 Vol.2, No.3, 188-194 (2010) doi:10.4236/health.2010.23027 Copyright © 2010 SciRes Openly accessible at http://www.scirp.org/journal/HEALTH/ Health Bayesian analysis of minimal model under the insulin-modified IVGTT Yi Wang1, Kent M. Eskridge2, Andrzej T. Galecki3 1,2Department of Statistics, University of Nebraska-Lincoln, Nebraska, USA 3Institute of Gerontology, University of Michigan, Michigan, USA Received 24 September 2009; revised 23 October 2009; accepted 26 October 2009. ABSTRACT A Bayesian analysis of the minimal model was proposed where both glucose and insulin were analyzed simultaneously under the insulin-mo- dified intravenous glucose tolerance test (IVG- TT). The resulting model was implemented with a nonlinear mixed-effects modeling setup using ordinary differential equations (ODEs), which leads to precise estimation of population pa- rameters by separating the inter- and intra-indi- vidual variability. The results indicated that the Bayesian method applied to the glucose-insulin minimal model provided a satisfactory solution with accurate parameter estimates which were numerically stable since the Bayesian method did not require ap proximation by linearization. Keywords: Minimal Model; Bayesian Analysis; IVGTT; Nonlinear Mixed-Effects Modeling; ODE 1. INTRODUCTION Mixed-ef fect s m odels that i nclude bo th fi xed and ra ndom effects to account for the inter-individual variability are becoming increasingly popular for analysis of population pharmacokinetic/pharmacodynamic (PK/PD) data [1-8, among others]. In these models it is assumed that all responses follow a similar functional form, but that pa- rameters vary a mong indivi du als. By separat ing the i nter- and intra-individual variability , mixed-effects m odels will often lead to more precise estimation of population pa- rameters. Continuous pharmacokinetic processes are often described by systems of ordinary differential equa- tions (ODE) which generally lead to models that are nonlinear in the parameters complicating estimation. However, closed form solutions for systems of differen- tial equations are not always possible therefore num erical solutions of differential equations are necessary in order to deal with many types of population PK/PD problems. The nlmeODE in R given by Tornoe et al. [9] and NLINMIX with ODE in SAS presented by Galecki et al. [10], which can handle the first-order ODE’s by com- bining odesolve with the NLME and NLINMIX respec- tively, are used for parameter estimation in nonlinear mixed-effects models. In addition, WBDiff (WinBUGS Differential Interface) given by Lunn [11] is a useful tool for dealing with pharmacokinetic models defined by ODEs in the Bayesian setting. The analysis based on ODEs may offer practical benefits in terms of easier PK/PD modeling, particularly when more complicated mechanistic models are used [12]. Diabetes is associated with a large number of abnor- malities in insulin metabo lism, ranging from an absolute deficiency to a combination of deficiency and resistance, causing an inability to dispose glucose from the blood stream. Three factors: Insulin sensitivity, Glucose effec- tiveness, and Pancreatic responsiveness, referred to in Pacini and Bergman [13], play an