Paper Menu >>
Journal Menu >>
 Simulation for chaos game representation of genomes by recurrent iterated function systems Simulation for chaos game representation of genomes by recurrent iterated function systems 1,2,*11 2 Zu-Guo Yu , Long Shi , Qian-Jun Xiao & Vo Anh 12 School of Mathematics and Computational Science, Xiangtan University, Hunan 411105, China. School of Mathematical Sciences, Queensland University of Technology, GPO Box 2434, Brisbane, Q 4001, Australia.* Correspondence should be addressed to Zu-Guo Yu (yuzg1970@yahoo.com). bases, the CGR would be a uniformly filled square, ABSTRACT conversely, any patterns visible in the CGR represent some pattern (information) in the DNA sequence Chaos game representation (CGR) of DNA (Goldman 1993). Goldman (1993) interpreted the sequences and linked protein sequences CGRs in a biologically meaningful way. All points from genomes was proposed by Jeffrey (1990) plotted within a quadrant must corresponding to sub- and Yu et al. (2004), respectively. In this sequences of the DNA sequence that end with the paper, we consider the CGR of three kinds of base labelling the corner of that quadrant. He also pro- sequences from complete genomes: whole posed a discrete time Markov Chain model to simu- genome DNA sequences, linked coding DNA late the CGR of DNA sequences and use the sequences and linked protein sequences. sequence's dinucleotide and trinucleotide frequen- Some fractal patterns are found in these cies to calculate the probabilities in these models. CGRs. A recurrent iterated function systems Goldman's Markov model can be calculated directly (RIFS) model is proposed to simulate the and easily from the raw DNA sequences, without ref- CGRs of these sequences from genomes and erence to the CGR. their induced measures. Numerical results Deschavanne et al. (1999) used CGR of genomes on 50 genomes show that the RIFS model can to discuss the classification of species. Almeida et al. simulate very well the CGRs and their (2001) showed the distribution of positions in the induced measures. The parameters esti-CGR plane is a generalization of Markov Chain prob- mated in the RIFS model reflect information ability tables that accommodates non-integer orders. on species classification.Joseph and Sasikumar (2006) proposed a fast algo- rithm for identifying all local alignments between two genome sequences using the sequence informa- tion contained in their CGR. Twenty different kinds of amino acids are found in 1. INTRODUCTIONproteins. The idea of CGR of DNA sequences pro- The hereditary information of organisms (except for posed by Jeffrey (1990) was generalized and applied RNA-viruses) is encoded in their DNA sequences for visualizing and analyzing protein databases by which are one-dimensional unbranched polymers Fiser et al. (1994). Generalization of CGR of DNA made up from four different kinds of monomers (nu-may take place in several ways. In the simplest case, cleotides): adenine (a), cytosine (c), guanine (g), and the square in CGR of DNA is replaced by an n-sided thymine (t). Based on a technique from chaotic regular polygon (n-gon), where n is the number of dif- dynamics, Jeffrey (1990) proposed a chaos game rep-ferent elements in the sequence to be represented. As resentation (CGR) of DNA sequences by using the proteins consist of 20 kinds of amino acids, a 20- four vertices of a square in the plane to represent sided regular polygon (regular 20-gon) is the most a,c,g and t. The method produces a plot of a DNA adequate for protein sequence representation. A few sequence which displays both local and global pat-thousand points