Advances in Biological Chemistry
Vol.2 No.2(2012), Article ID:18982,9 pages DOI:10.4236/abc.2012.22024
The vascular endothelial growth factor genes expression in glioma U87 cells is dependent from ERN1 signaling enzyme function
![]()
1Department of Molecular Biology, Palladin Institute of Biochemistry, National Academy of Sciences of Ukraine, Kiev, Ukraine
2Department of Pediatrics, National Bogomolets Medical University, Kiev, Ukraine
Email: *ominchenko@yahoo.com
Received 21 March 2012; revised 22 April 2012; accepted 30 April 2012
Keywords: VEGF Expression; ERN1; Hypoxia; Ischemic Conditions; Angiogenesis; Glioma U87 Cells
ABSTRACT
The expression of different vascular endothelial growth factor (VEGF) genes was studied in glioma U87 cells with endoplasmic reticulum-nuclei-1 (ERN1) loss of function and its regulation by hypoxia and glutamine or glucose deprivation conditions as model of ischemia. The blockade of function of the ERN1 enzyme, which is a major sensor of endoplasmic reticulum stress, leads to a decrease of the VEGFA, VEGFB and VEGFC mRNA expression level. The level of VEGFA proteins also decreases at this experimental condition in the cytosolic fraction, but increases in the nuclear fraction. Hypoxia does not affect VEGFC and increases the expression level of VEGFA and VEGFB mRNA in both used cell types, however, the change was much less profound in cells with suppressed function of ERN1. The expression level of VEGFC mRNA decreases in both used cell types in glutamine deprivation condition, however, the change was more profound in control glioma cells. At the same time, the expression level of VEGFA mRNA increases and VEGFB—decreases in glutamine deprivation condition in control glioma cells only. Exposure of glioma cells to glucose deprivation condition increases VEGFB mRNA expression level in both used cell types; however, VEGFA—in control glioma cells only and VEGFC—in cells with ERN1 signaling enzyme loss of function only. Thus, the results of this study clearly demonstrated the downregulation of the expression of all three VEGF genes in glioma cells with ERN1 loss of function which correlates to the suppressed angiogenesis and proliferation rate of these cells. Moreover, the effect of hypoxia and glutamine or glucose deprivation condition on the expression level of all VEGF genes is different and mainly depends on ERN1 signaling enzyme function.
1. INTRODUCTION
A high rate of glycolytic flux as well as an increase in angiogenesis, even in the presence of oxygen, is a central metabolic hallmark of most neoplastic tumors. The high glucose metabolism of cancer cells is caused by a combination of hypoxia-responsive transcription factors, activation of oncogenic proteins, and the loss of the tumor suppressor function. Over-expression of hypoxia inducible factor-1α (HIF-1α) or HIF-2α and v-myc myelocytomatosis viral oncogene homolog (MYC), activation of ras oncogene homolog, and the loss of tumor protein 53 (TP53) and/or other tumor suppressor functions each have been found to stimulate glycolysis and angiogenesis in part by activating a family of regulatory bifunctional 6- phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB), hexokinase and vascular endothelial growth factor (VEGF) gene expression as well as other pro-angiogenic factors [1-7]. Overexpression of VEGFA as well as PFKFB4, PFKFB3 and hexokinase 2 is an obligatory factor of tumor cell glycolysis and increased angiogenesis and proliferation [8-14].
Endoplasmic reticulum stress as well as hypoxia is an important factor of tumor growth. Endoplasmic reticulum to nuclei-1 signaling enzyme (ERN1) is an endoplasmic reticulum-resident transmembrane protein acting as a proximal sensor of a complex set of signaling pathways named the unfolded protein response. The endoplasmic reticulum is a key organelle in the cellular response to ischemia, hypoxia, and some chemicals which activate the unfolded protein response [15]. This adaptive response is activated upon the accumulation of misfolded proteins in the endoplasmic reticulum and is mediated by three endoplasmic reticulum-resident sensors named PERK (PRKlike ER kinase), IRE1/ERN1 (Inositol Requiring Enzyme-1/Endoplasmic Reticulum to Nuclei-1) and ATF6 (Activating Transcription Factor 6), however, endoplasmic reticulum to nuclei-1 is the dominant sensor [16-21]. Activation of the unfolded protein response tends to limit the de novo entry of proteins in to the endoplasmic reticulum and facilitate both the endoplasmic reticulum protein folding and degradation to adapt cells for survival or, alternatively, to enter cell death programs through endoplasmic reticulum-associated machineries [15,19, 22]. As such, it participates in the early cellular response to the accumulation of misfolded proteins in the lumen of the endoplasmic reticulum, occurring under both physiological and pathological conditions.
Two distinct catalytic domains of the bifunctional signaling enzyme endoplasmic reticulum to nuclei-1 were identified: a serine/threonine kinase and an endoribonuclease which contribute to ERN1 signalling. The ERN1- associated kinase activity autophosphorylates and dimerizes this enzyme, leading to the activation of its endoribonuclease domain, degradation of a specific subset of mRNA, and initiation of the pre-XBP1 (X-box binding protein 1) mRNA splicing [23-25]. Mature XBP1 mRNA splice variant encodes a transcription factor that has different C-terminus amino acid sequence and stimulates the expression of hundreds of unfolded protein responsespecific genes [24,26,27] . At the same time, it was shown that a kinase inhibitor activates the ERN1 endoribonuclease to confer cytoprotection against ER stress [28] . It is possible that this activation of the ERN1 endoribonuclease is a result of its interaction with other sensor-signalling systems of endoplasmic reticulum stress.