important role for glucose disposal. Failure in any of these may lead to impaired glucose tolerance, or, if severe, diabetes. Quan- titative assessment is possible by the minimal model [14], and may improve classification, prognosi s and therapy of the disease. The minimal model is based on an intrave- nous glucose tolerance test (IVG TT), where g lucose and insulin concentrations in plasma are sampled after an intravenous glucose injection. In patients with impaired glucose tolerant (IGT), the insulin response to glucose may be partially or totally suppressed. Of course, without the insulin response, the glucose disappearance model cannot provide an estimate of the metabolic parameters, since there is no input to the model. The insulin modifi- cation of IVGTT addressed the early problems with the minimal model ‘failures’ by insulin injection at 20 min- utes after glucose injection is given at time zero. Tradi- tionally, in the minimal model the glucose and insulin kinetics are described by two components, where the parameters traditionally have been estimated separately within each component by a nonlinear weighted least squares estim ation tec hnique in a t wo-step pr ocedure [13]. The use of population an alysis to extract all information, such as inter-subject variability, from experimental data  Y. Wang et al. / HEALTH 2 (2010) 188-194 Copyright © 2010 SciRes Openly accessible at http://www.scirp.org/journal/HEALTH/ 189 brings about an improvement in the estimates of popula- tion and individual characteristics. Previous work with the IVGTT focused only on glucose kinetics where insu- lin is treated as a known with no measurement errors for non-Bayesian analysis [10] and for Bayesian analysis [15]. However, the glucose-insulin system is an inte- grated system and could be considered as a whole. An assumption of the minimal model is that glucose and insulin constitute a single dynamical system and impor- tant information is lost in treating the insulin as known. For example, pancreatic responsiveness, one important factor of the individual metabolic portrait, cannot be estimated if insulin is treated as known. Therefore, a Bayesian analysis was adopted in combination with population mixed-effects modeling to estimate the four population and individual metabolic indices simultane- ously with the minimal model of glucose and insulin kinetics using data collected during insulin-modified intravenous glucose tolerance test (IVGTT). No other published wor k wa s ide nti fied t hat a naly zed bot h gluc ose and insulin simultaneously under the insulin-modified IVGTT. 2. METHODS 2.1. Bayesian Computational Algorithm for WBDiff Bayesian inference in WBDiff, which allows the nu- merical solution of arbitrary systems of ODEs within WinBUGS nonlinear mixed models, can be described based on the followin g hierarchical modeling. 1) Suppose a chemical or pharmacokinetic model is given by the systems of first-order ODE’s with the form ),,(/),( txgdttdx , , , (1) )(),( 00 xtx 0 tt where x is an k-dimensional dependent variable vector, g is the structural model while is a q-dimensi onal vector of unknown model parameters. 