Advances in Bioscience and Biotechnology
Vol.3 No.4(2012), Article ID:21848,6 pages DOI:10.4236/abb.2012.34056
Production and characterization of chitinase from Vibrio species, a head waste of shrimp Metapenaeus dobsonii (Miers, 1878) and chitin of Sepiella inermis Orbigny, 1848
![]()
1Centre of Advanced Study in Marine Biology, Faculty of Marine Sciences, Annamalai University, Parangipettai, India
2Department of Pharmacology, Chettinad University, Kanchipuram, India
Email: saran_prp@yahoo.com
Received 18 May 2012; revised 23 June 2012; accepted 5 July 2012
Keywords: Chitin; S. inermis; Chitnase; M. dobsonii; Vibrio sp.
ABSTRACT
The chitin is extracted from the cuttlebone of S. inermis and the mineral contents are predictable. The structure and degree of deacetylation of extracted cuttlebone chitin is dogged through Fourier Transform Infrared (FT-IR) spectroscopy. The extracted cuttlebone chitin is used as the substrate for the production of chitinase from Vibrio sp. The extra cellular proteins are concentrated by ammonium sulphate precipitation, dialysed and then purified by using gel (sephadex G-100) chromatography. Among the 40 fractions, only two fractions (active fractions) showed maximum absorbance at 280 nm, which are pooled, dialysed and free-dried. The enzyme activity (0.104 µmoles/ml) and molecular weight (50 - 60 kDa) of purified chitinase is also determined through SDSPAGE. The optimal condition for chitinase activity is pH between 6.0 - 6.5 and 45˚C.
1. INTRODUCTION
Chitin is one of the most abundant biopolymers in nature and possibly the most abundant in the marine environment, since it is produced by many marine organisms, including zooplankton and several phytoplankton species [1]. The other popular source is the mycelia (containing up to 20% chitin) from mushrooms and other fungi. About an average of 30% and up to 60% of the cuticles of insects are chitins [2]. Chitin production is estimated approximately 1 × 1011 tons annually and it is the second most abundant natural biopolymer. Because chitin has many useful biological properties such as biocompatibility, biodegradability, haemostatic activity and wound healing property, much attention has been paid to its biomedical applications; e.g. an absorbable suture, a drug carrier, an antitumor agent [3].
Chitin is composed of repeating N-acetyl D-glucosamine residues and promising biopolymer with numerous industrial, medical and commercial uses. However, it is difficult to purify and modify chemically. Hence identification of chitin modifying enzymes and elucidation of their activities could facilitate the efficient production of specific chitin products. The biodegradation of chitin requires the synergistic action of several hydrolytic enzymes for efficient and complete breakdown. The combined action of endo and exochitinases results in the degradation of chitin polymer into the soluble N-acetyl D-glucosamine [4].
Chitinases are hydrolyzing the chitin; these enzymes have been implicated in a wide range of biological events including host infection, growth and development, defense against pathogens and human diseases. The research of chitinases is most important, because of its biological controller in ecology, epidemiology of diseases and the preparation of medicines. In the marine environment, most chitin seems to be degraded by marine bacteria. Some chitinolytic bacteria, including Aeromonas, Pseudomonas, Clostridium and Vibrio have been isolated and their chitinase activity was also studied [5]. Hence an attempt has been made to isolate chitin from S. inermis and its structure is determined through FT-IR spectrum which is used for the production and characterization of chitinase from Vibrio sp.
2. MATERIALS AND METHODS
2.1. Extraction of Chitin
Chitin is prepared by following the method of Takiguchi [6].
2.2. Estimation of Minerals
The demineralized water of cuttlebone chitin is incubated with concentrated HNO3 for 24 hr after that mixture of HNO3: Perchloric acid mixture (4:1) is added and incubated. Then digested at 80˚C to the obtained white precipitate 10% (v/v) HNO3 is added and filtered. The total volume is made up to 20 ml and the minerals are estimated through ICP [7].
