activities of the human being. In China, Humulus scandens is also one of pollen allergy pathogenic source plants in autumn.
Humulus scandens is widely reported in DNA extraction, amplified fragments length polymorphism (AFLP), inter simple sequence repeat (ISSR), sequence characterized amplified regions (SCAR) marker, complementary DNA (cDNA) expression library, morphological microstructure, pharmaceutical composition, fodder value, biopesticides and viruses dip, particularly in the pharmaceutical composition, fodder value [2-13]. But there is few report on the system location and divergent time. In this article, we collected a H. scandens from Chongqing Botanical Garden, and 15 other Moraceae plants in East Asia. We also downloaded 19 internal transcribed spacer (ITS) sequences of Americas and Africa’s Moraceae plants from GenBank to analysis the system location and divergent time of Humulus scandens. In order to accumulate more data of Moraceae molecular phylogeny, we searched more reasonable Moraceae groups.
2. Materials and Methods
2.1. Materials
Humulus scandens is collected from Chongqing flowers and plants garden (Figure 1). Other moraceae material

Figure 1. Humulus scandens of Chongqing flowersand plants garden.
(Table 1), outgroup designation is the Ulmus parvifolia.
2.2. DNA Extraction
We used CTAB (cetyl trimethylammonium bromide) [14] method with a slightly modified to extract DNA. Briefly, DNAs were isolated from 0.15 g of silica gel-dried leaves, and the quality and consistency of DNAs were determined by 0.4% agarose gel eclectrophoresis in the presence of λ DNA/Hind—marker or by GeneQuant spectrophotometer (Eppendorf, Gemany).
2.3. PCR Amplification and Sequencing
The total volume of PCR amplification reaction is 20 μL, the reaction system including 10 ng DNA template, 50 ng forward and reverse primer, 2.5 mM of dNTPs, 2 μL 10 × PCR buffer and 5 U Taq DNA polymerase. The PCR amplification procedure is 94˚C degeneration for 4 min, then each circulate 94˚C degeneration for 1 min, 55˚C annealing (proposed) for 1 min, 72˚C extensions for 1 min, a total of 33 circulates, at last extension for 7 min at 72˚C, and the end control in 12˚C reactions. The amplification primers are designed based on GenBank, and adjusted sightly referring to the research of Mr. White etc. [15], then compounded by Shanghai Shenggong Sequencing Department. The primers are followed:
ITS5: 5’-GGAAGTAAAAGTCGTAACAAGG-3’
ITS4: 5’-TCCTC CGCTTATTGATAT GC-3’
which were synthesized by SSBETS (Shanghai Sangon Biological Engineering Technology and Services Co., Ltd; Shanghai). After purification with Microcon YM100 column (Millipore) and adjustment of the concentration, PCR products were sequenced with ABI Prism 3730 genetic analyzer by SSBETS using BigDye terminator v3.1 reagents. PCR primers were used as sequencing primers too, and the sequences were determined in both directions with Sanger dideoxy method.
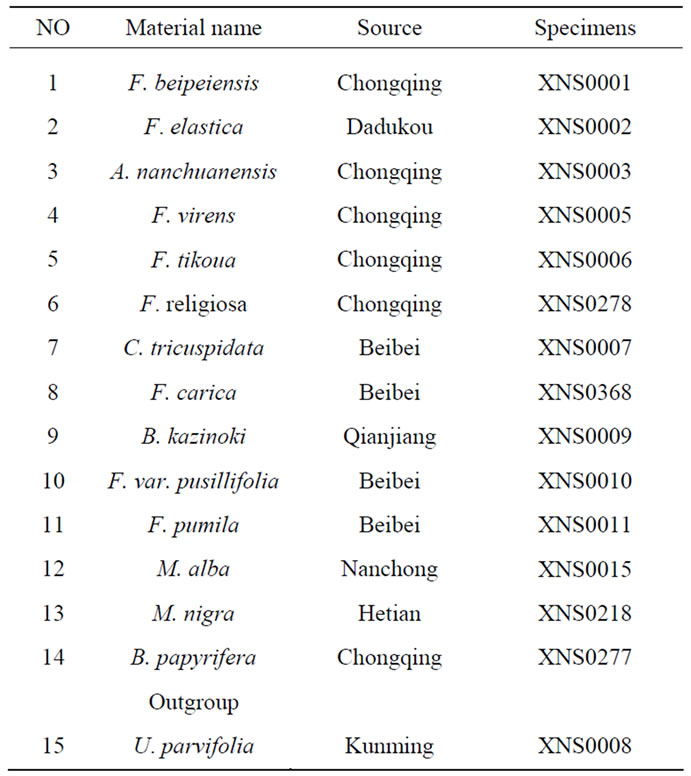
Table 1. Materials of the other moeaceae for the Molecular systematics research of Humulusscandens.