2) In nonlinear mixed-effects modeling, the within- group variability describing the difference between the observed response value and the predicted value can be modeled as 21 ~[() (,,);] ijijiji ij yNEyfx t (2) where i=1, m subjects and j=1, ni time points; is the solution of Eq.1 and the relationship between the ob- served response y and the predicted variable x is desig- nated by a nonl i near function f. ij x 3) The between-subject variability can be constructed by defining the subject-specific random effects as ),(~ pi MVN (3) where is a vector of mean population ph armacok inetic parameters and is the variance-covariance matrix of between-subj ect random variability. 4) In addition, hierarchical modeling in a Bayesian setting comprises the prior specification, where prior distributions are assigned to , , and . For instance, ~MVNq( , ), -1~Wishartp(R, ), ~Gamma(a, b) (4) The Bayesian inference is based on the following principle: posterior prior x likelihood. That is, the so-called “likelihood function” is used to update “prior beliefs” about some unknown parameters of interest to “posterior beliefs” in the light of observed data. To obtain the posterior estimators, using Monte Carlo approxima- tion we simulate values from the joint posterio r distribu- tion of all the model parameters given the observed data, more specifically, the full conditional distribution. For example, the logarithm of the full conditional distribution for the random effect i can be constructed as follows: m i n jiijii iyfff 11 ),|(log),|(log)|(log 1 11 log||( )( ) 22 T ii 2 11 1 log((, , )) 22 i n m ijiji ij ij nyfx t (5) where the dot notation in denotes the distribution of i conditional upon everything else in the model. The term )|( i f ),|(log i f refers to the logarithm of the prior distribution for i and this is specified to be a multivariate normal distribution with mean vector θ and variance -covariance matrix . The term ),|(log iij yf ,(iij xf refers to the log-likelihood of the jth observation fo r subject i under the model, and the concentration yij was assumed to fol- low a normal distribution with mean ), ij t and variance -1. Since we do not have a closed form for xij from Eq.1, the numerical solution xij has to be obtained from Eq.1 by fixing all the conditioning parameters so that the Gibbs sampler can generate a new value, say i(1) from the full conditional distribution (Eq.5) given the initial values to each unknown parameters (0), (0), and (0). After n such iterations, the algorithm yields a joint sample i(n), which can be used for statistical inference in WBDiff. The full conditional distributions for the other model par ame ters can be co nstru cted i n a si mi lar m anner. 2.2. Population Analysis on Glucose-Insulin Minimal Model In this section, Bayesian analysis in combination with population mixed-effects modeling was used to simulta- neously estimate the four population and individual metabolic indices: insulin sensitivity (SI), glucose effec- tiveness (SG) and pancreatic responsiveness ( 1 and 2), based on the integrated glucose-insulin minimal model  Y. Wang et al. / HEALTH 2 (2010) 188-194 Copyright © 2010 SciRes Openly accessible at http://www.scirp.org/journal/HEALTH/ 190 0 , using data collected during the insulin-modified intra- venous glucose tolerance test (IVGTT). 