result in an 'attractor' which gives a terns. Self-similarity or fractal structures were found visualization of the rare or frequent residues and in these plots. Some open questions from the biologi-sequence motifs. Fiser et al. (1994) pointed out that cal point of view based on the CGR were proposed the chaos game representation can also be used to (Jeffrey 1990). study 3D structures of proteins. If the DNA sequences were a random collection of Keywords: Genomes; Chaos game representa- tion; Recurrent iterated function systems J. Biomedical Science and Engineering, 2008, 1, 44-51Scientific Research Publishing JBiSE Published Online May 2008 in SciRes. http://www.srpublishing.org/journal/jbise SciRes Copyright © 2008  Basu et al. (1998) proposed a new method for the sequences and linked sequences of all protein chaos game representation of different families of sequences from complete genomes. proteins. Using concatenated amino acid sequences For DNA sequences, the CGR is obtained by using of proteins belonging to a particular family and a 12-the four vertices of a square in the plane to represent sided regular polygon, each vertex of which repre-a,c,g and t (Jeffrey 1990). The first point of the plot is sents a group of amino acid residues leading to con-placed half way between the center of the square and servative substitutions, the method generates the the vertex corresponding to the first letter, the ith CGR of the family and allows pictorial representa-point of the plot is placed half way between the (i- tion of the pattern characterizing the family. Basu et 1)th point and the vertex corresponding to the ith let- al. (1998) found that the CGRs of different protein ter in the DNA sequence. families exhibit distinct visually identifiable patterns. For linked protein sequences, we outline here the This implies that different functional classes of pro-way to get the CGR from Yu et al. (2004b). The pro- teins produce specific statistical biases in the distri-tein sequence is formed by twenty different kinds of bution of different mono-, di-, tri-, or higher order amino acids, namely Alanine (A), Arginine (R), peptides along their primary sequences.Asparagine (N), Aspartic acid (D), Cysteine (C), A well-known model of protein sequence analysis Glutamic acid (E), Glutamine (Q), Glycine (G), is the HP model proposed by Dill et al. (1985). In this Histidine (H), Isoleucine (I), Leucine (L), Lysine (K), model 20 kinds of amino acids are divided into two Methionine (M), Phenylalanine (F), Proline (P), types, hydrophobic (H) (or non-polar) and polar (P) Serine (S), Threonine (T), Tryptophan (W), Tyrosine (or hydrophilic). But the HP model may be too simple (Y) and Valine (V) (Brown 1998, page 109). In the and lacks sufficient information on the heterogeneity detailed HP model, they can be divided into four and the complexity of the natural set of residues classes: non-polar, negative polar, uncharged polar (Wang and Wang 2000). According to Brown (1998), and positive polar. The eight residues A, I, L, M, F, P, one can divide the polar class in the HP model into W, V designate the non-polar class; the two residues three classes: positive polar, uncharged polar and neg-D, E designate the negative polar class; the seven resi- ative polar. So 20 different kinds of amino acids can dues N, C, Q, G, S, T, Y designate the uncharged polar be divided into four classes: non-polar, negative class; and the remaining three residues R, H, K desig- polar, uncharged polar and positive polar. In this nate the positive polar class. model, one considers more details than in the HP For a given protein