2.4. Sequence Analysis Methods
DNA sequences were assembled with Sequencher 4.1.4 software (Gene Codes Corporation, Ann Arbor, MI, USA), while a few wrong bases in the sequences were corrected according to the base peak shape. And then, 19 ITS sequences that Milicia excelsa, Streblus glaber, Naucleopsis guianensis, Castilla elastica, Maquira calophylla, Helicostylis tomentosa, Poulsenia armata, Antiaropsis decipiens, Sparattosyce dioica, Ficus variegate, Ficus racemosa, Ficus irisana, Dorstenia roigii, Dorstenia africana, Trymatococcus oligandrus, Brosimum guianense Humulus scandens (GenBank: FJ980285), Humulus lupulus (GenBank: EF136401), Humulus lupulus (GenBank: DQ005990) was downloaded the from GenBank [16,17]. These were compared together with previous spliced sequences by using Clustalx 1.83 c software [18,19]. The determination of ITS sequencess’ range in this experiment’s was based on GenBank published data of Humulus lupulus cultivar Wye Target (GenBank: EF136401), 18S rRNA gene 3’, 26S rRNA gene 5’. Bioedit software was used to remove both ends of non-ITS sequences,and DNAstar software was used to analyze the length, G + C, percentage content, and mutation sites of ITS sequences. The phyletic series position of Humulus scandens was analyzed through the cooperation of PAUP Ver.4.0b10 and Modeltest V3.06 softwares [20-22]. Ulmus parvifolia was treated as outgroups, and divergent time (height) of Humulus scandens was calculated by BEAST software [23-25].
3. Results
3.1. Length, GC Percentage Content and Characters of Humulus scandens ITS Sequences
The boundary of Humulus scandens ITS sequences in this study was determined according to the published GenBank sequences of the 18S rRNA gene 3’ end, 28S rRNA gene 5’ end, and the 5.8S rDNA gene of Humulus lupulus cultivar Wye Target (GenBank: EF136401). Our results show that the full length of Humulus scandens ITS is 624 bp with a GC percentage content of 57.21%. The characters of Humulus scandens ITS sequence is shown in Figure 2.
3.2. The Systematic Position of Humulus scandens
Using the PAUP Ver.4.0b10 and Modeltest V3.06 software, and MP method to analyze the systematic position of Humulus scandens, we learn that the Tree length is 1467, Consistency index (CI) is 0.5794, retention index (RI) is 0.7666, rescaled consistency index is (RC) 0.4442. 704 total characters with 224 constant, 104 variable characters are in parsimony-uninformative. The number of parsimony-informative characters 376. The branching diagram sorted Cudrania tricuspidata out firstly, Bootstrap 100%, then Humulus, Bootstrap 100%. Humulus scandens system location is between Humulus lupulus (GenBank: EF136401) and Humulus scandens (GenBank: FJ980285), Humulus lupulus (GenBank: DQ005990), has a closer relationship with Humulus scandens (GenBank: FJ980285), Humulus lupulus (GenBank: DQ005990) (Figure 3).
