Circuits and Systems
Vol.07 No.04(2016), Article ID:66001,12 pages
10.4236/cs.2016.74028
Feature Selection Based on Enhanced Cuckoo Search for Breast Cancer Classification in Mammogram Image
M. N. Sudha1, S. Selvarajan2*
1Department of Master of Computer Applications, Institute of Road and Transport Technology, Erode, India
2Muthayammal College of Engineering, Rasipuram, India

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 9 March 2016; accepted 24 April 2016; published 27 April 2016
ABSTRACT
Proposed system has been developed to extract the optimal features from the breast tumors using Enhanced Cuckoo Search (ECS) and presented in this paper. The texture feature, intensity histogram feature, radial distance feature and shape features have been extracted and the optimal feature set has been obtained using ECS. The overall accuracy of a minimum distance classifier and k-Nearest Neighbor (k-NN) on validation samples is used as a fitness value for ECS. The new approach is carried out on the extracted feature dataset. The proposed system selects only the minimum number of features and performed the accuracy of 98.75% with Minimum Distance Classifier and 99.13% with k-NN Classifier. The performance of the new ECS is compared with the Cuckoo Search and Harmony Search. This result shows that the ECS algorithm is more accurate than the other algorithm. The proposed system can provide valuable information to the physician in medical pathology.
Keywords:
Breast Cancer Classification, Feature Extraction, Enhanced Cuckoo Search

1. Introduction
The occurrence of breast cancer in women has increased significantly in the recent years. In various types of cancer, breast cancer is one of the important causes of death among middle and old aged women. Throughout their lifetime [1] , more than 8% of women suffered from this disease. In worldwide, breast cancer caused 458,503 deaths (13.7% of cancer deaths in women) in the year 2008 [2] . Physical examination is one of the ways to detect the breast cancer and also the effectiveness of this method is limited by the subjective ability of doctors. Another way to detect breast cancer is screening mammography. It is one of the effective techniques for the early detection of breast cancer. As reported in [3] , screening mammography minimized the death rates from breast cancer among the women aged between 40 - 70 years [4] . To improve the accuracy, double reading of mammogram can be used. But it is cost effective and also it requires twice the radiologists’ reading time. It may not be possible to routinely perform a double reading by a radiologist. Hence, efforts have been made to develop a breast cancer classification system [5] [6] to help radiologist in the analysis of mammograms in hospitals, which increase the accuracy of diagnosis, as well as to improve the uniformity of interpretation of images by the use of the computer’s results as a reference [7] .
Background Study
Many researchers have developed various schemes for mammogram image classification. Hadjiiski et al. [8] used active contour method for segmentation purpose, stepwise feature selection and linear discriminant analysis for classification were used to select the useful features. Huo et al. [9] used a fully automated procedure to segment the masses using region growing algorithm, two-stage hybrid classifier includes a rule-based stage and an artificial neural network stage. Erkang et al. [10] proposed a histogram intersection based image classification. Hassanien et al. [11] proposed classification of the mammogram image based on rough set theory. Arod et al. [12] presented an AdaBost and Support vector machines for the mammogram image analysis. Zhang et al. [13] combines extracted features and human interpreted features from the mammogram, with the statistical classifier as other features in conjunction with the Genetic Neural Network. Eddaoudi et al. 2011 [14] presented a masses detection algorithm based on SVM classification and texture analysis. Islam et al. 2010 [15] proposed an efficient computer aided mass classification method in digitized mammograms using Artificial Neural Network (ANN). Pasquale Delogu et al. [16] used texture features and neural classifier for characterization of mammographic masses.