Single mutations were detected in endoplasmic reticulum to nuclei-1 gene of different human cancers and encoded by this gene enzyme was proposed as a major contributor to tumor (including glioblastoma) progression among protein kinases. The growing tumor requires the endoplasmic reticulum stress as well as hypoxia and ischemia for own neovascularization and growth [14,15] . Drogat et al. [14] have shown that the activation of the endoplasmic reticulum stress sensor IRE1 is a common determinant linking hypoxiaand hypoglycemia-dependent responses to the up-regulation of vascular endothelial growth factor-A (VEGF-A). Moreover, the blockade of main unfolded protein response sensor ERN1 in A549 human lung carcinoma cells and U87 glioma cells (from three to six independent clones were isolated and characterized for each cell line) were unable to trigger VEGF-A up-regulation upon either oxygen or glucose deprivation. Thus, the endoplasmic reticulum stress response-signalling ERN1 pathway is linked to the neovascularization process, tumor growth and cell death processes, because the complete blockade of ERN1 signal transduction pathway has anti-tumor effects preferentially via suppression of VEGFA [14,15,29]. It is important in studying the role of ERN1 signalling pathways in tumor progression, especially in malignant gliomas; it is important in developing a new understanding concerning molecular mechanisms of malignant tumors progression in relation to ischemia/hypoxia and it will help define the best targets for the design of specific inhibitors that could act as potent antitumor drugs.
There are three different genes encoded VEGF: VEGFA, VEGFB and VEGFC, however there are much more protein isoforms of VEGF, especially VEGFA, as a result of alternative splicing and VEGFC via complex proteolytic maturation, generating multiple processed forms which bind and activate VEGFR-3 receptors [12, 30-32]. VEGF genes are member of the PDGF/VEGF growth factor family and encode proteins that are often found as a disulfide linked homodimer. These growth factors are glycosylated signaling mitogens that specifically act on endothelial cells and have various effects, including mediating increased vascular permeability, inducing angiogenesis, vasculogenesis and endothelial cell growth, promoting cell migration, and inhibiting apoptosis as well as cell survival functions [33]. Recently, was found interaction between VEGF, IL-6, u-PA and oncostatin M in prostate cancer which is possible important for tumor neovascularization and growth [31]. Moreover, it was shown that VEGFC differentially regulates VEGFA expression in ocular and cancer cells and promotes angiogenesis via RhoA mediated pathway [32].
The objective of our study was to study the role of ERN1-signaling pathway in tumor angiogenesis and progression by investigation the expression of VEGFB and VEGFC genes as compared to VEGFA in hypoxic and glucose or glutamine deprivation conditions using ERN1-deficient glioma U87 cells.
2. MATERIALS AND METHODS
2.1. Cell Culture
The glioma cell line U87 was obtained from ATCC (USA) and grown in high glucose (4.5 g/l) Dulbecco’s modified Eagle’s minimum essential medium (DMEM; Gibco, Invitrogen, USA) supplemented with glutamine (2 mM), 10% fetal bovine serum (Equitech-Bio, Inc., USA), penicillin (100 units/ml; Gibco) and streptomycin (0.1 mg/ml; Gibco) at 37˚C in a 5% CO2 incubator. In this work we used two sublines of this glioma cell line. One subline was obtained by selection of stable transfected clones with overexpression of vector, which was used for creation of dnERN1. This untreated subline of glioma cells (control glioma cells) was used as control 1 in the study of the effect of hypoxia and glutamine or glucose deprivations on the expression level of different VEGF genes. Second subline was obtained by selection of stable transfected clones with overexpression of ERN1 dominant/negative constructs (dnERN1) and has suppressed both protein kinase and endoribonuclease activities of this signaling enzyme (clone 1C5) [14]. The expression level of VEGF genes in these cells was compared with cells, transfected by vector (control 1), but this subline was also used as control 2 for investigation the effect of hypoxia and glutamine or glucose deprivations on the expression level of VEGF genes under blockade ERN1 function.
Hypoxic conditions were created in special incubator with 3 % oxygen and 5 % carbon dioxide levels; culture plates with complete DMEM were exposed to these conditions for 16 hrs. For glucose or glutamine deprivation the growing medium in culture plates was replaced with a medium without glucose or without glutamine as well as without FBS and thus exposed for 16 hrs.
The suppression level of ERN1 enzymatic activity in glioma cells that over express a dominant-negative construct of endoplasmic reticulum-nuclei-1 (dnERN1) was estimated by analysis of the expression of XBP1 alternative splice variant (XBP1s), a key transcription factor in ERN1 signaling, and phosphorylated isoform ERN1 using cells treated by tunicamycin (0.01 mg/ml during 2 hours).
2.2. RNA Isolation
Total RNA was extracted from different tumor tissues and normal tissue counterparts as described [34]. RNA pellets were washed with 75% ethanol and dissolved in nuclease-free water.