3.3. The Divergent Time of Humulus scandens
According to the divergent time of Marijuana is 132 mya [26], using the sequence data file (NEXUS) which calculate the systematic position of Humulus scandens in the last section, input it into BEAST software package and do analysis. The loose molecular clock estimates the divergent time (height) of Humulus scandens, the divergent time of Humulus is 70.88 mya in Moraceae. Humulus scandens and Humulus lupulus (GenBank: EF136401) is 12.78 mya, and Humulus scandens (GenBank: FJ98- 0285). Humulus lupulus (GenBank: DQ005990) is 1.10 mya (Figure 4).
4. Discussion
4.1. Humulus scandens ITS Sequence’s Variation
This article downloaded the ITS sequence of Humulus scandens voucher PS0217MT01 (GenBank: FJ980285), Humulus lupulus cultivar Wye Target (GenBank: EF13- 6401), Humulus lupulus voucher Wells 5176 US (GenBank: DQ005990) from GenBank, and then compared them with ITS sequence of Humulus scandens. The results show that the variation (C to T) of Humulus scandens can only take place in the 585 location, but the variation of Humulus lupulus cultivar Wye Target (GenBank: EF136401) which is downloaded from GenBank is wide, because there are 44 nucleotide variations in it. the variation(T to A) of Humulus scandens voucher PS0217- MT01 (GenBank: FJ980285) takes place in the 567 location, and there is no variation in Humulus lupulus voucher Wells 5176 US (GenBank: DQ005990). However, the results of the cladogram show that they are in the same

Figure 2. The ITS sequence of Humulus scandens.
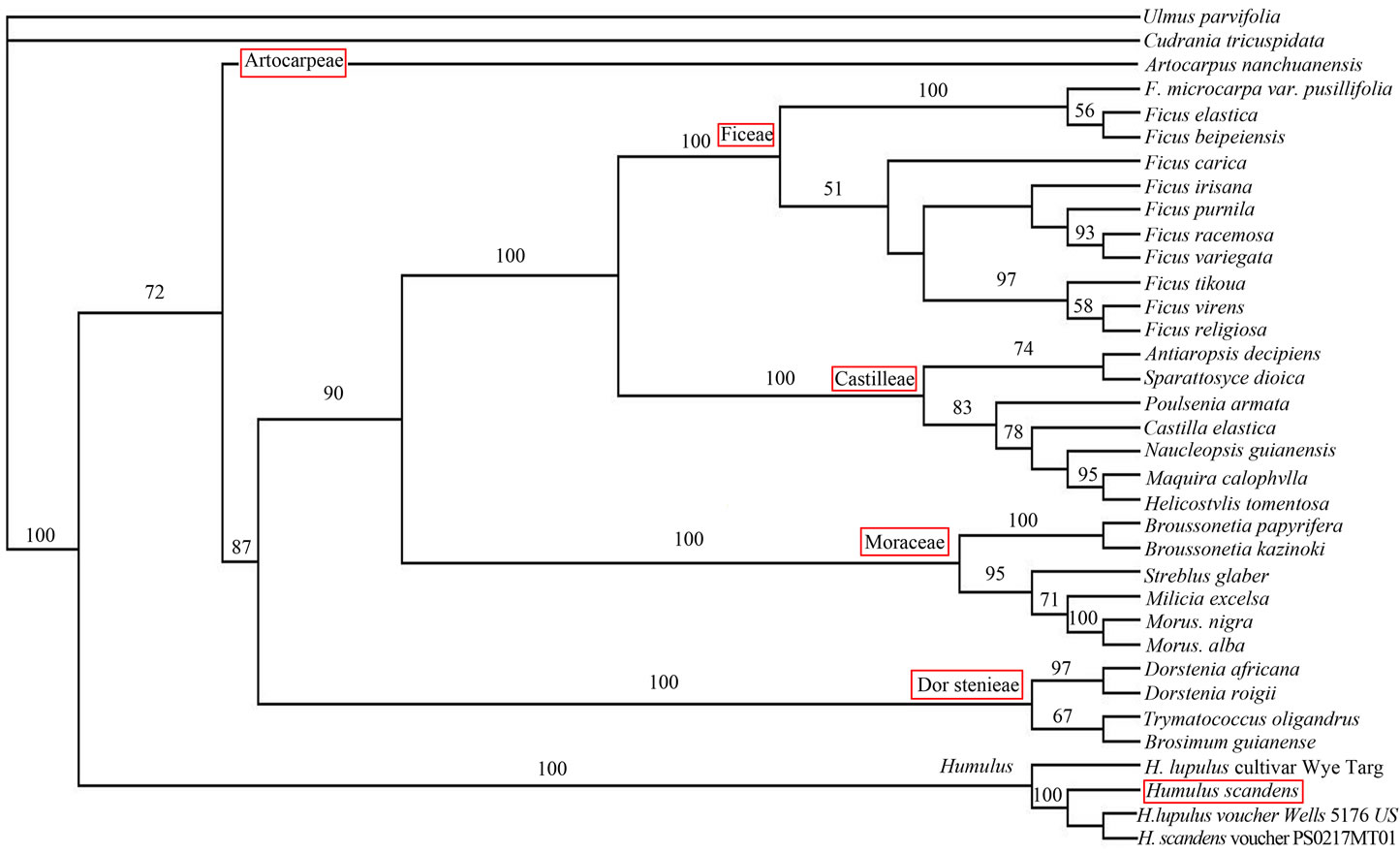
Figure 3. Systematic position of Humulus scandens based on ITS sequence analysis.
branch, which means this variation doesn’t break through the range of Humulus.
4.2. Comparison Based on the Systematical Position of Humulus scandens ITS and Classical Botanical Taxonomy