Kinoshita et al. [17] have used a texture features and shape features combination, three-layer feed-forward neural network as a classifier for and malignant classification system. Rangayyan et al. [18] developed a region-based edge profile acutance measure for evaluating the sharpness of mass boundaries. They reported that 92% of classification accuracy in classification of benign and malignant mass. Priebe et al. [19] estimated system based on the fractal features and texture for the detection of abnormalities. Sahiner et al. [20] introduced the rubber band straightening transform (RBST) for classification of malignant or benign mass. Chitre et al. [21] used texture measures for the classification of benign or malignant regions on mammogram. Furuya et al. [22] used different features like, first-order statistics features, second-order statistic (co-occurrence) features, density features, and shape features and introduced feature selection algorithm and evaluation of feature selection criteria to discriminate malignant tumors from normal on mammograms. Bruce et al. [23] demonstrated the techniques to extract multiresolution features to quantify mass shapes and the shapes were segmented manually by a radiologist. Pohlman et al. [24] created a system for segmenting a mass. It dealt with the adaptive region growing algorithm for segmenting of the masses. Schroeter et al. [25] have investigated the method based on the parameters of the Gaussian mixture by the genetic algorithm (GA) in the context of MR data segmentation. Bazi and Melgani [26] have proposed a support vector machine (SVM) classification system for detecting the most unique features and estimating the SVM parameters by using a Genetic Algorithm (GA). Daamouche et al. [27] presented the technique for feature extraction using Particle Swarm Optimization (PSO) to select the most informative features obtained by morphological profiles for classification.
Nandi et al. proposed a novel mass classification method using genetic programming (GP) and feature selection in [28] . Mavroforakis et al. [29] analyzed at various clinical features for benign/malignant classification and performed statistical analysis tests on those features. The authors used four different classifiers namely linear discriminant analysis, artificial neural networks, SVMs and k-nearest-neighbor to assess the discriminative power of each of the features.
The functional block diagram of the proposed system for classifying breast tumor in the mammogram is shown in Figure 1. The mammogram has been obtained and processed using various techniques. The features obtained are expected to provide valuable information to analyze the nature of the mammogram for further decision making in the clinical pathology. The current work includes preprocessing, Mass segmentation, feature extraction, classification based on feature selection. The techniques are presented in the following sections.
Figure 1. Flow Graph for breast cancer classification system.
2. Preprocessing
Images in the Digital Database for Screening Mammography (DDSM) are noisy, containing background information and identifiable label which need to be eliminated before classification. Low pass filter is efficient [30] , which is applied to the image that maintains the mass portion while suppressing noise. Cropping process that eliminates image label and background information [31] . Histogram equalization is a method in image process- ing, which is used for contrast adjustment using the image histogram’s [32] . Through this adjustment, the intensity values can be better distributed on the histogram. Through this process the image abnormalities will be better visible.
Figure 2 Shows the preprocessing of mammogram image.
3. Segmentation
To analyze the mammogram image, it is essential to differentiate the suspicious area from its surroundings. The main idea of segmentation is to extract Region of Interest (ROI) contain mass portions and locate the suspicious mass portion of the ROI. The ROI portion that contains masses was manually extracted from the original mammography images.
Region-Growing Algorithm
This algorithm starts from a seed pixel which manually pointed inside the suspicious region and it expands the area around the seed to include nearby pixels falling within a threshold range. For the similar condition, two thresholds Th1 and Th2 have been used and are given in Equations (1) & (2).
 (1)
(1)
Figure 2. Preprocessing of mammogram image.
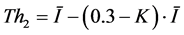 (2)
(2)
where 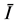 the average value of luminance in the segmented region and it is changing at each growing step, and K depends on a factor F. It is given in Equation (3)
the average value of luminance in the segmented region and it is changing at each growing step, and K depends on a factor F. It is given in Equation (3)
 (3)
(3)
where d is the distance of the pixel with maximum intensity from the initial seed, Iseed is the intensity value of the seed, Imax is the maximum intensity value in the ROI, and N × N is the size of the ROI. Further details can be found in [33] . Figure 3(a) & Figure 3(b) illustrate the results of ROI from mammogram image & results of mass segmentation from ROI.