2.3. Reverse Transcription and Quantitative PCR Analysis
The expression levels of different vascular endothelial growth factor (VEGF) mRNA were measured in glioma cell line U87 and its subline (clone 1C5) with a deficiency of endoplasmic reticulum—nuclei-1 by quantitative polymerase chain reaction of complementary DNA (cDNA) using, Mx 3000P QPCR” (Stratagene, USA) and SYBRGreen Mix (AB gene, Great Britain). QuaniTect Reverse Transcription Kit (QIAGEN, Germany) was used for cDNA synthesis as described previously [34]. Polymerase chain reaction was performed in triplicate. For amplification of VEGFA cDNA we used forward (5’-ccttgctgctctacctccac-3’ and reverse (5’-atctgcatggtgatgttgga-3’) primers. The nucleotide sequences of these primers correspond to sequences 1076 - 1095 and 1355 - 1336 of human VEGFA cDNA (GenBank accession number NM_003376). The amplification of VEGFB cDNA was performed using forward primer (5’-gacagtgctgtgaagccaga-3’) and reverse primer (5’-ccctgctgagtctgaaaagc-3’). These oligonucleotides correspond to sequences 428 - 447 and 734 - 715 of human VEGFB cDNA (GenBank accession number NM_003377). The amplification of VEGFC cDNA for real time RCR analysis was performed using two oligonucleotides primers: forward—5’-tgaacaccagcacgagctac-3’ and reverse—5’-catccagctccttgtttggt-3’. The nucleotide sequences of these primers correspond to sequences 951 - 970 and 1292 - 1273 of human VEGFC cDNA (GenBank accession number NM_005429). The amplification of betaactin cDNA was performed using forward—5’-CGTACCACTGGCATCGTGAT-3’ and reverse—5’-GTGTTGGCGTACAGGTCTTT-3’ primers. The expression of beta-actin mRNA was used as control of analyzed RNA quantity. The primers were received from “Sigma” (USA).
The amplification of XBP1 cDNA was performed using HotStarTaq Master Mix Kit (“QIAGEN”, Germany), “MasterCycler Personal” (“Eppendorf”, Germany) and primers: forward—5’-ggagttaagacagcgcttgg -3’ and reverse—5’-tcacccctccagaacatctc-3’. The nucleotide sequences of these primers correspond to sequences 441 - 460 and 608 - 589 of XBP1 mRNA (GenBank accession number NM_005080). The size of amplified fragment is 168 bp for non spliced variant and 142 bp for alternative splice variant (XBP1s). The phosphorylated isoform of ERN1 was measured by Western blot analysis using IRE1p, phosphorylated (Ser724), US Biological.
The ribonuclease protection assay of VEGF and 18 S rRNA levels were mainly measured as described previously [4,12].
The cytosol and nuclear extract fractions from control glioma cells and cells with ERN1 enzyme loss of function were received as described previously [35]. VEGFA protein was measured by Western blot analysis using polyclonal anti-VEGFA (Santa Cruz Biotechnology) both in cytosol and nuclear extract from control cells and cells with ERN1 enzyme loss of function. ACTB (beta-actin) and Lamin B1 (Santa Cruz Biotechnology) were used as control of analyzed protein quantity in cytosol and nuclear extract of glioma cells.
An analysis of quantitative PCR was performed using special computer program “Differential expression calculator” and statistic analysis – in Excel program. The values of VEGFA, VEGFB and VEGFC mRNA expressions were normalized to the expression of beta-actin mRNA and represent as percent of control (100%). All values are expressed as mean ± SEM from triplicate measurements performed in 4 independent experiments. The amplified DNA fragments were separated on a 2% agarose gel and that visualized by 5x Sight DNA Stain (EUROMEDEA).
3. RESULTS AND DISCUSSION
3.1. Characteristics of U87 Glioma Cells with ERN1 Loss of Function
The analysis of the suppression level of the function of endoplasmic reticulum stress response-signalling endoplasmic reticulum to nuclei-1 enzyme in glioma cells that over express a dominant-negative construct of ERN1 (dnERN1) was estimated by determining the expression of XBP1 alternative splice variant (XBP1s), a key transcription factor in ERN1 signaling, and by the level of the phosphorylated isoform ERN1 using cells treated by tunicamycin (0.01 mg/ml during 2 hrs). Alternative splice variant of XBP1 mRNA is a shorter variant which encodes a longer protein isoform of the transcription factor.
As shown in Figure 1, treatment of control (transfected by vector) glioma U87 cells with tunicamycin leads to the formation of an alternative splice variant of XBP1 while in cells transfected by dnERN1 the alternative splicing of XBP1 is completely blocked. Moreover, endoplasmic reticulum stress induced by tunicamycin enhances phosphorylation of ERN1, this effect of tunicamycin, however, is completely eliminated in glioma cells transfected by dnERN1 as is demonstrated by a Western blot analysis using antibody against phosphorylated IRE1(Ser724). Thus, in fact, the U87 glioma cells that over express a dominant-negative construct of ERN1 used in this study are cells with an ERN1 enzyme loss of function, because the ERN1 enzyme is not phosphorylated and XBP1 cannot create the alternative splice variant, which is a key transcription factor in ERN1 signaling in endoplasmic reticulum stress condition.

Figure 1. Effect of tunicamycin (0.01 mg/ml during 2 hr) on the level of phosphoERN1 (Western blot analysis) and expression level of transcription factor XBP1 and its alternative splice variant (XBP1s) mRNA (reversetranscriptase-mediated PCR analysis) in glioma U87 cells stable transfected with vector and dnERN1.
Thus, this data demonstrates that the blockade of the ERN1 signaling enzyme function by dnERN1 construct completely suppresses the formation of alternative splice variant of XBP1 and the main biological function of ERN1.
3.2. Expression of Different VEGF Genes in U87 Glioma Cells with ERN1 Loss of Function
We have studied the effect of the blockade the ERN1 enzyme function on the expression of different VEGF genes in glioma cells using qPCR analysis. As shown in Figure 2, the expression level of VEGFA, VEGFB and VEGFC mRNA in glioma cells, deficient in the response-signaling enzyme ERN1, decreases compared with the control U87 cells, transfected by vector: −27%, −12% and −20%, correspondingly. Decrease of VEGFA mRNA expression in glioma U87 cells with suppressed function of signaling enzyme ERN1 (clone 1C5) correlate with data Drogat et al. [14] which have also found reduced level of VEGFA mRNA and protein in other clone of glioma U87 cells without ERN1 function—C6.