sequence s=ss with length l, 1l model. We call this model a detailed HP model (Yu et where s is one of the twenty kinds of amino acids for i al.2004a). Based on the detailed HP model, we pro-i=1,,l ,we define posed a CGR for the linked protein sequences from the genomes (Yu et al. 2004b). The recurrent iterated function system in fractal theory (Barnsley and Demko, 1985; Falconer, 1997) has been applied successfully to fractal image con- struction (Barnsley and Demko, 1985; Vrscay, 1991), one dimensional measure representation of genomes (Anh et al. 2002; Yu et al. 2001, 2003) and magnetic We then obtain a sequence X(s)=aa , where a is field data (Wanliss et al. 2005; Anh et al. 2005) for 1l i example. Yu et al. (2007) proposed a CGR for the a letter of the alphabet {0,1,2,3}. We next define the magnetic field data and used the RIFS model to simu-CRG for a sequence X(s) in a square [0,1][ 0,1], late the CGR.where the four vertices correspond to the four letters Although we proposed the CGR for linked protein 0,1,2,3. The first point of the plot is placed half way sequences from genomes (Yu et al. 2004b), we did between the center of the square and the vertex corre- not consider how to simulate the CGRs. In this paper, sponding to the first letter of the sequence X(s); the we extend the CGR to the study of whole-genome ith point of the plot is then placed half way between DNA sequences and linked coding DNA sequences the (i-1)th point and the vertex corresponding to the from genomes. Then we use the RIFS model to simu-ith letter. We then call the obtained plot the CGR of late the CGR of these 3 kinds of data from genomes the protein sequence s based on the detailed HP and their induced measures. The probability matrix in model. our RIFS model is similar to the one in Markov model Usually whole-genome DNA sequences and linked used by Goldman (1993), but the way to estimate this coding DNA sequences are relatively long, hence the matrix is different. resulting CGRs are too dense to visualize any pattern directly. The linked protein sequences are 3 times shorter than the linked coding DNA sequences, and 2. CHAOS GAME REPRESENTATION OF their CGRs produce clearer self-similar patterns. For GENOMES example, we show the CGR of the linked protein Three kinds of sequences from complete genomes are sequence of the bacterium Mycobacterium tuberculo- considered, namely, whole-genome DNA sequences sis CDC1551 (MtubC) in . (including protein-coding and non-coding regions), Considering the points in a CGR of an organism, linked sequences of all protein-coding DNA Figure 1 (1) SciRes JBiSE Copyright © 2008 Z.G. Yu et al./J. Biomedical Science and Engineering 1 (2008) 44-51 45 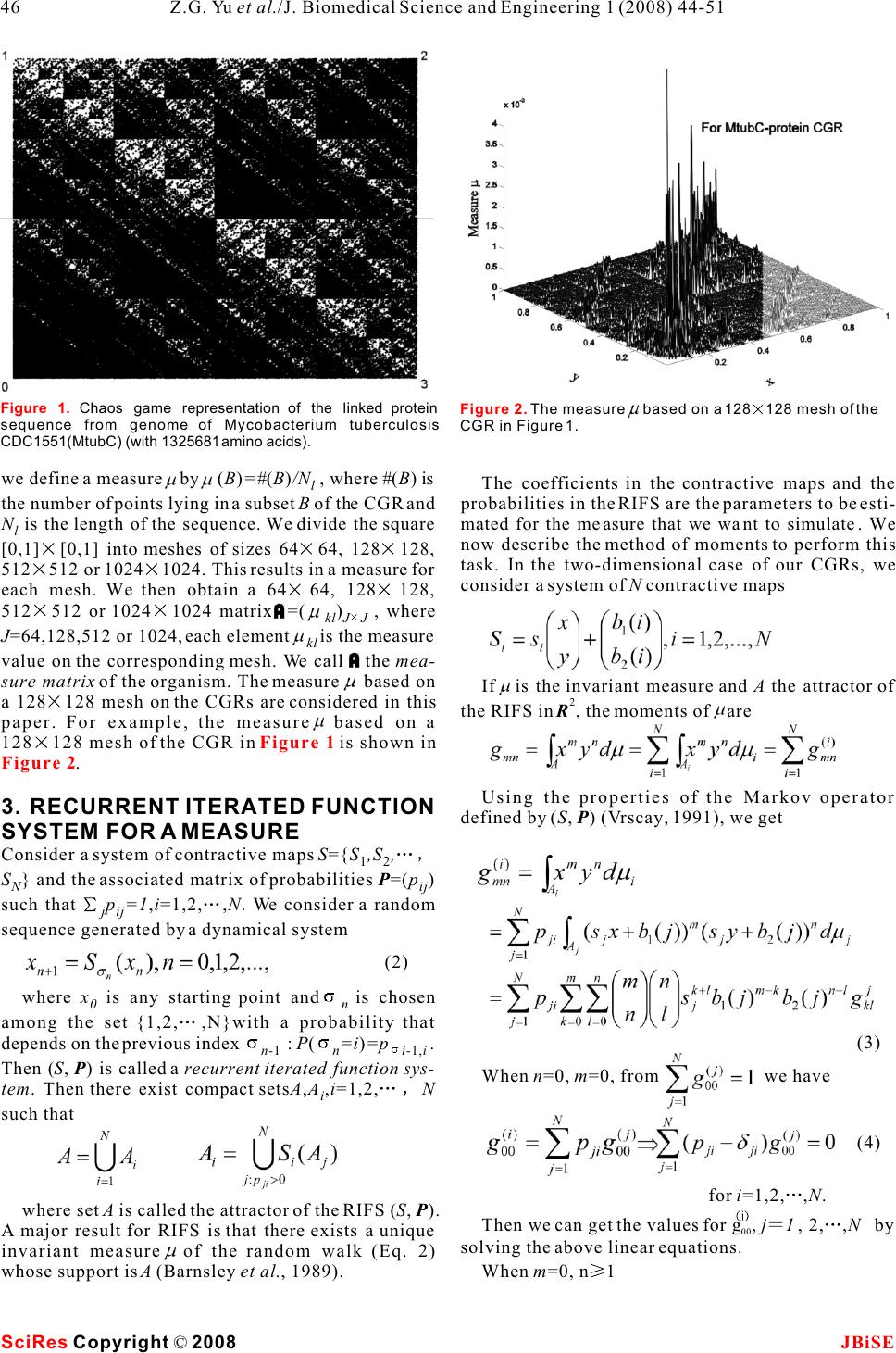 we define a measureby(B)=#(B)/N , where #(B) is The coefficients in the contractive maps and the l the number of points lying in a subset B of the CGR and probabilities in the RIFS are the parameters to be esti- N is the length of the sequence. We divide the square mated for the measure that we want to simulate. We lnow describe the method of moments to perform this [0,1][0,1] into meshes of sizes 6464, 128128, task. In the two-dimensional case of our CGRs, we 512512 or 10241024. This results in a measure for consider a system of N contractive maps each mesh. We then obtain a 6464, 128128, 512512 or 10241024 matrixA=() , where kl JJ J=64,128,512 or 1024, each elementis the measure kl value on the corresponding mesh. We call A the mea- sure matrix of the organism. The measure based on Ifis the invariant measure and A the attractor of 2 a 128128 mesh on the CGRs are considered in