Humulus scandens has a large controversy in the taxonomy. In Engler system, Humulus scandens belongs to Humulus of the subfam.Cannabiodeae, which is under the Moraceae. From 1926 to 1934, Hutchinson and Rendle upgraded the subfam. Cannabiodeae to Cannabinaceae, so some taxonomists such as Hu Xian-su, Tahtaian, Cronquist all classified Humulus scandens to Humulus, Cannabinaceae [27]. “Eight-Class System” [28] supported by Wu Zheng-yi and Flora of China classified the Humulus in subfam. Cannabiodeae, Moraceae as same as in Engler system. In this study, according to the cladogram, the Cudrania tricuspidata can be firstly separated from the outgroup Ulmus parvifolia, and then the rest will become a major branch, and the bootstrap is 100%. This is followed by Humulus whose bootstrap is also 100%. The 5 groups of Moraceae that are Artocarpeae, Dorstenieae, Moreae, Castilleae and Ficeae, and they have almost the same with the on 26S, ndhF genes based by Nyree J. C., 2005. Because Humulus is not divided into the 5 groups of Moraceae. Thus, Hutchinson and Rendle point of view, Humulus from Moraceae Separate. The results are as same as the APF’s researchful views Consistency [29].
4.3. Humulus scandens Can Be the Moraceae Outgroup in the Study of Molecular Systems
Nyree et al., used 26S and ndhF gene to research the Moraceae molecular System in 2005. He used Celtidaceae, Celtis philippinensis, Cannabaceae, Cannabis sativa, Humulus lupulus, Cecropiaceae, Coussapoa villosa, Cecropia, Cecropia peltal, Urticaeae, ourouma sp., Leucosyke sp., Boehmeria nivea, Pilea, Pikea fobtana, Debregeasia Debregeasia longifolia, Poikilospermum, Poikilospermum suaveolens on outgroups selection, but he didn’t analyze which outgroups is best. In this study, The Cudrania tricuspidata first separates from the Moraceae, then the Humulus. According to Sang Tao, 1996 , Maddison, et al., 1984; Watrous et al., 1981 study, cladistics study on the principle of outgroup selection principle, multiple outgroup, the closer the better within the outgroup comparable [30]. If you know the earliest group which separates from others in the intraspecific groups, this group can be used as the outgroup of the other members of the outgroup [31]. So, in Moraceae molecular system, Humulus scandens is better as the Moraceae outgroups.