2.2.1. Bergman’s Modified Minimal Model In an IVGTT study a dose of glucose was administered intravenously over a 60 seconds period to overnight- fasted subjects, 20 min after the glucose bolus, insulin was injected over 1-2 min either into the portal vein or into the femoral vein, and subsequently the glucose and insulin concentrations in plasma were frequently sampled (usually 30 ti mes) over a period of 180 minutes. Pr ofiles of the 10 patients are displayed in Figure 1 [10]. Based on Figure 1, the intravenous glucose dose im- mediately elevates the glucose concentration in the plasma forcing the pancreatic β-cells to secrete insulin. The insulin in the plasma is hereby increased, and the glucose uptake in muscles, liver and tissue is raised by the remote insulin in action. This lowers the glucose con- centration in plasma, implying the β-cells to secrete less insulin, from which a feedback effect arises [16]. The integrate d gluc ose-ins ulin sy stem can be des cribe d by t he following non-linearly coupled system of differential equations, but this approach is not exactly the same model as used in [17]. Here I1 is introduced to account for the injection of insulin at t1 after the glucose bolus during the IVGTT: 1 23 01 1 /(())()(),(0), /( )(( )),(0)0, /(())(()),(0),() b b b dGdtpG tGXt GtGG dXdtp XtpItIX dIdtn ItIG thtIIItI (6) where t=0 is the glucose injection time, + denotes posi- tive reflection, namely, () ,(), (() ) 0, . Gt hGth Gt h otherwise and the model parameters are as explained in [13]. 2.2.2. Bayesian Analysis of Bergman’s Modified Minimal Model Using WBDiff A Bayesian framework for modeling the time-varying glucose and insulin profiles during the IVGTT and in- ter-individual variability requires a three-stage hierar- chical model. At the first stage, glucose values G(tj,θi) and insulin values I(tj,θi) in subject i at time tj were ob- tained as the solutions to Eq.6. The model we consider assumes Gij = G(tj,θi) + εij1 and Iij = I(tj,θi) + εij2, where εijk is a mean zero norm ally and independently distributed error term. An additional assumption about within- subject errors will be forthcoming in due course. Stacking the two response variables Gij and Iij into a single re- sponse vector, an indicator variable for the two res po nses can be used to construct the model function. By combin- ing G() and I(), we obtain (, ), ijkjiij Yft where (,), T ijij ij YGI(, ),1 (, ) (, ),2 ji kji ji Gt k ft It k and θi is a vector of parameters of this model for subject i denoted by 12 (, ). T ij ij ij 1230 011 (,, ,,,,,,,). T iiiiiiiiii i pppGn hItI Here it is further assumed that the time when insulin injection occurs after glucose injection at time zero is also an unknown parameter denoted as t1 which must be es- timated. To account for correlation between the two re- sponse variables m easured on the sam e occasion, we m ay assume the elements of εij are correlated, with vari- ance-covariance matrix τ-1 2. Here for simplicity it is assumed that 2=I2, since the intent in this paper is to introduce the random effects to account for in- ter-individual variability rather than define a vari- ance-covariance structure for random errors to address intra-individual variability. However, the