Thus, the mRNA expression level of these genes depends upon the function of the ERN1 signaling enzyme. These results correlate with the anti-tumor effects (decreased neovascularization process and tumor growth) of complete blockade of ERN1 signal transduction pathway [14,15,29].
3.3. Effect of Hypoxia and Ischemic Conditions on the mRNA Expression Level of Different VEGF Genes in U87 Glioma Cells and Its Dependence upon the ERN1 Response-Signalling Enzyme Function
We have also studied the effect of hypoxia and glutamine
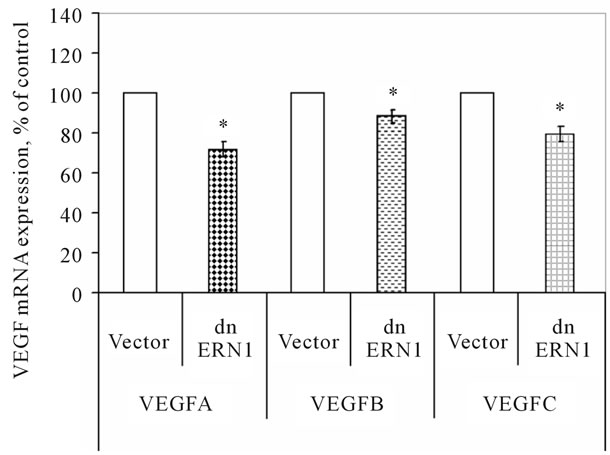
Figure 2. Effect of signaling enzyme ERN1 blockade (dnERN1) on the expression levels of VEGFA, VEGFB and VEGFC mRNA in glioma U87cells measured by qPCR. *P < 0.05 as compared to control (Vector).
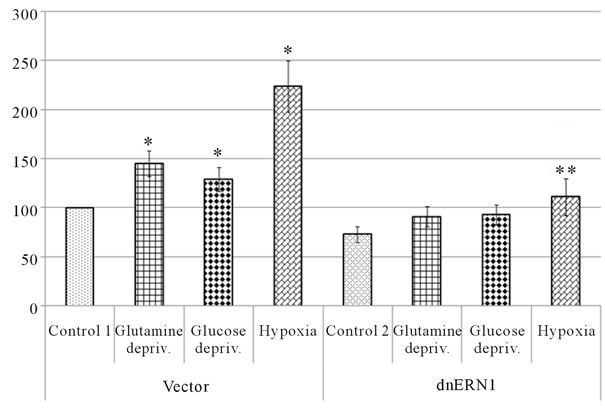
or glucose deprivation conditions (for 16 hrs) on the mRNA expression level of different VEGF genes in U87 glioma cells as well as the dependence of hypoxic and ischemic regulation of VEGF transcripts upon the ERN1 response-signalling enzyme function. As shown in Figure 3(a), the expression level of VEGFA mRNA, measured by qPCR, significantly increases under hypoxia in both used glioma cell types: 2.2 fold in control glioma cells and only +52% in cells with ERN1 enzyme loss of function. Similar results were received using ribonuclease protection assay (Figures 3(b) and (c)). Thus, hypoxia strongly induces VEGFA mRNA expression in U87 control glioma cells growing in DMEM containing 10 % FBS; however, the change was much less profound in cells with suppressed function of ERN1 (clone 1C5).
Similar experiments were provided by Drogat et al. [14], but they received less profound induction of VEGFA mRNA expression by hypoxia in control U87 cells without any significant changes in cells, stably transfected with dnERN1; however, experiments were carried out using C6 clone of modified U87 glioma cells growing in DMEM containing 1% FBS. This discrepancy in the induction of VEGFA expression level under hypoxia is probably due to differences in experimental conditions: different clones of used cells (C6 and 1C5) and different levels of FBS (1% and 10%) as well as glucose (1 and 4.5 g/l) in the medium.
We have also found that glutamine and glucose deprivation conditions lead to an increase of the expression level of VEGFA mRNA in control glioma cells only (+45% and +29%, correspondingly) as compared to control 1 (Figure 3(a)). Similar changes in VEGFA mRNA expression in control glioma U87 cells we received using ribonuclease protection assay (+42% and +31%, correspondingly, for glutamine and glucose deprivation conditions; Figure 3(c)). Thus, VEGFA gene expression is also sensitive to the glutamine and glucose deprivation conditions, as model of ischemia; however, the suppression of the ERN1 signaling enzyme function reduces effect of glutamine and glucose deprivation on the expression of VEGFA transcript in glioma cells.
The strong induction of VEGFA mRNA expression in U87 glioma cells under glucose deprivation condition was previously shown by Drogat et al. [14]. At the same time, no significant changes in VEGFA expression were found in cells expressing a dominant-negative IRE1 transgene. Thus, glioma cells with suppressed IRN1 enzyme function were unable to trigger VEGFA up-regulation upon glucose deprivation [14]. It is possible that less profound induction of VEGFA mRNA expression in control U87 cells under glucose deprivation condition in our experiments is probably due to differences in experimental conditions in experiments of Drogat et al. [14] and our experiments: different clones of used cells (C6
 (a)
(a)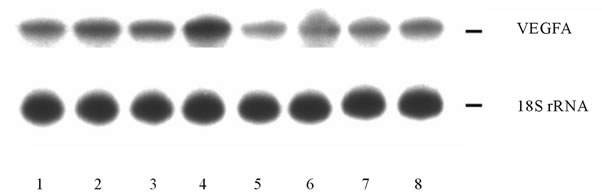 (b)
(b) (c)
(c)
Figure 3. Effect of hypoxia and on the expression of VEGFA mRNA in control (Vector) and signaling enzyme ERN1 loss of function (dnERN1) glioma U87 cells measured by qPCR (a) and ribonuclease protection assay (b). *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2. 1: Control 1; 2 and 6: Glutamine deprivation condition; 3 and 7: Glucose deprivation condition; 4 and 8: Hypoxia; 5: Control 2. (c) Quantification of ribonuclease protection assay; n = 4.
and 1C5) and different levels of glucose (1 and 4.5 g/l) in the DMEM medium during cells growing as well as different levels of FBS (1% and 0%, correspondingly) under glucose deprivation condition.