this the RIFS in R, the moments ofare paper. For example, the measurebased on a 128128 mesh of the CGR in is shown in . Using the properties of the Markov operator 3. RECURRENT ITERATED FUNCTION defined by (S, P) (Vrscay, 1991), we get SYSTEM FOR A MEASURE Consider a system of contractive maps S={S,S, 12 S} and the associated matrix of probabilities P=(p) Nij such that p=1,i=1,2, ,N. We consider a random jij sequence generated by a dynamical system where x is any starting point andis chosen 0n among the set {1,2,,N}with a probability that depends on the previous index : P(=i)=p . n-1ni-1,i Then (S, P) is called a recurrent iterated function sys-When n=0, m=0, from we have tem. Then there exist compact setsA,A,i=1,2, N i such that for i=1,2, ,N. where set A is called the attractor of the RIFS (S, P). j Then we can get the values for g,j12,,Nby , 00 A major result for RIFS is that there exists a unique solving the above linear equations. invariant measureof the random walk (Eq. 2) When m=0, n1 whose support is A (Barnsley et al., 1989). Figure 1 Figure 2 Figure 1. Chaos game representation of the linked protein sequence from genome of Mycobacterium tuberculosis CDC1551(MtubC) (with 1325681 amino acids). Figure 2. The measurebased on a 128128 mesh of the CGR in Figure 1. (2) (3) (4) SciRes JBiSE Copyright © 2008 46Z.G. Yu et al./J. Biomedical Science and Engineering 1 (2008) 44-51  If we denote by G the moments obtained directly mn from a given measure, and g the formal expression mn of moments obtained from the above formulae, then solving the optimization problem hence the moments are given by the solution of the linear equations will provide the estimates of the parameters of the RIFS. Once the RIFS (S(x),p,i,j=1,2, ,N ) has been esti- iij mated, its invariant measure can be simulated in the following way: Generate the attractor of the RIFS via the random walk (Eq. 2). Letbe the indicator func- B When n=0,m1 tion of a subset B of the attractor A. From the ergodic theorem for RIFS (Barnsley et al., 1989), the invari- ant measure is then given by hence the moments are given by the solution of the linear equations By definition, an RIFS describes the scale invariance of a measure. Hence a comparison of the given measure with the invariant measure simulated from the RIFS will confirm whether the given mea- sure has this scaling behaviour. This comparison can be undertaken by computing the cumulative walk of a measure visualized as intensity values on a JJ mesh; here J=128 in this paper. When m,n1If we convert the two-dimensional matrix A=()to an one dimensional vector by concate- kl JJ nate every row in A at the end of previous row. We denote the one-dimensional vector as f=(f,f,,f). 12 JJ The cumulative walk is defined as Where f is the average value of all element in vec- tor f. Returning to the CGR, an RIFS with 4 contractive hence the moments are given by the solution of the maps {S,S,S,S} is fitted to the measure obtained linear equations1234 from the CGR using the method of moments. Here we can fix Hence the parameters needed to be estimated are the probabilities in the matrix P. Once we have estimated the probability matrix in the RIFS, we can start from the point (0.5, 0.5) and use the chaos game algorithm Eq. (2) to generate a random point sequence{x} with i the same length N of the whole- genome DNA l for i=1,2, ,N.sequence, linked coding DNA sequence or the linked (6) (7) SciRes JBiSE Copyright © 2008 47 Z.G. Yu et al./J. Biomedical Science and Engineering 1 (2008) 44-51 (5) 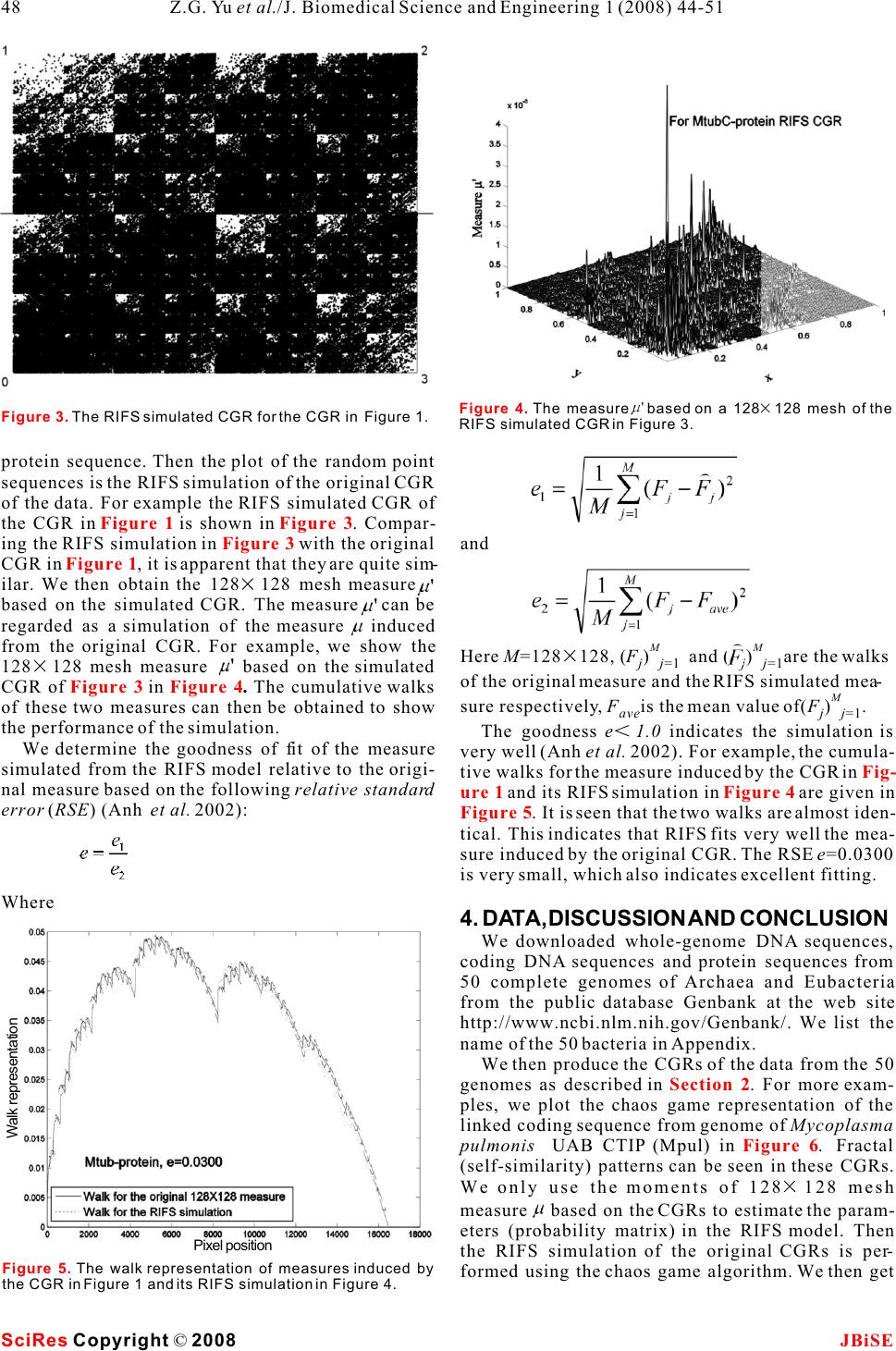 protein sequence. Then the plot of the random point