variance func- tion fk2(tj, ) and the weight wj are specified for hetero- geneous within-subject error (εijk) variance for two rea- sons: 1) the glucose and insulin concentration points before 8 minu tes can then be zero-weighted to account for the time taken by the injected glucose to diffuse in its distribution space, which can be ac hieved by setting wj=0 at any tim e points bef ore 8 minutes and wj=1 for others; 2) it is commonly recognized that intra-subject variation of this kind tends to increase with plasma concentration level. That is, the higher the concentration level, the lar- ger the variation so that less weight should be assigned. Thus, we assume the following covariance matrix for the within-subject errors εijk. 21 1 230 (,) 0 |~ 0, 0( k ik k ft N ft ,) . The second stage is characterized by making assump- tions about individual parameters. In particular, it was assumed that the individual parameters were drawn from a multivariate log-normal distribution guaranteeing non- negativity of parameters 1230 0111010 (,,,,,,,,, )~(,), T iiiiiiiii ix pppGnh ItILnormal where is an unknown population mean vector and is an unknown covariance matrix. At the third stage, prior distributions for population parameters , , and τ were specified. These prior distributions were vague repre- senting a ‘lack’ of prior knowledge with: 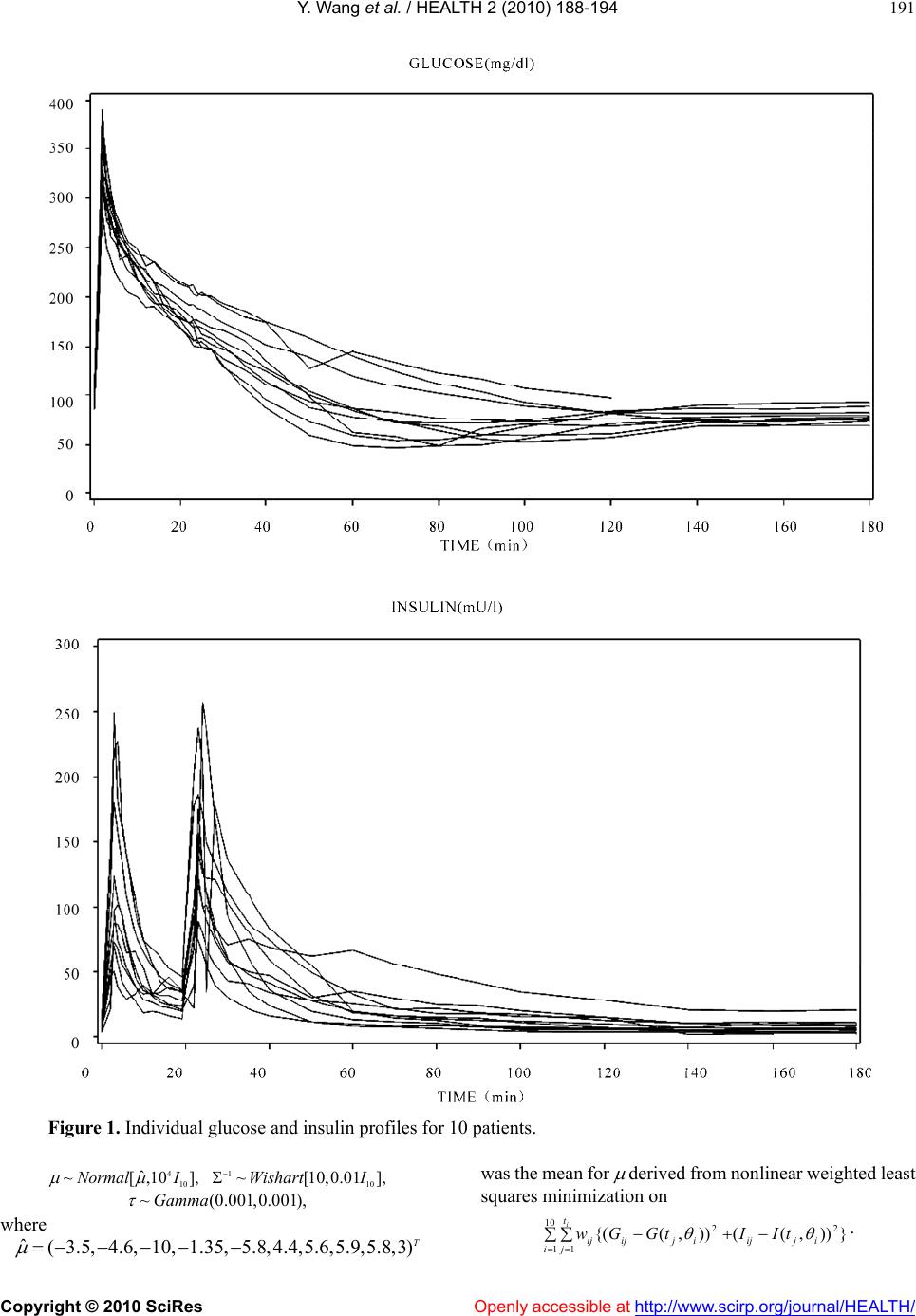 Y. Wang et al. / HEALTH 2 (2010) 188-194 Copyright © 2010 SciRes http://www.scirp.org/journal/HEALTH/Openly accessible at 191 Figur e 1. Individual glucose and insulin profiles for 10 patients. was the mean for derived from nonl inear wei ghted least squares minimization on 410 ˆ ~[,10NormalI ], ~Gamma 110 ~[10,0.01Wishart I (0.001,0.001), ], where })),(()),({( 2 10 11 2ijij i t jijijijtIItGGw i . ˆ(3.5, 4.6, 10,1.35, 5.8,4.4,5.6,5.9,5.8,3)T 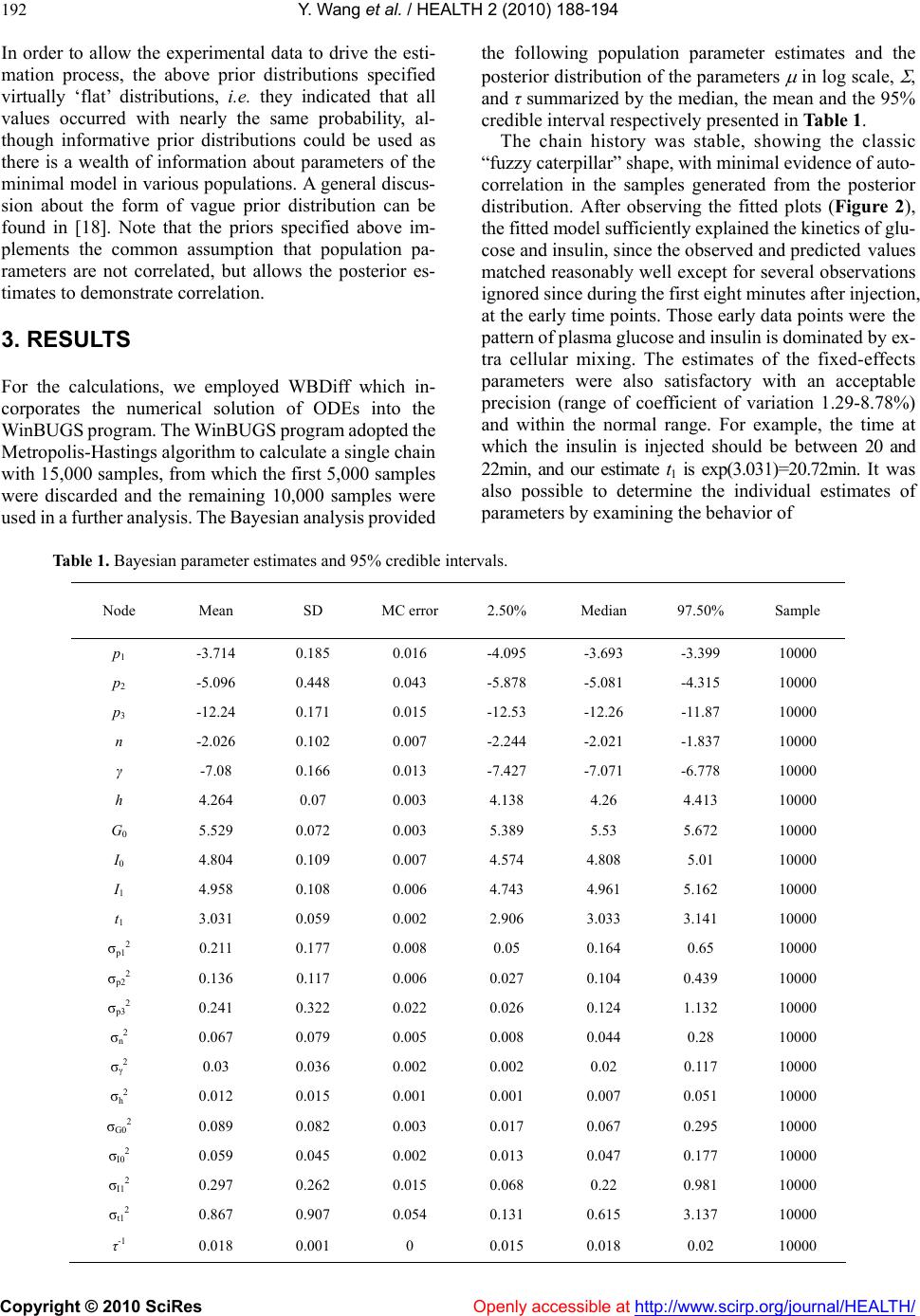 Y. Wang et al. / HEALTH 2 (2010) 188-194 Copyright © 2010 SciRes Openly accessible at http://www.scirp.org/journal/HEALTH/ 192 In order to allow the experimental data to drive the esti- mation process, the above prior distributions specified virtually ‘flat’ distributions, i.e. they indicated that all values occurred with nearly the same probability, al- though informative prior distributions could be used as there is a wealth of information about parameters of the minimal model in v arious populations. A general discus- sion about the form of vague prior distribution can be found in [18]. Note that the priors specified above im- plements the common assumption that population pa- rameters are not correlated, but allows the posterior es- timates to demonstrate correlation. 3. RESULTS For the calculations, we employed WBDiff which in- corporates the numerical solution of ODEs into the W inBUGS program. The W inBUGS program adopted the Metropolis-Hastings algorithm to calculate a single chain with 15,000 samples, from which the first 5,000 samples were discarded and the remaining 10,000 samples were used in a further analysis. The Bayesian analysis provided the following population parameter estimates and the posterior distr ibution of the parameters in log scale, , and τ summarized by the median, the mean and the 95% credible interval