2.3. FT-IR Spectral Analysis
The quality parameter of chitin is analyzed by FT-IR spectroscopy [8]. FT-IR spectrum of solid samples of chitin is relied on a Bio-Rad FTS-40 model, USA and its average degree of acetylation is calculated by following the method of Baxter et al. [9].
2.4. Isolation of Microorganism
Heads of soft shell shrimps (M. dobsonii) are collected from the Cuddalore landing center (Lat.11˚24'N; Long 79˚49'E), Tamil Nadu, India. Samples are homogenized and diluted 10−1 to 10−5 in sterile distilled water and spread on chitin—containing minimal agar plates. After incubation for 2 weeks at room temperature, clear zone forming bacteria are selected as chitinase producer [5].
2.5. Production of Chitinase Enzyme
For the production of chitinase from Vibrio sp. is grown in 100 ml of medium consisting of 0.35% K2HPO4, 0.15% KH2PO4, 0.1% MgSO4·7H2O, 0.1% tryptone, 0.1% (w/v) yeast extract, and 1% (w/v) extracted cuttlebone chitin powder in a 250 ml Erlenmeyer flasks for a further culture at 28˚C for 4 days. Cells induced by using powdered chitin are harvested by centrifuged at 12,000 g for 20 min at 4˚C. The supernatant is precipitated by adding 90% (w/v) ammonia sulfate and then dialyzed against 50 mM sodium phosphate buffer (pH 6.0) to remove salts and the crude enzyme was thus obtained [10].
2.6. Preparation of Colloidal Chitin
Colloidal cuttlebone chitin is prepared by the method of Rodriguez-Kabana et al. [11] by partial hydrolysis of chitin with 10 N HCl for 2 hr at room temperature. The colloidal chitin is washed several times with large volumes of distilled water to adjust the pH to 7.0.
2.7. Assay of Chitinase Activity
Chitinase activity is assayed by detecting the reducing groups produced by the chitinase reaction using colloidal extracted cuttlebone chitin as the substrate [11]. The assay is performed according to the procedure previously reported [12]. One unit (U) of chitinase activity is defined as the amount of enzyme that produced 1 µmol of reducing equivalent per minutes at 37˚C. The monomer of chitin, GlcNAc, is taken as the standard for the reduce sugar [13]. Briefly, a 0.13 ml enzyme preparation is incubated with 0.87 ml of 0.2% (w/v) colloidal chitin azure solution at 37˚C for 30 min. The reaction mixture is then centrifuged and the absorbance of the supernatant is measured at 550 nm.
2.8. Effect of pH and Temperature Optima on the Activity
The effect of pH on the purified chitinase is determined at different pH values (4.5 to 10.0) under standard assay conditions. The buffer systems used are as follows: 0.1 M citrate-phosphate buffer, pH 4.5 - 6.5; 0.1 M phosphate buffer, pH 6.5 - 8.5; 0.1 M glycine-NaOH buffer, pH 8.5 - 10.0. The optimal temperature for the chitinase activity is determined in the range of 20˚C to 50˚C under standard assay conditions. The purified enzyme is preincubated for 5 min at the temperature tested. The reaction was started with the addition of para nitrophenly β-Dacetylglycosaminide.
2.9. Estimation of Protein
Protein concentrations are measured according to Bradford’s [14] method using BSA as the standard.
2.10. Purification and Molecular Weight Determination of Chitinase
After production, the cells are removed by centrifugation at 5000 × g and 4˚C for 20 min. Protein in the cell-free culture broth (900 ml) are precipitated with ammonium sulfate (30%, w/v). The precipitate is obtained by centrifugation (16,000 × g, 30 min, 4˚C) and suspended in 3 ml of 0.1 M citrate-phosphate buffer (pH 6.0). The suspension is eluted through sephadex G-100 (Sigma) in a glassy column (100 × 1.0 cm). The elution buffer is 50 mM Tris-HCl and the volumetric flow rate is 0.33 ml/ min. Every 4 ml from the effluent of gel filtration is collected in a fraction [15]. SDS-PAGE analysis of proteins is carried out by using 12.5% (w/v) polyacrylamide gel at 120 V for 90 min. Solutions for preparing 12.5% (w/v) resolving gel included H2O (6.4 ml), 30% acrylamide mix (8.3 ml), 1.5 M Tris (pH 8.8, 5 ml), 10% (w/v) SDS (0.2 ml), 10% (w/v) ammonium persulfate (0.1 ml), and N,N,N,N’-tetramethylethylenediamine (0.008 ml). The gel separation was performed based on a suggested protocol [16] except for eliminating the step of heating sample to 100˚C for 3 min prior to electrophoresis.