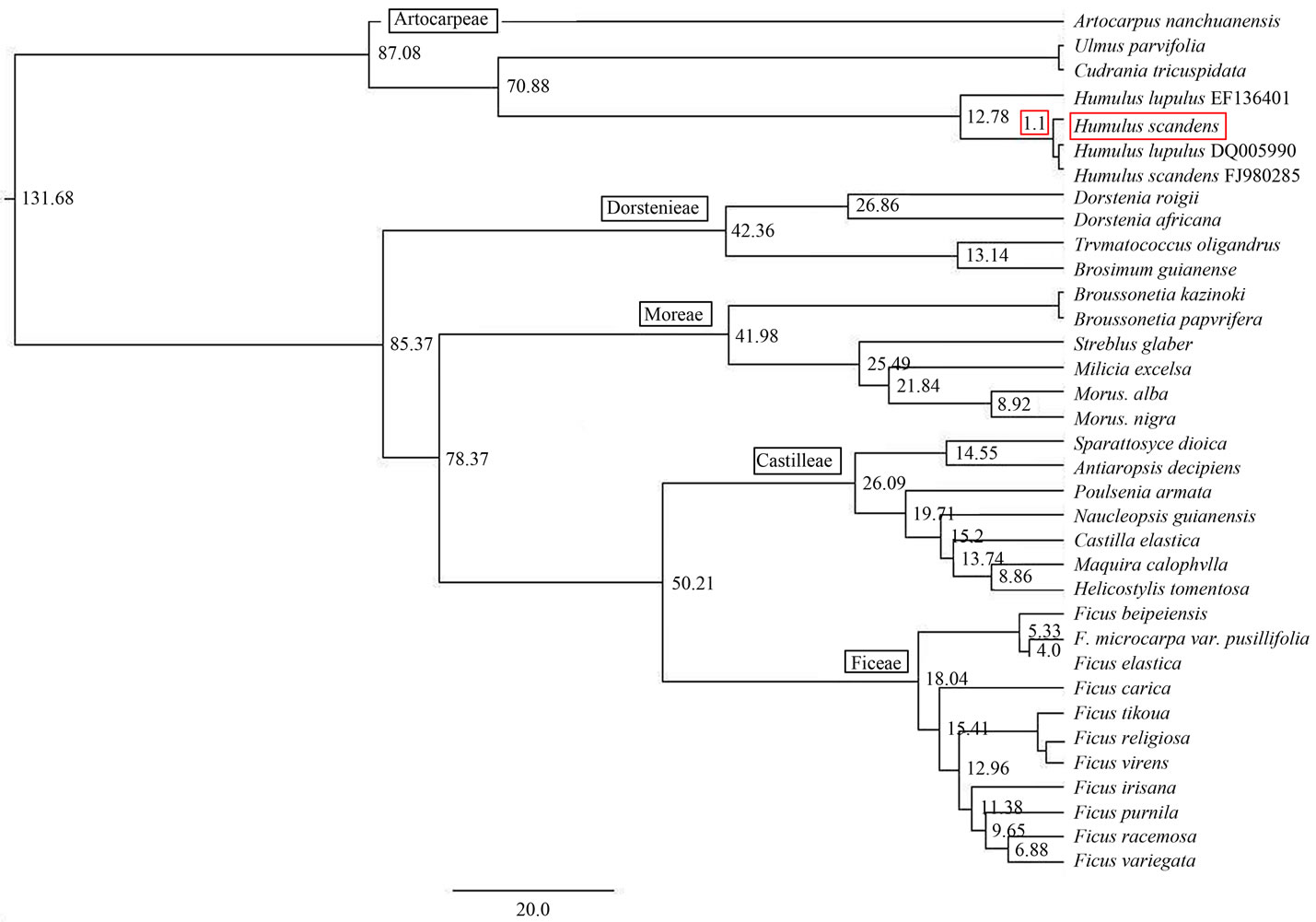
Figure 4. Humulus scandens divergent time of based on ITS sequences.