sequences is the RIFS simulation of the original CGR of the data. For example the RIFS simulated CGR of the CGR in is shown in . Compar- ing the RIFS simulation in with the original and CGR in , it is apparent that they are quite sim- ilar. We then obtain the 128128 mesh measure based on the simulated CGR. The measure can be regarded as a simulation of the measure induced MM from the original CGR. For example, we show the Here M=128128, (F)and (F)are the walks jj=1 jj=1 128128 mesh measure based on the simulated of the original measure and the RIFS simulated mea- CGR of in . The cumulative walks M sure respectively, Fis the mean value of(F). of these two measures can then be obtained to show avej j=1 the performance of the simulation.The goodness e1.0 indicates the simulation is We determine the goodness of fit of the measure very well (Anh et al. 2002). For example, the cumula- simulated from the RIFS model relative to the origi-tive walks for the measure induced by the CGR in nal measure based on the following relative standard and its RIFS simulation in are given in error (RSE) (Anh et al. 2002):. It is seen that the two walks are almost iden- tical. This indicates that RIFS fits very well the mea- sure induced by the original CGR. The RSE e=0.0300 is very small, which also indicates excellent fitting. Where 4. DATA, DISCUSSION AND CONCLUSION We downloaded whole-genome DNA sequences, coding DNA sequences and protein sequences from 50 complete genomes of Archaea and Eubacteria from the public database Genbank at the web site http://www.ncbi.nlm.nih.gov/Genbank/. We list the name of the 50 bacteria in Appendix. We then produce the CGRs of the data from the 50 genomes as described in . For more exam- ples, we plot the chaos game representation of the linked coding sequence from genome of Mycoplasma pulmonis UAB CTIP (Mpul) in .Fractal (self-similarity) patterns can be seen in these CGRs. We only use the moments of 128128 mesh measurebased on the CGRs to estimate the param- eters (probability matrix) in the RIFS model. Then the RIFS simulation of the original CGRs is per- formed using the chaos game algorithm. We then get Figure 1Figure 3 Figure 3 Figure 1 Figure 3Figure 4 Fig- ure 1 Figure 4 Figure 5 Section 2 Figure 6 Figure 3. The RIFS simulated CGR for the CGR in Figure 1.Figure 4. The measure based on a 128128 mesh of the RIFS simulated CGR in Figure 3. Figure 5. The walk representation of measures induced by the CGR in Figure 1 and its RIFS simulation in Figure 4. SciRes JBiSE Copyright © 2008 48Z.G. Yu et al./J. Biomedical Science and Engineering 1 (2008) 44-51 Pixel position Walk representation 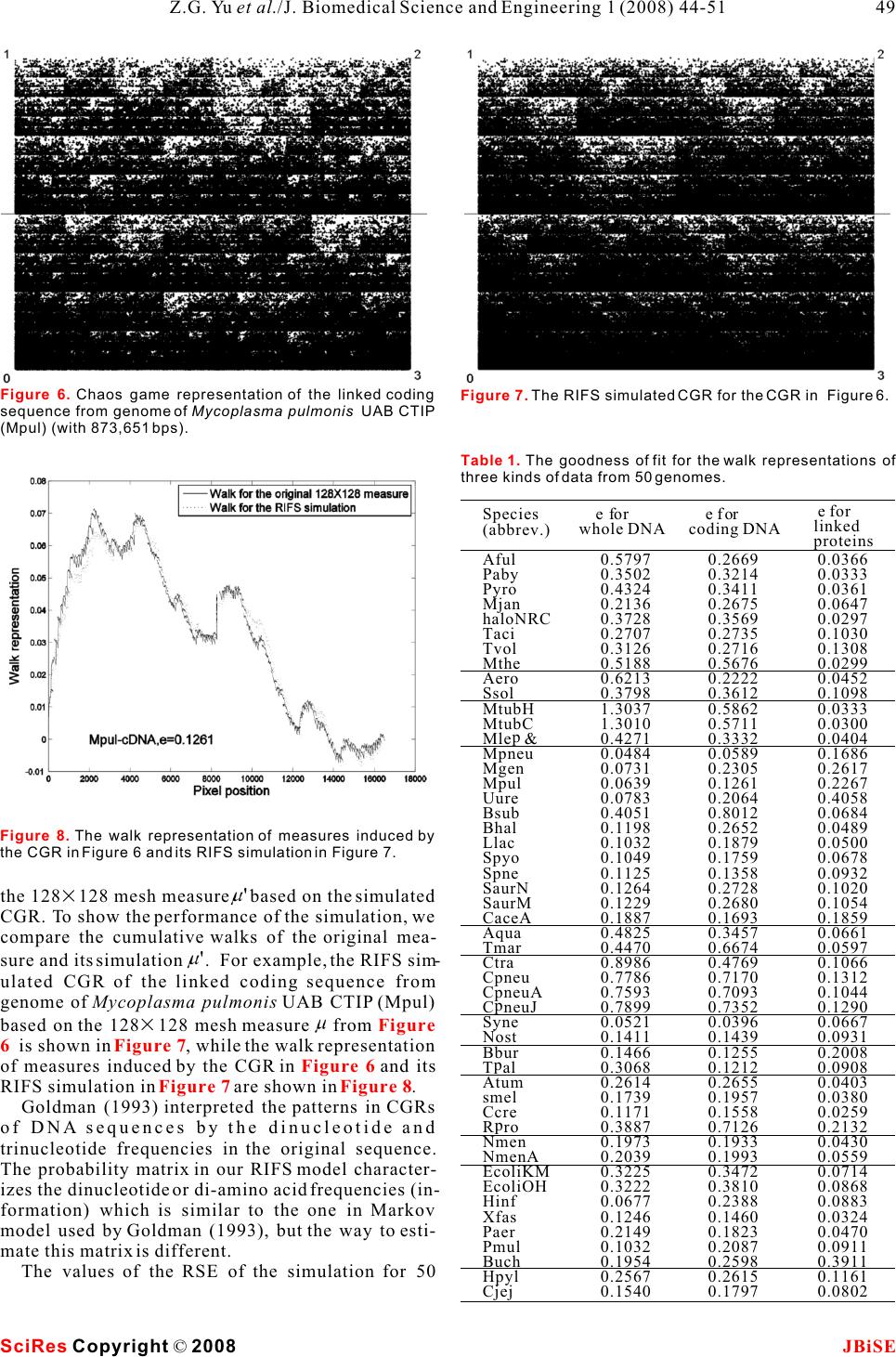 the 128128 mesh measurebased on the simulated CGR. To show the performance of the simulation, we compare the cumulative walks of the original mea- sure and its simulation. For example, the RIFS sim- ulated CGR of the linked coding sequence from genome of Mycoplasma pulmonis UAB CTIP (Mpul) based on the 128128 mesh measurefrom is shown in , while the walk representation of measures induced by the CGR in and its RIFS simulation in are shown in . Goldman (1993) interpreted the patterns in CGRs of DNA sequences by the dinucleotide and trinucleotide frequencies in the original sequence. The probability matrix in our RIFS model character- izes the dinucleotide or di-amino acid frequencies (in- formation) which is similar to the one in Markov model used by Goldman (1993), but the way to esti- mate this matrix is different. The values of the RSE of the simulation for 50 Figure 6Figure 7 Figure 6 Figure 7Figure 8 Figure 6. Chaos game representation of