3. RESULTS AND DISCUSSION
The percentage yield of chitin was found to be 22%.
3.1. Mineral Study
Minerals such as Ca (5.90%), P (0.05%), Cu (0.002%), Fe (0.5%), Mg (004%) and Zn (007%) of cuttlebone of S. inermis were recorded.
3.2. FT-IR Spectral Analysis
The FT-IR spectrum of the standard and cuttlebone chitin was obtained and the effective peaks were compared with the standard chitin (Table 1).
The FT-IR spectrum of the standard chitin showed twelve major peaks at the range of 897.41, 1026.63, 1077.93, 1154.64, 1259.54, 1422.73, 1587.94, 1660.55, 2361.41, 2922.85, 2922.85 and 3377.95 cm−1; whereas the cuttlebone chitin from S. inermis recorded the peaks in the range of 470.40 to 3753.13 cm−1 (Figures not shown) [17].
3.3. Degree of Deacetylation
The degree of deacetylation was calculated as 55.95%.
3.4. Isolation of Microorganism
On the chitin agar clear zone producing organism was selected for further chitinase study and the organism was identified as Vibrio sp. (Figure 1) through the biochemical test. Golden yellow colonies were produced on the TCBS agar [18].
3.5. Production of Chitinases
Vibrio sp. was grown aerobically in 100 ml of the optimum medium in a 250 ml Erlenmeyer flask at 30˚C. During the process of incubation, cell growth (as judged by OD660) and pH were measured. Consequently it was inferred that chitinase enzymes produced by Vibrio sp.
3.6. Purification of Enzyme
Extra cellular chitinase was partially purified by gel chromatography using sephadex G-100 as column, 40 fractions were collected. Among the 40 fractions two fractions (6 and 14) showed maximum absorbances (Figure 2), which were pooled and used for further studies.
3.7. Activity of Chitinase
The conventional method using colloidal chitin as the substrate is based on the evaluation of reducing sugars released due to the action of chitinase. Incubation of GlcNAc with the colour reagent solution that was made by dissolving 0.5 g potassium ferriccyanide in 1 lit of 0.5 M sodium carbonate led to the decrease in the absorbance at 420 nm, due to the reducing power of GlcNAc. For the determination of enzymatic activity, a calibration curve up to GlcNAc concentration of 0.104 µmol/ml showing the release of reduce sugar (in 30 min) from the cleavage of chitin versus the chitinase concentration was thus obtained as shown in Figure 3.
3.8. Determination of Molecular Weight
The molecular weight of purified chitinase was found to be 50 - 60 kDa. It was confirmed through SDS-PAGE by compared with protein molecular marker (Genei) (20 -

Table 1. FT-IR spectrum of standard and cuttlebone chitin of S. inermis.