respectively presented in Table 1. The chain history was stable, showing the classic “fuzzy caterpillar” shape, with minimal evidence of auto- correlation in the samples generated from the posterior distribution. After observing the fitted plots (Figure 2), the fitted model sufficiently explained the kinetics of glu- cose and insulin, since the observed and predicted values matched reasonably well except for several observations ignored since during the first eight minutes after injection, at the early time points. Those early data points were the pattern of plasma gluc ose and i nsulin is dominated by ex- tra cellular mixing. The estimates of the fixed-effects parameters were also satisfactory with an acceptable precision (range of coefficient of variation 1.29-8.78%) and within the normal range. For example, the time at which the insulin is injected should be between 20 and 22min, and our estimate t1 is exp(3.031)=20.72min. It was also possible to determine the individual estimates of paramete rs by examining the behavi o r o f Table 1. Bayesian parameter estimates and 95% credible intervals. Node Mean SD MC error 2.50% Median 97.50% Sample p1 -3.714 0.185 0.016 -4.095 -3.693 -3.399 10000 p2 -5.096 0.448 0.043 -5.878 -5.081 -4.315 10000 p3 -12.24 0.171 0.015 -12.53 -12.26 -11.87 10000 n -2.026 0.102 0.007 -2.244 -2.021 -1.837 10000 γ -7.08 0.166 0.013 -7.427 -7.071 -6.778 10000 h 4.264 0.07 0.003 4.138 4.26 4.413 10000 G0 5.529 0.072 0.003 5.389 5.53 5.672 10000 I0 4.804 0.109 0.007 4.574 4.808 5.01 10000 I1 4.958 0.108 0.006 4.743 4.961 5.162 10000 t1 3.031 0.059 0.002 2.906 3.033 3.141 10000 σp12 0.211 0.177 0.008 0.05 0.164 0.65 10000 σp22 0.136 0.117 0.006 0.027 0.104 0.439 10000 σp32 0.241 0.322 0.022 0.026 0.124 1.132 10000 σn2 0.067 0.079 0.005 0.008 0.044 0.28 10000 σγ2 0.03 0.036 0.002 0.002 0.02 0.117 10000 σh2 0.012 0.015 0.001 0.001 0.007 0.051 10000 σG02 0.089 0.082 0.003 0.017 0.067 0.295 10000 σI02 0.059 0.045 0.002 0.013 0.047 0.177 10000 σI12 0.297 0.262 0.015 0.068 0.22 0.981 10000 σt12 0.867 0.907 0.054 0.131 0.615 3.137 10000 τ-1 0.018 0.001 0 0.015 0.018 0.02 10000 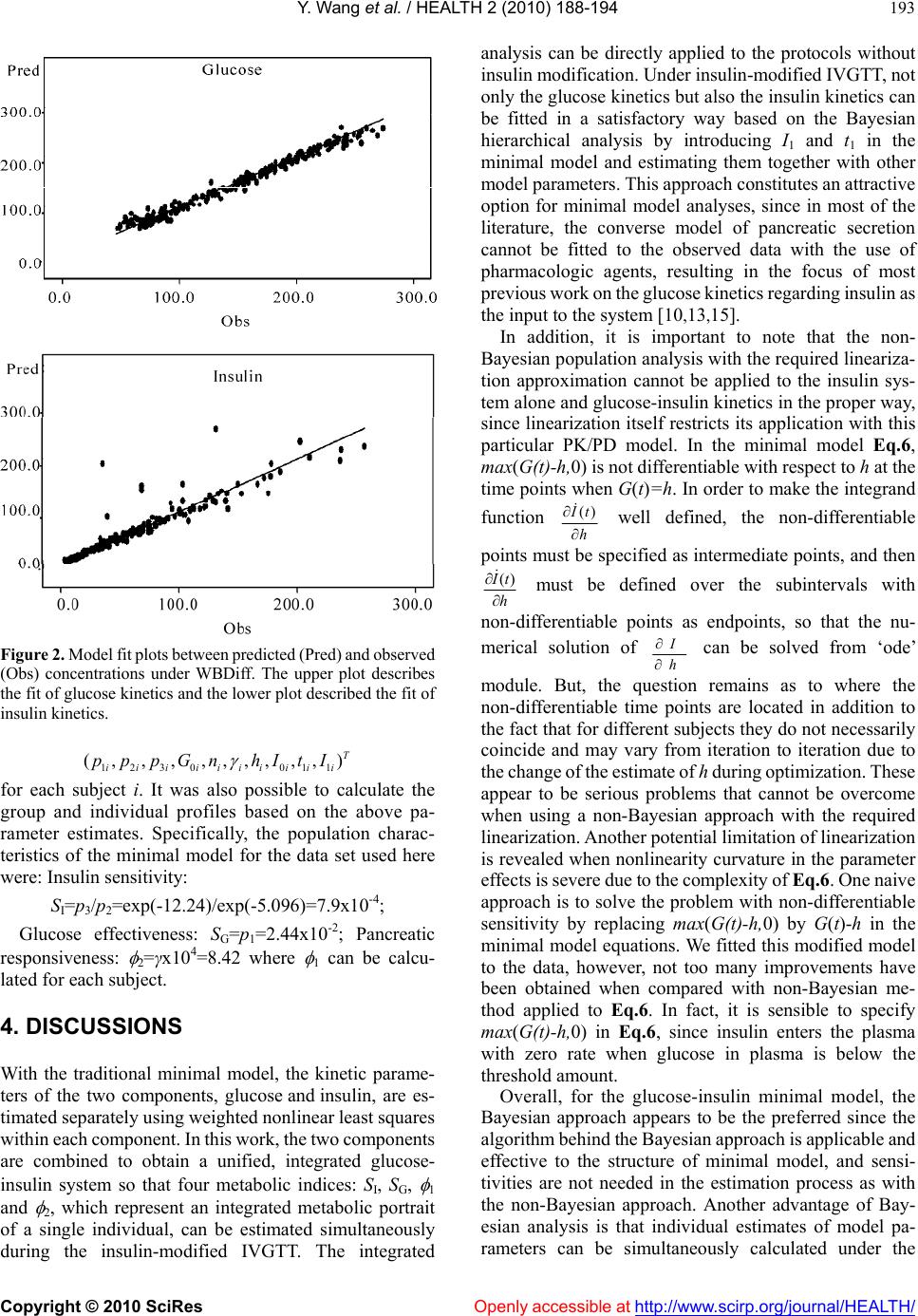 Y. Wang et al. / HEALTH 2 (2010) 188-194 Copyright © 2010 SciRes Openly accessible at http://www.scirp.org/journal/HEALTH/ 193 Figure 2. Model fit plots between predicted (Pred) and observ ed (Obs) concentrations under WBDiff. The upper plot describes the fit of glucose kinetics and the lower plot described the fit of insulin kinetics. 1230 011 (,,,,,,,,, ) T iiiiiiiiii pppGn hItI for each subject i. It was also possible to calculate the group and individual profiles based on the above pa- rameter estimates. Specifically, the population charac- teristics of the minimal model for the data set used here were: Insulin sensitivity: SI=p3/p2=exp(-12.24)/exp(-5.096) =7.9x10-4; Glucose effectiveness: SG=p1=2.44x10-2; Pancreatic responsiveness: 2=γx104=8.42 where 1 can be calcu- lated for each subject. 