5. Conclusion
This Humulus scandens’ ITS sequence length is 624 bp with a GC percentage content of 57.21%, and systematic position in Humulus lupulus (GenBank: EF136401) and Humulus scandens (GenBank: FJ980285), Humulus lupulus (GenBank: DQ005990), with Humulus scandens (GenBank: FJ980285), Humulus lupulus (GenBank: DQ- 005990) has the near sibship. Humulus in Moraceae family system position between Cudrania tricuspidata and Artocarpeae. The divergent time of Humulus is 70.88 mya in Moraceae; Humulus scandens and Humulus lupulus (GenBank: EF136401) is 12.78 mya; with Humulus scandens (GenBank: FJ980285), Humulus lupulus (GenBank: DQ005990) is 1.10 mya. In the Moraceae branch’s molecular systematics research, the suitable choice is to choose Humulus scandens as Moraceae’s outgroup.
REFERENCES
- Flora of China Editorial Committee, “Flora of China,” Science Press, Beijing, 1998, pp. 220-225.
- S.-H. Zou and T.-Y. Liu, “Trend of Nutrient Components and Flavonoids of Humulus scandens. (Lour.) Merr at Different Growth Stages,” China Animal Husbandry and Veterinary, Vol. 37, No. 5, 2010, pp. 21-25.
- S.-Q. Tao, Q.-J. Zhang, Y. Zhang and F.-A. Wu, “Humulus scandens Different Solvent of Extracts the Virulence of Tetranychus cinnabarinus,” Agricultural Science of Jiangsu, Vol. 39, No. 3, 2011, pp. 158-159.
- L. Qing, S.-Y. Yang, X.-C. Sun, J.-J. Wang and C.-Y. Zhou, “Detection of Several Co-Infected Viruses in Humulus scandens,” Plant Protection, Vol. 35, No. 4, 2009, pp. 138-139.
- L.-H. Xiao, W.-J. Gao, X.-L. Zhang, L.-D. Lu, C.-L. Deng and S.-X. Lu, “DNA Extraction and Establishment of AFLP Marker Reaction System of Humulus scandens,” Molecular Plant Breeding, Vol. 6, No. 2, 2008, pp. 393- 398.
- Z.-X. Chen, S. Meng, X. Zhu, L. Wang and F.-H. Meng, “Determination of Luteolin7-o B-D Gluco,” Molecular Plant Breeding, Vol. 25, No. 6, 2011, pp. 547-549.
- J.-J. Wang, C.-L. Deng, L.-D. Lu and W.-J. Gao, “Humulus scandens Nuclear Type and Sex Chromosome Research,” Henan Agricultural Science, No. 10, 2007, pp. 61-63.
- J.-S. Wang, F.-F. Huang and K. Chen, “Studied on the Karyomorphology of Humulus scandens,” Journal of Hubei Normal University (Natural Science), Vol. 19, No. 4, 1999, pp. 71-74.
- S.-Y. Ni and X.-H. Song, “Studies on Ldentification of Humulus scandens,” Pharmaceutical and Clinical Research, Vol. 18, No. 1, 2010, pp. 90-91.
- Y. Liu, X.-Z. Sun, W.-G. Zhang and X.-L. Feng, “Construction and Primary Characterization of Cdna Expression Library of Humulus scandens Pollen,” Journal of Xi’an Jiaotong University (Medical Sciences), Vol. 27, No. 4, 2006, pp. 327-329.
- J.-M. Lu, X.-Z. Sun, M.-X. Li, Y. Liu, J. Xu and Y.-H. Zhang, “Eukaryotic Expression and Immunization Analysis of the DNA Vaccine of the Major Grass Pollen Allergen of Humulus scandens,” Journal of Xi’an Jiaotong University (Medical Sciences), Vol. 32, No. 2, 2011, pp. 154-160.
- W.-J. Gao, T. Sha, Y.-K. Ji, C.-L. Deng and L.-D. Lu, “Clone and Development of ISSR and SCARMarkers Linked to Male Humulus scandens,” Journal of Tropical and Subtropical Botany, Vol. 18, No. 3, 2010, pp. 283- 287.
- Z.-X. Chen and S. Meng, “Research Progress of Humulus scandens,” Chinese Medicine Matter, Vol. 25, No. 2, 2011, pp. 175-179.