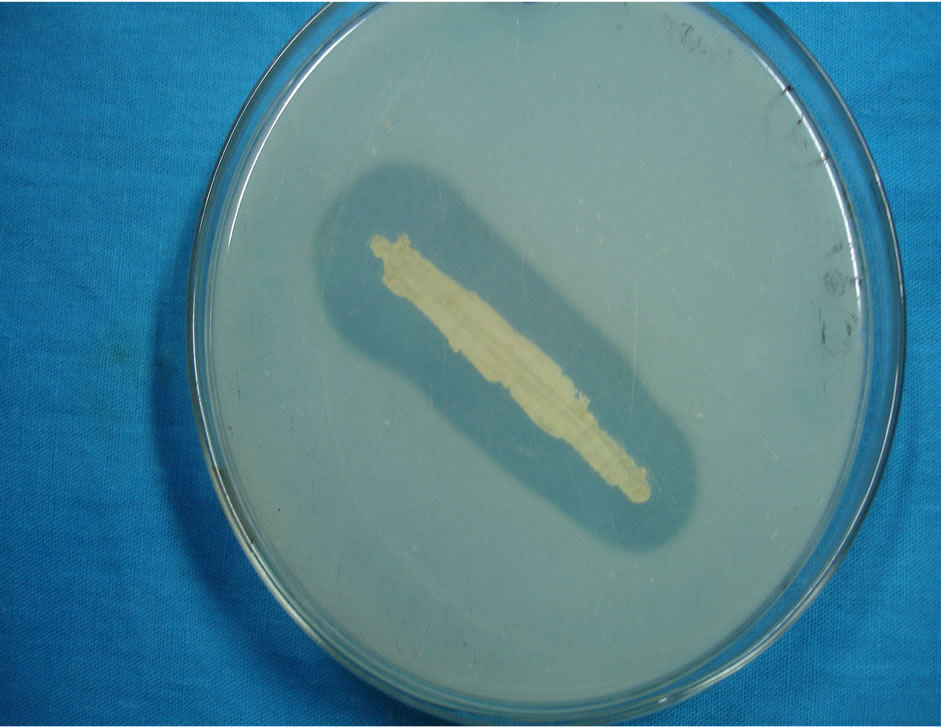
Figure 1. Vibrio sp. in TCBS agar medium.
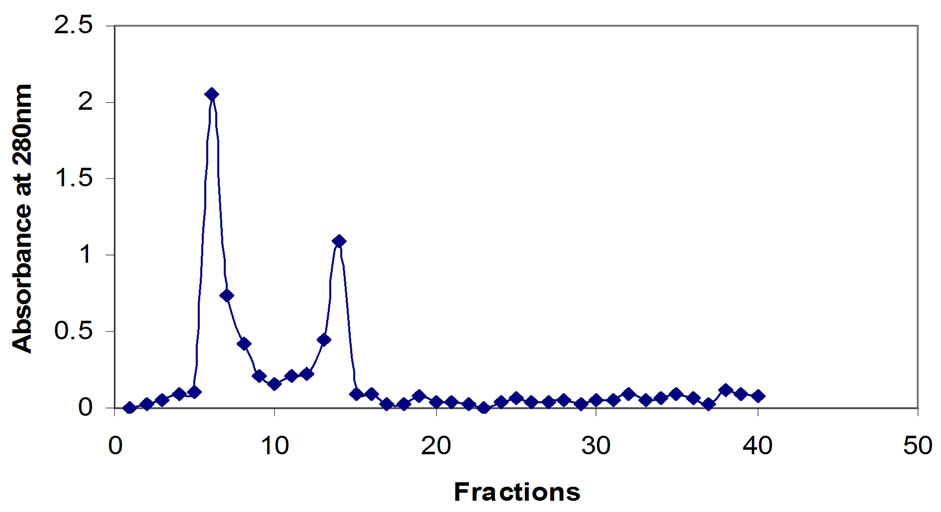
Figure 2. Gel chromatography of chitinase.