Induction of VEGFA mRNA expression in U87 glioma cells under glutamine deprivation condition correlates with data Drogat et al. [36] which received similar results using A549/8 human lung carcinoma cells [36]; however, they did not studied the VEGFA mRNA expression in glioma cells and in cells with suppressed function of ERN1. Thus, we have shown for the first time that blockade the ERN1 enzyme function eliminate the induction of VEGFA mRNA expression in U87 glioma cells under glutamine deprivation condition.
Investigation of VEGFB mRNA expression shows that exposure of cells to hypoxia also leads to an increase of this mRNA expression level, but in glioma cells with suppressed function of the ERN1 signaling enzyme effect of hypoxia was significantly less: +55% in control glioma cells, as compared to control 1, and only +16% in cells with ERN1 loss of function, as compared to control 2 (Figure 4). Moreover, these results clearly demonstrated that VEGFA gene expression is more sensitive to hypoxia as compared to VEGFB gene.
As shown in Figure 4, exposure of cells to glucose deprivation condition also leads to an increase in the expression level of VEGFB transcript in both used glioma cell types: +77% in control glioma cells, as compared to control 1, and +61% in cells with suppressed function of the ERN1 signaling enzyme, as compared to control 2. At the same time, the expression level of VEGFB transcript decreases in control glioma cells exposed to the glutamine deprivation condition (−25%); however, no significant changes were found in glioma cells with ERN1 signaling enzyme loss of function. Thus, effect of glutamine and glucose deprivation conditions on the expression level of VEGFB mRNA was different in control glioma cells and in cells with ERN1 enzyme loss of function, while VEGFA mRNA expression level changes in the same way at these experimental conditions.
The expression level of VEGFC mRNA does not change significantly under hypoxia both in control glioma cells and in cells with ERN1 response-signalling enzyme loss of function (Figure 5). It was also shown that the exposure of glioma cells to glucose deprivation condition does not change significantly the expression level of VEGFC mRNA in control glioma cells, as compared to control 1, but increases (+48%) in cells with suppressed function of the response-signalling enzyme ERN1, as compared to control 2. At the same time, exposure of cells to glutamine deprivation condition leads to significant decrease the expression level of VEGFC transcript in both used glioma cell types: almost 2 fold in control glioma cells, as compared to control 1, and only −15% in cells with suppressed function of the ERN1 signaling enzyme, as compared to control 2 (Figure 5). Thus, hypoxia does not affect the expression of VEGFC gene in both used cell types; however, glutamine deprivation condition leads to suppression of the expression level of VEGFC mRNA in both used cell types, but the change was more profound in control glioma cells. At the same time, the exposure of cells to glucose deprivation condition increases VEGFC mRNA expression level in cells with ERN1 signaling enzyme loss of function only.
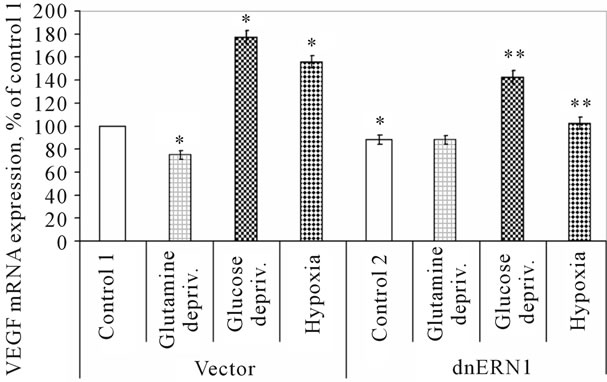
Figure 4. Effect of hypoxia and glutamine or glucose deprivation on the expression of VEGFB mRNA in control (Vector) and signaling enzyme ERN1 loss of function (dnERN1) glioma U87 cells measured by qPCR. *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
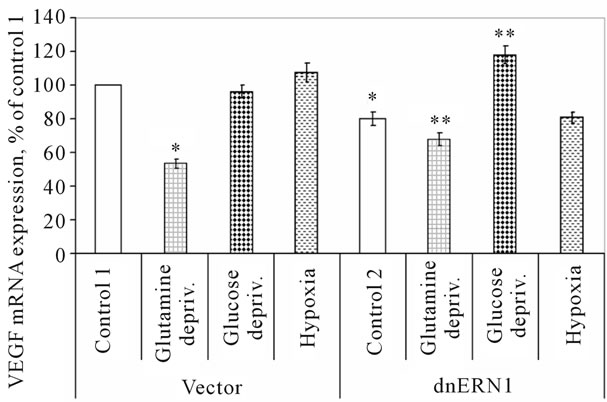
Figure 5. Effect of hypoxia and glucose or glutamine deprivation on the expression of VEGFC mRNA in control (Vector) and signaling enzyme ERN1 loss of function (dnERN1) glioma U87 cells measured by qPCR. *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
The results of this study clearly demonstrated the downregulation of the expression of all three VEGF genes in glioma cells with ERN1 loss of function which correlates to the suppressed angiogenesis and proliferation rate of these cells. Moreover, the effect of hypoxia and glutamine or glucose deprivation condition on the expression level of all VEGF genes is different and mainly depends on ERN1 signaling enzyme function.