the linked coding sequence from genome of Mycoplasma pulmonis UAB CTIP (Mpul) (with 873,651 bps). Figure 7. The RIFS simulated CGR for the CGR in Figure 6. Figure 8. The walk representation of measures induced by the CGR in Figure 6 and its RIFS simulation in Figure 7. Table 1. The goodness of fit for the walk representations of three kinds of data from 50 genomes. Species (abbrev.) Aful Paby Pyro Mjan haloNRC Taci Tvol Mthe Aero Ssol MtubH MtubC p Mle & Mpneu Mgen Mpul Uure Bsub Bhal Llac Spyo Spne SaurN SaurM CaceA Aqua Tmar Ctra Cpneu CpneuA p CneuJ Syne Nost Bbur p Tal Atum smel Ccre p Rro Nmen NmenA EcoliKM EcoliOH Hinf Xfas Paer Pmul Buch Hpyl Cjej e f or wholeDN A 0.5797 0.3502 0.4324 0.2136 0.3728 0.2707 0.3126 0.5188 0.6213 0.3798 1.3037 1.3010 0.4271 0.0484 0.0731 0.0639 0.0783 0.4051 0.1198 0.1032 0.1049 0.1125 0.1264 0.1229 0.1887 0.4825 0.4470 0.8986 0.7786 0.7593 0.7899 0.0521 0.1411 0.1466 0.3068 0.2614 0.1739 0.1171 0.3887 0.1973 0.2039 0.3225 0.3222 0.0677 0.1246 0.2149 0.1032 0.1954 0.2567 0.1540 e f o r codingDN A 0.2669 0.3214 0.3411 0.2675 0.3569 0.2735 0.2716 0.5676 0.2222 0.3612 0.5862 0.5711 0.3332 0.0589 0.2305 0.1261 0.2064 0.8012 0.2652 0.1879 0.1759 0.1358 0.2728 0.2680 0.1693 0.3457 0.6674 0.4769 0.7170 0.7093 0.7352 0.0396 0.1439 0.1255 0.1212 0.2655 0.1957 0.1558 0.7126 0.1933 0.1993 0.3472 0.3810 0.2388 0.1460 0.1823 0.2087 0.2598 0.2615 0.1797 e for linked proteins 0.0366 0.0333 0.0361 0.0647 0.0297 0.1030 0.1308 0.0299 0.0452 0.1098 0.0333 0.0300 0.0404 0.1686 0.2617 0.2267 0.4058 0.0684 0.0489 0.0500 0.0678 0.0932 0.1020 0.1054 0.1859 0.0661 0.0597 0.1066 0.1312 0.1044 0.1290 0.0667 0.0931 0.2008 0.0908 0.0403 0.0380 0.0259 0.2132 0.0430 0.0559 0.0714 0.0868 0.0883 0.0324 0.0470 0.0911 0.3911 0.1161 0.0802 SciRes JBiSECopyright © 2008 49 Z.G. Yu et al./J. Biomedical Science and Engineering 1 (2008) 44-51  genomes and their induced measures. Third, the RIFS genomes are listed in .simulation of measures for linked protein data is It is seen that all the values of the RES except two better than that of measures for whole-genome DNA are much less than 1.0, confirming that the RIFS data and linked coding DNA data. Finally, the esti- model can simulate very well the measures of three mated parameters in the RIFS models for the linked kinds of data. The values of e for whole-genome DNA protein data from genomes can be used to character- data are generally larger than those for linked coding ize the phylogenetic relationships of the genomes. DNA data, which in turn are larger than those for linked protein data. In other words, the RIFS model can simulate the measures for linked protein data better than the measures for linked coding DNA data, ACKNOWLEDGEMENTS and can simulate measures for linked coding DNA Financial support was provided by the Chinese National Natural data better than the measures for whole-genome DNA Science Foundation (grant no. 30570426), Fok Ying Tung Educa- tion Foundation (grant no. 101004) and the Youth foundation of data. We notice that the linked protein sequence is Educational Department of Hunan province in China (grant no. shorter than the corresponding linked coding DNA 05B007) (Z.