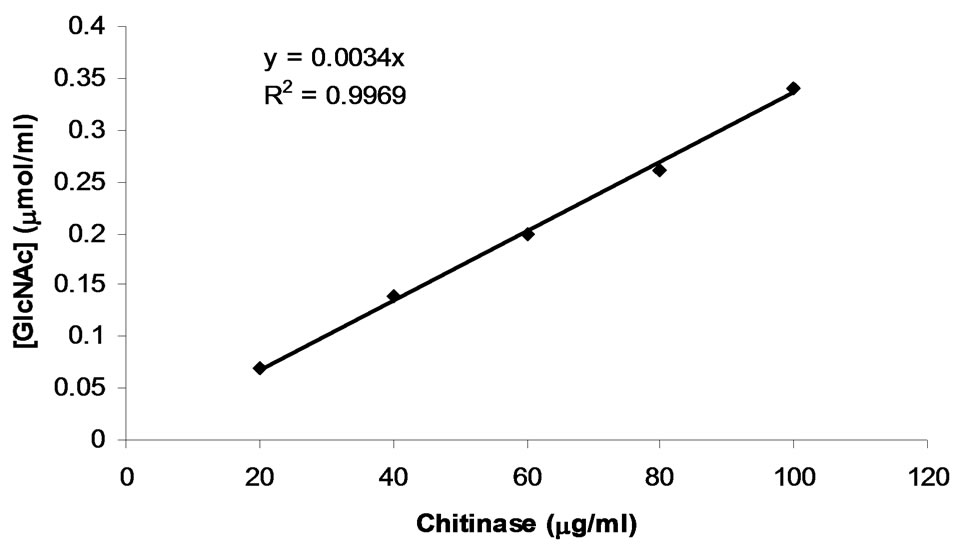
Figure 3. Determination of chitinase activity by the method of standard reducing sugar.
200 kDa) and chitinase (Sigma) (Figure 4).
3.9. pH Optimum of Chitinase
The effect of pH of the chitinase was studied with citratephosphate buffer (pH 4.5 - 6.5), phosphate buffer (pH 6.5 - 8.5), and glycine-NaOH buffer (pH 8.5 - 10.0) under the standard conditions. The pH activity of chitinase was maximum between pH 6.0 - 6.5 (Figure 5). Similarly the other marine bacterial chitinases showed broader pH optima or were more active in neutral or slightly alkaline
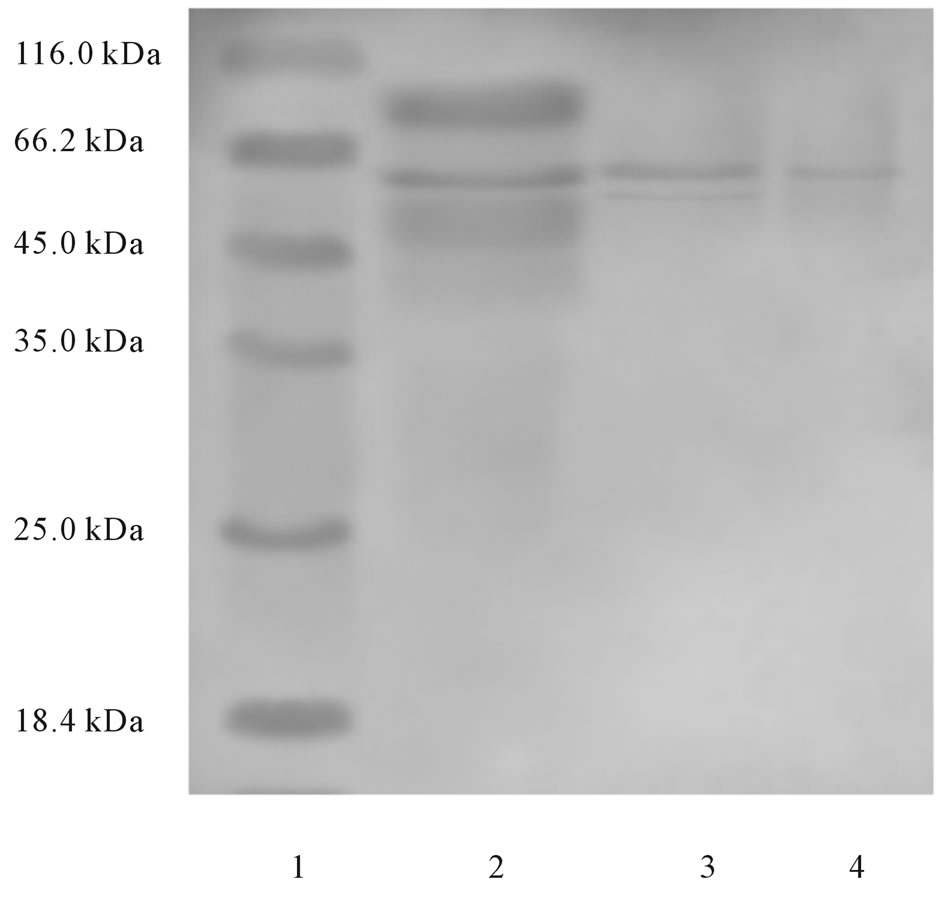
Figure 4. SDS-PAGE of chitinase. Lane 1: Molecular weight markers; Lane 2: Ammonium sulphate precipitated proteins; Lane 3: Sephadex G-100 purified chitinase; Lane 4: Standard chitinase.