3.4. Effect of Signaling Enzyme ERN1 Blockade on the Level of VEGFA Protein in Glioma Cells
We have also studied the VEGFA protein level in control glioma U87 cells and glioma cells with ERN1 loss of function. As shown in Figure 6, two major bands of VEGFA protein are present in both control glioma cells and glioma cells with ERN1 loss of function, but there is a
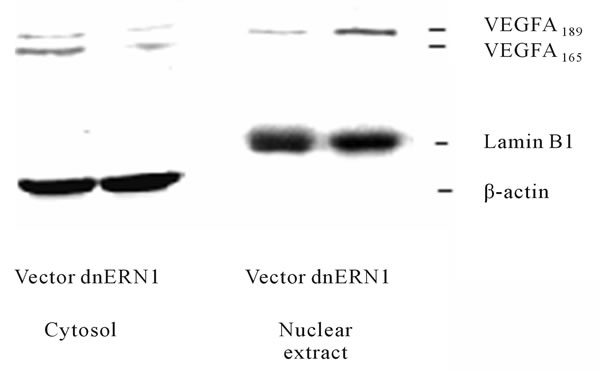
Figure 6. Effect of signaling enzyme ERN1 blockade on the expression levels of VEGFA protein in the cytosol and nuclear extract fractions of control (Vector) and ERN1 loss of function (dnERN1) glioma U87 cells measured by Western blot analysis. The ACTB (β-actin) and lamin B1 proteins were used as control of loaded protein quantity of cytosol and nuclear extract fractions, respectively.
significant decrease of both VEGFA proteins in cytoplasmic fraction after blockade of ERN1 signaling enzyme function that correlates with the downregulation of VEGFA mRNA. These two isoforms of VEGFA proteins correspond to VEGFA-189 and VEGFA-165 which contains 189 and 165 amino acid residues, respectively. However, VEGFA-189 variant was also identified in the nuclear fraction of glioma cells. It was also shown that blockade the ERN1 signaling enzyme function leads to increase VEGFA-189 protein level in the nuclei of glioma cells. Significant decrease of VEGFA protein under suppression of the ERN1 signaling enzyme function was previously demonstrated in U87 glioma cells and A549 human lung carcinoma cells [14]. This data correlate with our results, but we studied subcellular distribution of VEGFA isoforms and demonstrate its cytoplasmic localization as well as nuclear localization of VEGFA-189, although the biological significance of nuclear VEGFA- 189 remains to be determined.
It is possible that nuclear localization of VEGFA-189 protein is related to its specific role in the nuclei, probably associated with the regulation of transcription in glioma cells with suppressed function of the ERN1 signaling enzyme. This phenomenon is probably due to regulation of angiogenesis and proliferation. Recently, similar results were received for PFKFB3 enzyme, which controls glycolysis via regulation the level of fructose- 2,6-bisphosphate, a key regulator of phosphofructokinase-1 [3]. This enzyme was found in the nuclei and shown that nuclear targeting of PFKFB3 increases proliferation via cyclin-dependent kinase [10]. However, the detailed molecular mechanisms of regulation of different VEGFA protein isoforms by ERN1 signaling system and its biological significance warrants further study.
Results of these investigations demonstrated that the expression of genes encoding different VEGF proteins in glioma cells is mainly regulated by hypoxia, glutamine or glucose deprivation and depends upon the activity of the signaling enzyme endoplasmic reticulum to nuclei-1. We have also shown the different sensitivity of different VEGF genes to hypoxia as well as to glutamine or glucose deprivation conditions both in control glioma cells and cells with ERN1 loss of function. Thus, VEGFA gene is more sensitive to hypoxia as compared to VEGFB gene while VEGFC gene is resistant to this experimental condition.
4. CONCLUSION
The major finding reported here is that the expression of VEGFA, VEGFB and VEGFC genes are dependent on the function of ERN1 signaling enzyme, a major sensor of endoplasmic reticulum stress, in normal conditions, under hypoxia and under nutrient (glutamine and glucose) deprivation conditions and that all three VEGF variants possibly participate in glioma angiogenesis and proliferation. The decrease of VEGFA mRNA in glioma cells with ERN1 loss of function correlates with significant decrease of the VEGFA protein in the cytosolic fraction. Moreover, we have identified VEGFA-189 in the nuclear fraction of glioma cells and have shown that the blockade the ERN1 signaling enzyme function leads to an increase its level; although, the biological significance of this data warrants further study. It is possible that different variants of VEGF proteins play different roles in the ERN1 signaling system associated with endoplasmic reticulum stress in hypoxic and nutrient deprivation conditions as well as in tumor angiogenesis and proliferation. The detailed molecular mechanisms of the regulation of different VEGF genes by ERN1 signaling system under hypoxia and ischemic conditions are complex and warrant further study.
5. ACKNOWLEDGEMENTS
This work was supported by a grant from the National Academy of Sciences of Ukraine (Grant No. 0111U002234). We thank Prof. Michel Moenner (INSERM U1029, France) for permanent support and interest in this work as well as the gift of IRE-1alpha knockdown U87 glioma cells.