- K. H. Weising, K. Wolff and W. Meyer, “DNA Fingerprinting in Plants and Fungi,” CRC Press, Boca Raton, 1995, pp. 50-54.
- T. J. White, Tbruns and J. T. Sleeand, “Amplification Anddirectse Quencingoffun Galribosomal RNA Genes for Phylogenetics,” In: M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White, Eds., PCR Protocols: A to Methods and Applications, Academic Press, San Diego, pp. 351-322.
- D. Y. Peng, “Draft of Index to Codes of Vascular Plants of Taiwan,” Council of Agriculture Press, Taiwan, 1997, pp. 136-194.
- M. M. Apetorgbor, E. Turco, J. R. Obbinah and C. A. Ragazzi, “Potential Factors Limiting Viability of Milicia excelsa (Welw.) C. C. Berg Seeds in Plantation Establishment in West Africa,”Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz, No. 3, 2004, pp. 238-246.
- J. D. Thompson, T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, “The Clustal X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Research, Vol. 25, No. 24, 1997, pp. 4876-4882. doi:10.1093/nar/25.24.4876
- D. L. Swofford, “PAUP Phylogenetic Analysis Using Parsimony (and Other Methods),” Version 4.0b10, 2002.
- D. Posada and K. A. Crandall, “Modeltest: Testing the Model of DNA Substitution,” Bioinformatics, Vol. 14, No. 9, 1998, pp. 817-818. doi:10.1093/bioinformatics/14.9.817
- A. J. Drummond and A. Rambaut, “Relaxed Phylogenetics and Dating with Confidence,” 19 January 2010. http://www.wenku.baidu.com/view/cd28037f5acfa1c7aa00cc62.htm
- J. P. Huelsenbeck and F. Ronquist, “MRBAYES: Bayesian Inference of Phylogenetic Trees,” Bioinformatics, Vol. 17, No. 8, 2001, pp. 754-755. doi:10.1093/bioinformatics/17.8.754
- A. J. Drummond, G. K. Nicholls, A. G. Rodrigo, et al., “Estimating Mutation Parameters, Population History and Genealogy Simultaneously from Temporally Spaced Sequence Data,” Genetics, Vol. 161, No. 3, 2002, pp. 1307- 1320.
- I. J. Wilson, M. E. Weale and D. J. Balding, “Inferences from DNA Data: Population Histories, Evolutionary Processes and Forensic Match Probabilities,” Journal of the Royal Statistical Society Series A: Statistics in Society, Vol. 166, 2003, pp. 155-188.
- N. J. C. Zerega, W. L. Clement, S. L. Datwyler and G. D. Weiblen, “Biogeography and Divergence Times in the Mulberry Family (Moraceae),” Molecular Phylogenetics and Evolution, Vol. 39, 2005, pp. 402-416.
- M. Sun, “The Hemp Branch Plant Has the Milk,” Botany Notification, Vol. 8, No. 1, 1991, pp. 63-64.
- Z.-Y. Wu, A.-M. Lu and Y.-C. Tang, “Chinese Angiosperms Division Is a Comprehensive Theory,” Science Press, Beijing, 2003, pp. 560-575.
- Angiosperm Phylogenetic Study Group, “White, Gironniera, Parasponia, Pteroceltis Maxim, Trema Taxonomic Studies.” http://zh.wikipedia.org/wiki
- T. Sang and B.-S. Xu, “A Review of Current Theories and Methods in Cladistics and a Cladistic Study of Twelve Lindera Species in Eastern China,” Acta Phytotaxonomica Sinica, Vol. 34, No. 1, 1996, pp. 12-28.
- W. P. Maddison, M. J. Donoghue and D. E. Maddison, “Outgroup Analysis and Parsimony,” Systematic Zoology, Vol. 33, No. 1, 1984, p. 83. doi:10.2307/2413134
- L. E. Watrous and Q. D. Wheeler, “The Out-Group Comparison Method of Character Analysis,” Systematic Zoology, Vol. 30, No. 1, 1981, pp. 1-11. doi:10.2307/2992297