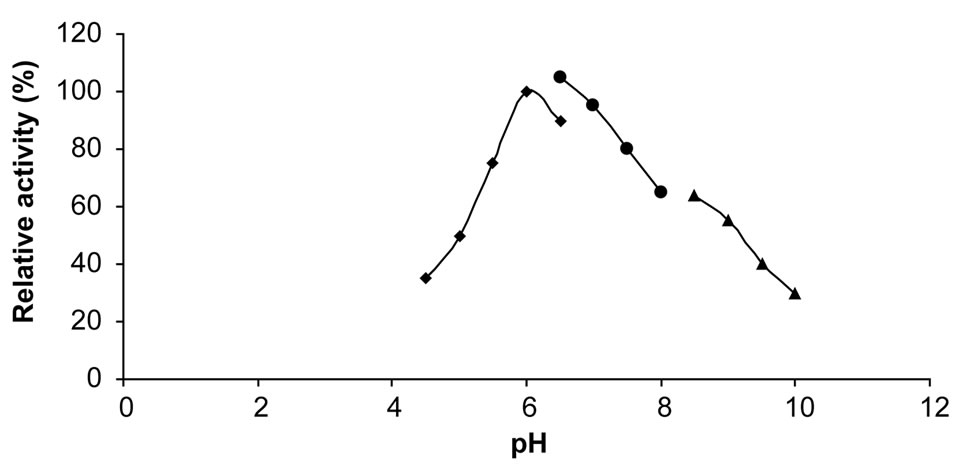
Figure 5. Effect of pH on activity of chitinase (■: citrate-phosphate buffer; ●: phosphate buffer; ▲: glycine-NaOH buffer).
conditions [19].
3.10. Effect of Temperature of Chitinase
The optimum temperature of chitinase was 45˚C (Figure 6). Chitinase I of strain 98CJ11027 showed a similar temperature optimum to those of other marine bacteria (Aeromonas hydrophila H-2330) [20], Alteromonas sp. and Pseudomonas aeroginosa K-187 [19].
In the present investigation chitin extracted from the cuttlebone of S. inermis was found to be 22%, this is higher than the result of Tolaimate et al. [7], reported 20% of chitin was found in the cuttlebone of S. officinalis. Whereas, in general, the squid/ octopus contain 3% - 20% and dry Squilla contains 15% of chitin [21].