![]()
![]()
REFERENCES
- Yalcin, A., Telang, S., Clem, B. and Chesney, J. (2009) Regulation of glucose metabolism by 6-phosphofructo-2- kinase/fructose-2,6-bisphosphatases in cancer. Experimental and Molecular Pathology, 86, 174-179. doi:10.1016/j.yexmp.2009.01.003
- Wolf, A., Agnihotri, S., Micallef, J., Mukherjee, J., Sabha, N., Cairns, R., Hawkins, C. and Guha, A. (2011) Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. Journal of Experimental Medicine, 208, 313-326. doi:10.1084/jem.20101470
- Rider, M. H., Bertrand, L., Vertommen, D., Michels, P.A., Rousseau, G.G. and Hue, L. (2004) 6-phosphofructo-2- kinase/fructose-2,6-biphosphatase: Head-head with a bifunctional enzyme that controls glycolysis. Biochemical Journal, 381, 561-579. doi:10.1042/BJ20040752
- Minchenko, A.G., Leshchinsky, I., Opentanova, I.L., Sang, N., Srinivas, V., Armstead, V.E. and Caro, J. (2002) Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. The Journal of Biological Chemistry, 277, 6183-6187. doi:10.1074/jbc.M110978200
- Minchenko, O., Opentanova, I. and Caro, J. (2003) Hypoxic regulation of the 6-phosphofructo-2-kinase/fructose- 2,6-bisphosphatase gene family (PFKFB-1-4) expression in vivo. FEBS Letters, 554, 264-270. doi:10.1016/S0014-5793(03)01179-7
- Minchenko, O.H., Opentanova, I.L., Minchenko, D.O., Ogura, T. and Esumi, H. (2004) Hypoxia induces transcription of 6-phosphofructo-2-kinase/fructose-2,6-bis-phosphatase 4 gene via hypoxia-inducible factor-1alpha activation. FEBS Letters, 576, 14-20. doi:10.1016/j.febslet.2004.08.053
- Minchenko, O.H., Ochiai, A., Opentanova, I.L., Ogura, T., Minchenko, D.O., Caro, J., Komisarenko, S.V. and Esumi, H. (2005) Overexpression of 6-phosphofructo-2-kinase/ fructose-2,6-bisphosphatase-4 in the human breast and colon malignant tumors. Biochimie, 87, 1005-1010. doi:10.1016/j.biochi.2005.04.007
- Chesney, J. (2006) 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase and tumor cell glycolysis. Current Opinion in Clinical Nutrition & Metabolic Care, 9, 535-539. doi:10.1097/01.mco.0000241661.15514.fb
- Bartrons, R. and Caro, J. (2007) Hypoxia, glucose metabolism and the Warburg’s effect. Journal of Bioenergetics and Biomembranes, 39, 223-229. doi:10.1007/s10863-007-9080-3
- Yalcin, A., Clem, B.F., Simmons, A., Lane, A., Nelson, K., Clem, A.L., Brock, E., Siow, D., Wattenberg, B., Telang, S. and Chesney, J. (2009) Nuclear targeting of 6-phosphofructo-2-kinase (PFKFB3) increases proliferation via cyclin-dependent kinase. The Journal of Biological Chemistry, 284, 24223-24232. doi:10.1074/jbc.M109.016816
- Denko, N.C. (2008) Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nature Reviews Cancer, 8, 705- 713. doi:10.1038/nrc2468
- Minchenko, A., Bauer, T., Salceda, S. and Caro, J. (1994) Hypoxic stimulation of vascular endothelial growth factor expression in vitro and in vivo. Laboratory Investigation, 71, 374-379.
- Minchenko, D.O., Bobarykina, A.Y., Senchenko, T.Y., Hubenya, O.V., Tsuchihara, K., Ochiai, A., Moenner, M., Esumi, H. and Minchenko, O.H. (2009) Expression of the VEGF, Glut1 and 6-phosphofructo-2-kinase/fructose-2,6- bisphosphatase-3 and -4 in human cancers of the lung, colon and stomach. Studia Biologica, 1, 25-34.
- Drogat, B., Auguste, P., Nguyen, D.T., Bouchecareilh, M., Pineau, R., Nalbantoglu, J., Kaufman, R.J., Chevet, E., Bikfalvi, A. and Moenner, M. (2007) IRE1 signaling is essential for ischemia-induced vascular endothelial growth factor-A expression and contributes to angiogenesis and tumor growth in vivo. Cancer Research, 67, 6700- 6707. doi:10.1158/0008-5472.CAN-06-3235
- Moenner, M., Pluquet, O., Bouchecareilh, M. and Chevet, E. (2007) Integrated endoplasmic reticulum stress responses in cancer. Cancer Research, 67, 10631-10634. doi:10.1158/0008-5472.CAN-07-1705
- Hetz, C. and Glimcher, L.H. (2009) Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Molecular Cell, 35, 551-561. doi:10.1016/j.molcel.2009.08.021
- Aragón, T., van Anken, E., Pincus, D., Serafimova, I.M., Korennykh, A.V., Rubio, C.A. and Walter, P. (2009) Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature, 457, 736-740. doi:10.1038/nature07641
- Saito, A., Ochiai, K., Kondo, S., Tsumagari, K., Murakami, T., Cavener, D.R. and Imaizumi, K. (2011) Endoplasmic reticulum stress response mediated by the PERKeIF2-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. The Journal of Biological Chemistry, 286, 4809-4818. doi:10.1074/jbc.M110.152900