4. DISCUSSIONS With the traditional minimal model, the kinetic parame- ters of the two components, glucose and insulin, are es- timated separately using weighted nonlinear least squares within each c omponent. In thi s work, the two com ponents are combined to obtain a unified, integrated glucose- insulin system so that four metabolic indices: SI, SG, 1 and 2, which represent an integrated metabolic portrait of a single individual, can be estimated simultaneously during the insulin-modified IVGTT. The integrated analysis can be directly applied to the protocols without insulin m odificat ion. Unde r insul in-m odifie d IVGT T, not only the glucose kinetics but also the insulin kinetics can be fitted in a satisfactory way based on the Bayesian hierarchical analysis by introducing I1 and t1 in the minimal model and estimating them together with other model parameters. This approach constitutes an attractive option for minimal model analyses, since in most of the literature, the converse model of pancreatic secretion cannot be fitted to the observed data with the use of pharmacologic agents, resulting in the focus of most previous wor k on the glucose ki netics regarding insulin as the input to the system [10,13,15]. In addition, it is important to note that the non- Bayesian population an alysis with the required lin eariza- tion approximation cannot be applied to the insulin sys- tem alone and glucose-insulin kinetics in the proper way, since linearization itself restricts its application with th is particular PK/PD model. In the minimal model Eq.6, max(G(t)-h,0) is not differentiable with respect to h at the time points when G(t)=h. In order to make the integrand function () t h well defined, the non-differentiable points must be specified as intermediate points, and then () t h must be defined over the subintervals with non-differentiable points as endpoints, so that the nu- merical solution of h can be solved from ‘ode’ module. But, the question remains as to where the non-differentiable time points are located in addition to the fact that for different subjects they do not necessarily coincide and may vary from iteration to iteration due to the change of the estim ate of h during optimization. These appear to be serious problems that cannot be overcome when using a non-Bayesian approach with the required linearization. Another potential limitation of linearization is revealed when nonlinearity curvature in the parameter effects is se vere due to the com plexity of Eq.6. One naive approach is to solve the problem with non-differentiable sensitivity by replacing max(G(t)-h,0) by G(t)-h in the minimal model equations. We fitted this modified model to the data, however, not too many improvements have been obtained when compared with non-Bayesian me- thod applied to Eq.6. In fact, it is sensible to specify max(G(t)-h,0) in Eq.6, since insulin enters the plasma with zero rate when glucose in plasma is below the threshold amount. Overall, for the glucose-insulin minimal model, the Bayesian approach appears to be the preferred since the algorithm behind the Bayesian app roach is applicable and effective to the structure of minimal model, and sensi- tivities are not needed in the estimation process as with the non-Bayesian approach. Another advantage of Bay- esian analysis is that individual estimates of model pa- rameters can be simultaneously calculated under the  Y. Wang et al. / HEALTH 2 (2010) 188-194 Copyright © 2010 SciRes Openly accessible at http://www.scirp.org/journal/HEALTH/ 194 Bayesian process. Therefore, Bayesian appro ach should be considered as an additional tool for data a nalysis, since it enables the analysis of systems of ODEs by nonlinear mixed-effects modeling and provide precise parameter estimates, make them useful tools for population PK/PD analysis of complicated systems of ODEs with and with- out an explicit form solution. 