Zhou et al. [22] found that the chitinase enzyme showed optimum activity pH at 6.5 and optimum temperature at 50˚C. Further it was stable in the pH range of 4 to 9 and temperature below 40˚C, during 60 and 120 min treat-
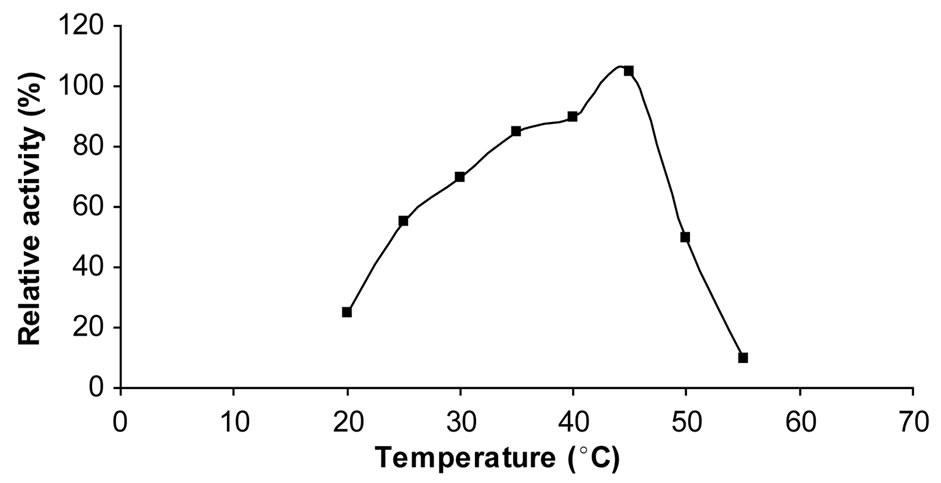
Figure 6. Effect of temperature on chitinase activity.
ment respectively. The enzyme produced by Vibrio sp. was found to be represented by a single protein band of molecular weight about 30 kDa, on SDS-PAGE. In terms of molecular mass comparison, the chitinase from Vibrio sp. 1211 seems to be different from those of other microbes. Almost all of the chitinases from microorganisms have higher molecular weights: 42 kDa for the enzyme from fungi Piromyces sp.; 59 - 89 kDa for that from Bacillus lichiniformis; 41 - 44 kDa for that from Streptomyces sp. The chitinases from the marine bacterium Aeromonas hydrophila also reported a higher molecular weight of 62 kDa. The present study, the pH (6.0 - 6.5), temperature (45˚C) and the molecular weight of the chitinase produced by Vibrio sp. was found to be higher and it was estimated as 50 - 60 kDa.
Dahiya et al. [4] observed that Enterobacter sp. NRG4 secretes extracellular chitinase and molecular weight was found to be about 60 kDa. Chtinase from Enterobacter sp. NRG4 was active over a broad pH range i.e. from pH 4.5 to 8.0, optimum being 5.5. Wang et al. [10] found that the chitinase activity increased along with the cell growth (as judged by OD660) and reached maximum when the cell growth reached mid-exponential phase at 3 days of incubation and the molecular mass of Bacillus subtilis W-118 chitinase was estimated as 20.6 kDa. Park et al. [5] isolated twenty eight different chitinase producing bacteria and they identified through the morphological, physiological, biochemical characteristics and 16s rDNA sequence as all are member of the genus Vibrio.
4. CONCLUSION
The chitin yield was 22% in the cuttlebone of S. inermis and the molecular weight of the purified extra cellular chitinase was found to be 50 - 60 kDa, optimum pH between 6.0 - 6.5 and the optimum temperature was found to be 45˚C. Further study in this line is required to purify the enzyme by HPLC, sequenced, determine the kinetics and synthesized by rDNA technology.
REFERENCES
- Svitil, L.A., Ni-Chandhain, M.S., Moore, A.J. and Kirchman, L.D. (1997) Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Applied Environmental Microbiology, 63, 408-413.
- Tombs, E. and Harding, A. (1998) An introduction to polysaccharide biotechnology. Taylor and Francis Publications, London, 144-151.
- Ravikumar, M.N.V. (2000) A review of chitin and chitosan applications. Reaction and Functional polymer, 46, 1-27.
- Dahiya, N., Tewari, R., Tiwari, P.R. and Hoondal, S.G. (2005) Chitinase from Enterobacter sp. NRG4: Its purification, characterization and reaction pattern. Electronic Journal of Biotechnology, 8, 134-145. doi:10.2225/vol8-issue2-fulltext-6
- Park, S.H., Lee, J. and Lee, W.C. (2000) Purification and characterization of chitinase from a marine bacterium Vibrio sp. 98CJ11027. Journal of Microbiology, 38, 224- 229.