-G. Yu), and the Australian Research Council (grant no. sequence, while the linked coding DNA is shorter DP0559807) (V.V. Anh). than the whole-genome sequence. We guess the length of the data reflects the information complexity REFERENCE of the data and the RIFS model is still simple model [1]J.S. Almeida, J.A. Carrico, A. Maretzek, P.A. Noble & M. which simulates simpler data better. This result indi-Fletcher. Analysis of genomic sequences by Chaos Game Rep- cates that we can use the estimated parameters in the resentation. Bioinformatics 2001,17:429-437. RIFS model for linked protein data from genomes to [2]V.V. Anh, K.S. Lau, & Z.G. Yu. Recognition of an organism from fragments of its complete genome. Phys. Rev. E 2002, characterize the genomes. We find that the estimated 66(031910):1-9. probability matrices in the RIFS model for species [3]V.V. Anh, Z.G. Yu, J.A. Wanliss, & S.M. Watson. Prediction of from the same category are similar to each other. For magnetic storm events using the Dst index. Nonlin. Processes example, the estimated probability matrices for the Geophys. 2005 , 12:799-806. [4]M.F. Barnley, J.H. Elton & D.P. Hardin. Recurrent iterated func- measures of linked protein sequences from the three tion systems. Constr. Approx. B 1989, 5: 3-31. Gram-positive Eubacteria (high G+C) Mycobacte- [5]M.F. Barnsley & S. Demko. Iterated function systems and the rium tuberculosis H37Rv (MtubH), Mycobacterium global construction of fractals. Proc. R. Soc. London, Ser. A tuberculosis CDC1551 (MtubC) and Mycobacterium 1985, 399:243-275. [6]S. Basu, A. Pan, C. Dutta & J. Das. Chaos game representation leprae TN (Mlep) are:of proteins. J. Mol. Graphics and Modelling 1998, 15:279-289. [7]T.A. Brown. Genetics (3rd Edition) 1998. CHAPMAN & HALL, London. [8]P.J. Deschavanne, A Giron, J. Vilain, G. Fagot & B. Fertil. Genomics signature: Characterization and classification of spe- cies assessed by chaos game representation of sequences. Mol. Biol. Evol 1999, 16:1391-1399. [9]K.A.Dill. Theory for the folding and stability of globular Proteins. Biochemistry 1985, 24:1501-1509. [10]K. Falconer. Techniques in Fractal Geometry 1997, Wiley. [11]A. Fiser, GE Tusnady & I. Simon. Chaos game representation of protein structures. J. Mol. Graphics 1994, 12:302-304. [12]N. Goldman. Nucleotide, dinucleotide and trinucleotide fre- quencies explain patterns observed in chaos game representa- tions of DNA sequences. [13]H.J. Jeffrey. Chaos game representation of gene structure. Nucleic Acids Research 1990, 18(8): 2163-2170. [14]J.Joseph & R. Sasikumar. Chaos game representation for comparision of whole genomes. BMC Bioinformatics 2006, 7(243): 1-10. [15]E.R. Vrscay. Iterated function systems: theory, applications and inverse problem. Fractal Geometry and Analysis 1991, pages 405-468. [16]J. Wang & W. Wang. Modeling study on the validity of a possi- bly simplified representation of proteins. Phys. Rev. E 2000, 61:6981-6986. [17]J.A. Wanliss, V.V. Anh, Z.G. Yu & S. Watson. Multifractal mod- Hence we can use the RIFS estimated probability elling of magnetic storms via symbolic dynamics analysis. J. matrices of the linked protein sequences from Geophys. Res. 2005, 110(A08214):1-11. genomes to define a distance metric between two spe-[18]Z.G. Yu, V.V. Anh & K.S. Lau. Measure representation and cies for the purpose of construction of phylogenetic multifractal analysis of complete genomes. Phys. Rev. E 2001, 64(031903):1-9. tree. This work is being undertaken. [19]Z.G. Yu, V.V. Anh & K.S. Lau. Iterated function system and We can now draw some conclusions. First, the multifractal analysis of biological sequences. International J. chaos game representation of the three kinds of data Modern Physics B 2003, 17: 4367-4375. from genomes can give a visualization of the [20]Z.G. Yu, V.V. Anh, and K.S. Lau, “Fractal analysis of large pro- teins based on the Detailed HP model”, Physica A, 337 (2004a), genomes and produce some fractal patterns. Second, pp. 171-184. the RIFS model can be used to simulate CGRs of [21]Z.G. Yu, V.V. Anh & K.S. Lau. Chaos game