- Bi, M., Naczki, C., Koritzinsky, M., Fels, D., Blais, J., Hu, N., Harding, H., Novoa, I., Varia, M., Raleigh, J., Scheuner, D., Kaufman, R.J., Bell, J., Ron, D., Wouters, B.G. and Koumenis, C. (2005) ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO Journal, 24, 3470-3481. doi:10.1038/sj.emboj.7600777
- Blais, J.D., Filipenko, V., Bi, M., Harding, H.P., Ron, D., Koumenis, C., Wouters, B.G. and Bell, J.C. (2004) Transcription factor 4 is translationally regulated by hypoxic stress. Molecular and Cellular Biology, 24, 7469-7482. doi:10.1128/MCB.24.17.7469-7482.2004
- Fels, D.R. and Koumenis, C. (2006) The PERK/eIF2- a/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biology and Therapy, 5, 723-728. doi:10.4161/cbt.5.7.2967
- Luo, D., He, Y., Zhang, H., Yu, L., Chen, H., Xu, Z., Tang, S., Urano, F. and Min, W. (2010) AIP1 is critical in transducing IRE1-mediated endoplasmic reticulum stress response. The Journal of Biological Chemistry, 283, 11905- 11912. doi:10.1074/jbc.M710557200
- Korennykh, A.V., Egea, P.F., Korostelev, A.A., FinerMoore, J., Zhang, C., Shokat, K.M., Stroud, R.M. and Walter, P. (2009) The unfolded protein response signals through high-order assembly of Ire1. Nature, 457, 687-693. doi:10.1038/nature07661
- Romero-Ramirez, L., Cao, H., Nelson, D., Hammond, E., Lee, A.H., Yoshida, H., Mori, K., Glimcher, L.H., Denko, N.C., Giaccia, A.J., Le, Q.T. and Koong, A.C. (2004) XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Research, 64, 5943-5947. doi:10.1158/0008-5472.CAN-04-1606
- Lin, J.H., Li, H., Yasumura, D., Cohen, H.R., Zhang, C., Pannin, B., Shokat, K.M., Lavail, M.M. and Walter, P. (2007). IRE1 signaling affects cell fate during the unfolded protein response. Science, 318, 944-949. doi:10.1126/science.1146361
- J. Hollien, J.H. Lin, H. Li, N. Stevens, P. and Walter, J.S. (2009) Weissman, regulated Ire1-dependent decay of messenger RNAs in mammalian cells. Journal of Cell Biology, 186, 323-331. doi:10.1083/jcb.200903014
- Acosta-Alvear, D., Zhou, Y., Blais, A., Tsikitis, M., Lents, N.H., Arias, C., Lennon, C.J., Kluger, Y. and Dynlacht, D.D. (2007) XBP1 controls diverse cell typeand condition-specific transcriptional regulatory networks. Molecular Cell, 27, 53-66. doi:10.1016/j.molcel.2007.06.011
- Han, D., Upton, J.-P., Hagen, A., Callahan, J., Oakes, S.A. and Papa, F.R. (2008) A kinase inhibitor activates the IRE1alpha RNase to confer cytoprotection against ER stress. Biochemical and Biophysical Research Communications, 365, 777-783. doi:10.1016/j.bbrc.2007.11.040
- Auf, G., Jabouille, A., Guérit, S., Pineau, R., Delugin, M., Bouchecareilh, M., Favereaux, A., Maitre, M., Gaiser, T., von Deimling, A., Czabanka, M., Vajkoczy, P., Chevet, E., Bikfalvi, A. and Moenner, M. (2010) A shift from an angiogenic to invasive phenotype induced in malignant glioma by inhibition of the unfolded protein response sensor IRE1. The Proceeding of the National Academy of Sciences of the United States of America, 107, 15555-15558.
- Ferrara, N., Gerber, H.P. and LeCouter, J. (2003) The biology of VEGF and its receptors. Nature Medicine, 9, 669-676. doi:10.1038/nm0603-669
- Weiss, T.W., Simak, R., Kaun, C., Rega, G., Pfluger, H., Maurer, G., Huber, K. and Wojta, J. (2011) Oncostatin M and IL-6 induce u-PA and VEGF in prostate cancer cells and correlate in vivo. Anticancer Research, 31, 3273-3278.
- Kumar, B., Chile, S.A., Ray, K.B., Reddy, G.E., Addepalli, M.K., Kumar, A.S., Ramana, V. and Rajagopal, V. (2011) VEGF-C differentially regulates VEGF-A expression in ocular and cancer cells; promotes angiogenesis via RhoA mediated pathway. Angiogenesis, 14, 371-380. doi:10.1007/s10456-011-9221-5
- Byrne, A.M., Bouchier-Hayes, D.J. and Harmey J.H. (2005) Angiogenic and cell survival functions of vascular endothelial growth factor (VEGF). Journal of Cellular and Molecular Medicine, 9, 777-794. doi:10.1111/j.1582-4934.2005.tb00379.x
- Minchenko, D.O., Karbovskyi, L.L., Danilovskyi, S.V., Moenner, M. and Minchenko, O.H. (2012) Effect of hypoxia and glutamine or glucose deprivation on the expression of retinoblastoma and retinoblastoma-related genes in ERN1 knockdown glioma U87 cell line. American Journal of Molecular Biology, 2, 142-152. doi:10.4236/ajmb.2012.21003
- Armstead, V.E., Minchenko, A.G., Campbell, B. and Lefer, A.M. (1997) P-selectin is up-regulated in vital organs during murine traumatic shock. FASEB Journal, 11, 1271- 1279.
- Drogat, B., Bouchecareilh, M., North, S., Petibois, C., Deleris, G., Chevet, E., Bikfalvi, A. and Moenner, M. (2007) Acute L-glutamine deprivation compromises VEGF-A upregulation in A549/8 human carcinoma cells. Journal of Cellular Physiology, 212, 463-472. doi:10.1002/jcp.21044
NOTES
*Corresponding author.