- Takiguchi, Y. (1991) Preparation of chiotosan and partially deacetylated chitin. In: Otakara, A. and Yabuki, M., Eds., Chitin, Chitosan-Jikken Manual, Gihodou Shupan Kabushki Kasisha, Japan, 9-17.
- Tolaimate, A., Debrieres, J., Rhazi, M. and Alagui, A. (2003) Contribution to the preparation of chitins and chitosans with controlled physico-chemical properties. Polymers, 44, 7939-7952.
- Shigemasa, Y., Matsuura, H., Sashwa, H. and Saimoto, H. (1996) Evaluation of different absorbance ratios from infrared spectroscopy for analyzing the degree of deacetylation in chitin. International Journal of Biological Macromolecule, 18, 237-242. doi:10.1016/0141-8130(95)01079-3
- Baxter, A., Dillon, M., Antony Taylor, K.D. and Roberts, G.A.F. (1992) Improved method for determination of the degree of N-acetylation of chitosan. International Journal of Biological Macromolecule, 14, 166-169. doi:10.1016/S0141-8130(05)80007-8
- Wang, S.L., Lin, T.Y., Yen, Y.H., Liao, H.F. and Chen, Y.J. (2006) Bioconversion of shellfish chitin wastes for the production of Bacillus subtilis W-118 chitinase. Carbohydrate Research, 341, 2507-2515. doi:10.1016/j.carres.2006.06.027
- Rodriguez-Kabana, R., Godoy, G., Morgan-Jones, G. and Shelby, R.A. (1983) Effects of chitin amendments to soil on Heterodera glycines, microbial populations and colonization of cysts by fungi. Plant Soil, 75, 95-106. doi:10.1007/BF02178617
- Imoto, T. and Yagishita, K. (1971) A simple activity measurement of lysozyme. Agriculture Biological Chemistry, 35, 1154-1156. doi:10.1271/bbb1961.35.1154
- Imanaka, T., Fukui, T. and Fujiwara, S. (2001). Chitinase from Thermococcus kodakaraensis KOD1. Methods of Enzyme, 330, 319-329. doi:10.1016/S0076-6879(01)30385-3
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248-254. doi:10.1016/0003-2697(76)90527-3
- Guo, S.H., Chen, J.K. and Lee, W.C. (2004) Purification and characterization of extracellular chitinase from Aeromonas schubertii. Enzyme Microbial Technology, 35, 550- 556. doi:10.1016/j.enzmictec.2004.08.025
- Sambrook, J. and Russell, D.W. (2001) Molecular cloning: A laboratory manual. Cold Spring Habor Lab Press, Long Island, A8.40-A8.51.
- Pearson, F.G., Marchessault, R.H. and Liang, C.Y. (1960) Infra red spectra of crystalline polysaccharides V chitin. Journal of Polymer Science, 13, 101-106. doi:10.1002/pol.1960.1204314109
- Baumano, P., Furaiss, A.L. and Lee, J.V. (1984) Bergey’s manual of systematic bacteriology. Williams & Wilkins Co., London.
- Wang, S.L. and Chang, W.T. (1997) Purification and characterization of two bifunctional chitinases/lysozymes extracellularly produced by Pseudomonas aeruginosa K- 187 in a shrimp and crab shell powder medium. Applied Environmental Microbiology, 63, 380-386.
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 277, 680-685. doi:10.1038/227680a0
- Patil, Y.T. and Satam, S.B. (2002) Chitin and chitosan: Treasure from crustacean shell waste. Sea Food Export Journal, 7, 31-38.
- Zhou, N.S., Yang, Y.C., Huang, L., Cai, C.H. and Lin, Y.C. (1999) Isolation and characterization of chitinase from marine bacterium Vibrio sp. World Journal of Microbiology and Biotechnology, 15, 745-746. doi:10.1023/A:1008932217926
NOTES
*Deparment of Pharmacology, Chettinad Hospital and Research Institute, Kelambakkam, Tamil Nadu, India.