representation, and Table 1 SciRes JBiSE Copyright © 2008 50Z.G. Yu et al./J. Biomedical Science and Engineering 1 (2008) 44-51  multifractal and correlation analysis of protein sequences from complete genome based on detailed HP model. J. Theor. Biol. 2004, 226(3): 341-348. [22]Z.G. Yu, V.V. Anh, J.A. Wanliss & S.M. Watson. Chaos game representation of the Dst index and prediction of geomagnetic storm events. Chaos, Solitons & Fractals 2007, 31:736-746. NC002745), Staphylococcus aureus subsp. aureus Mu50 (SaurM, NC002758), and Clostridium acetobutylicum ATCC 824 (CaceA, NC003030). The others are Gram-negative Eubacteria, which consist of two hyperthermophilic bacteria: Aquifex aeolicus VF5 (Aqua, NC000918) and Thermotoga maritima MSB8 (Tmar, NC000853); four Chlamydia: Chlamydia trachomatis D/UW-3/CX (Ctra, NC000117), Chlamydia pneumoniae CWL029 (Cpneu, NC000922), Chlamydia pneumoniae AR39 (CpneuA, NC002179) and Chlamydia pneumoniae J138 (CpneuJ, APPENDIX NC002491); two Cyanobacterium: Synechocystis sp. PCC6803 (Syne, These 50 bacteria include eight Archae Euryarchaeota: Archaeoglobus NC000911) and Nostoc sp. PCC 7120 (Nost, NC003272); two Spirochaete: fulgidus DSM 4304 (Aful, NC000917), Pyrococcus abyssi GE5 (Paby, Borrelia burgdorferi B31 (Bbur, NC001318) and Treponema pallidum NC000868), Pyrococcus horikoshii OT3 (Pyro, NC000961), Nichols (Tpal, NC000919); and fifteen Proteobacteria. The fifteen Methanococcus jannaschii DSM 2661 (Mjan, NC000909), Halobacterium Proteobacteria are divided into four subdivisions, namely alpha subdivision: sp. NRC-1 (haloNRC, NC002607), Thermoplasma acidophilum DSM 1728 Agrobacterium tumefaciens strain C58 (Atum, NC003062), Sinorhizobium (Taci, NC002578), Thermoplasma volcanium GSS1 (Tvol,NC002689), and meliloti 1021 (smel, NC003047), Caulobacter crescentus CB15 (Ccre, Methanobacterium thermoautotrophicum deltaH (Mthe, NC000916); two NC002696) and Rickettsia prowazekii Madrid (Rpro, NC000963); beta sub- Archae Crenarchaeota: Aeropyrum pernix K1 (Aero, NC000854) and division: Neisseria meningitidis MC58 (Nmen, NC003112) and Neisseria Sulfolobus solfataricus P2 (Ssol, NC002754); three Gram-positive meningitidis Z2491 (NmenA, NC003116); gamma subdivision: Esche- Eubacteria (high G+C): Mycobacterium tuberculosis H37Rv (MtubH, richia coli K-12 MG1655 (EcoliKM, NC000913), Escherichia coli O157:H7 NC000962), Mycobacterium tuberculosis CDC1551 (MtubC, NC002755) EDL933 (EcoliOH, NC002695), Haemophilus influenzae Rd (Hinf, and Mycobacterium leprae TN (Mlep, NC002677); twelve Gram-positive NC000907), Xylella fastidiosa 9a5c (Xfas, NC002488), Pseudomonas Eubacteria (low G+C): Mycoplasma pneumoniae M129 (Mpneu, aeruginosa PA01 (Paer, NC002516), Pasteurella multocida subsp. NC000912), Mycoplasma genitalium G37 (Mgen, NC000908), Mycoplasma multocida str. Pm70 (Pmul, NC002663) and Buchnera str. APS (Buch, pulmonis UAB CTIP (Mpul, NC002771), Ureaplasma urealyticum serovar NC002528); and epsilon subdivision: Helicobacter pylori 26695 (Hpyl, 3 str. ATCC 700970 (Uure, NC002162), Bacillus subtilis subsp. subtilis str. NC000915) and Campylobacter jejuni subsp. jejuni NCTC 11168 (Cjej, 168 (Bsub, NC000964), Bacillus halodurans C-125 (Bhal, NC002570), NC002163). The abbreviations in the brackets stand for the names of these Lactococcus lactis subsp. lactis Il1403 (Llac, NC002662), Streptococcus species and their NCBI accession numbers. pyogenes M1 GAS (Spyo, NC002737), Streptococcus pneumoniae TIGR4 (Spne, NC003028), Staphylococcus aureus subsp. aureus N315 (SaurN, SciRes JBiSECopyright © 2008 51 Z.G. Yu et al./J. Biomedical Science and Engineering 1 (2008) 44-51 |