5. ACKNOWLEDGEMENTS We gratefully acknowledge support from the Claude Pepper Center Grants AG08808 and AG024824 from the National Institute of Aging. REFERENCES [1] Beal, S.L. and S heiner, L.B. (1982) Estimating population kinetics. CRC Critical Reviews in Bioengineering, 8, 195-222. [2] Lindstrom, M.J. and Bates, D.M . (1990) Nonlinear mixed effects models for rep eated m easures data. Biometrics, 46, 673-687. [3] Lindsey, J.K. (1993) Models for repeated measurements. Oxford. [4] Davidian, M. and Giltinan, D.M. (1995) Nonlinear models for repeated measurement data. Chapman and Hall. [5] Vonesh, E.F. and Chinchilli, V.M. (1996) Linear and nonlinear models for the analysis of repeated measure- ments. Marcel Dekker. [6] Pinheiro, J.C. and Bates, D.M. (2000) Mixed-effects models in S and Splus. Springer. [7] Diggle, P.J., Heagerty, P., Liang, K.Y. and Zeger, S.L. (2002) Analysis of longitudinal data. Oxford. [8] Demidenko, E. (2004) Mixed models: Theory and appli- cations. John Wiley & Sons, Canada. [9] Tornoe, C.W., Agerso, H. , Jonsson, E.N., Madsen, H. and Nielsen, H.A. (2004) Non-linear mixed-effects pharma- cokinetic/pharmacodynamic modeling in NLME using differential equations. Computer Methods and Programs in Biomedicine, 76, 31-40. [10] Galecki, A.T., Wolfinger, R.D., Linares, O.A ., Smith, M.J. and Halter, J.B. (2004) Ordinary differential equation PK/PD using the SAS mac ro NLINMIX. Journal of Bio- pharmaceutical Statistics, 14(2), 483-503. [11] Lunn, D.J. (2004) WinBUGS differential interface- worked examples. www.winbugs-development.org.uk. [12] Wang, Y., Eskridge, K.M. and Zhang, S. (2008) Semi- parametric m ixe d-effects anal ysi s of PK /PD m odels u sing differential equations. Journal Pharmacokinetics and Pharmacodynamics, 35, 443-463. [13] Pacini, G. and Bergman, R.N. (1986) MINMOD: A com- puter program to calculate insulin sensitivity and pancre- atic responsivity from the frequently sampled intravenous glucose tolerance test. Computer Methods and Programs in Biomedicine, 23, 113-122. [14] Bergman, R.N., Ider, Y.Z., Bowden, C.R. and Cobelli, C. (1979) Quantitative estimation of insulin sensitivity. American Journal of Physiology, 236, E667-E677. [15] Agbaje, O.F., Luzio, S.D., Albarrak, A.I.S., Lunn, D.J., Owens, D.R. and Hovorka, R. (2003) Bayesian hierar- chical approach to estimate insulin sensitivity by minimal model. Clinical Science, 105, 551-560. [16] Topp, B., Promislow, K. and De Vries, G. (2000) A model of β-cell mass, insulin, and glucose kinetics: Pathway to diabetes. Journal of Theoretical Biology, 206, 615-619. [17] De Gaetano, A. and Arino, O. (2000) Mathematical mod- eling of the intravenous glucose tolerance test. Journal of Mathematical Biology, 40(2), 136-168. [18] Gamerman, D. (1997) Markov chain monte carlo: Sto- chastic simulation for bayesian inference. Chapman & Hall.
|