4. Feature Extraction
Once the mass boundary is identified, it is essential to extract various features from segmented image, in order to generate feature vector to be used in classification stage. The various features extracted are Texture feature, Intensity histogram feature, Shape features, Radial distance features. Table 1 shows the summary of newly computed mammogram image features according to feature type, feature number and description.
5. Feature Selection Based on an Enhanced Cuckoo Search for Breast Cancer Classification
The main aim of feature selection (FS) is to identify important features from original feature vector. This is done by removing irrelevant, redundant and noisy features which will be helpful for the best performance in terms of accuracy [40] . Search procedures are mainly used for feature selection. A number of search procedures have been proposed and popularly used feature selection algorithms are genetic algorithm (GA) [41] , Particle swarm optimization (PSO) [42] and metaheuristic algorithms.
Metaheuristic algorithms are capable of extracting information from a set of features and often generate the best features in practice. The metaheuristic algorithms, namely Bat Algorithm (BA) [43] , Ant Colony Optimization (ACO) [44] , Harmony Search (HS) [45] [46] and recently the Cuckoo Search (CS) algorithm [47] . The proposed feature selection approach called Enhanced cuckoo search, having different types of host nest with multiple eggs. It searches for the best feature subsets that maximize the classification performance.


Figure 3. (a) ROI from mammogram image; (b) Results of mass segmentation from ROI.
Table 1. Summary of Computed mammogram image features.
5.1. Cuckoo Search
Cuckoo search (CS) is one of the recent Nature-Inspired metaheuristic algorithm developed by Xin-She Yang and Suash Deb in 2009 [47] . Cuckoos are attractive birds, because of the beautiful sounds they can make and their aggressive reproductive strategy. A few species such as the Ani and Guira cuckoos lay their eggs in communal nests and they may remove others’ eggs to increase the hatching probability of their own eggs. CS is based on the brood parasitism of few cuckoo species, by laying their eggs in the nest of other host birds. If a host bird identifies the eggs, which are not their own, it will throw these unknown eggs away or simply abandon its nest and built a new nest elsewhere [48] . Each egg in the nest represents a solution, and a cuckoo eggs represents a new solution. If the new solution (cuckoo) is better than old then replaces the solution which is so good in the nest. This algorithm is enhanced by the Levy flights [47] rather than by isotropic random walks.
The following three idealized rules for describing standard cuckoo search have been used
o At a time each cuckoo lays one egg and dumps at a randomly chosen nest.
o The best nests with high quality eggs will be passed over to the next generations.
o Number of available host nests are fixed and the cuckoo’s egg is discovered by the host bird with a probability pa ɛ (0,1). The host bird can do either get rid of the egg or just abandon the nest and build a completely new nest.
This last assumption can be approximated by a fraction pa of the n host nests that are replaced by new nests (with new random solutions). Further detail can be found in [47] . Cuckoo search is very simple and has extensive search space. It uses the levy flight to global search instead of standard random walk, which makes CS to explore the search space more efficiently.
5.2. Enhanced Cuckoo Search
Cuckoo search is modified to Enhanced Cuckoo Search with different types of host nest with multiple eggs [49] . In general cuckoo selects three types of nests for laying their eggs.
o The common cuckoo selects a group of host nests with egg characteristics similar to their own [50] .
o Others cuckoo selects a group of host nest with egg characteristics dissimilar to their own.
o Some other species of cuckoo lay cryptic egg, which are dark in color when the eggs of their host birds are light. This trick is used to hide the eggs from the host and evolve in cuckoos that parasite host with dark, domed nests.
Pseudo code is shown in Figure 4, for Enhanced Cuckoo Search based on egg laying behavior of cuckoo and multiple eggs in the nest.
5.2.1. Egg Representation
In the process of feature selection using ECS, each egg contains the solution it represents. The eggs are
Figure 4. Pseudo code for enhanced cuckoo search.
represented by 123 features. The mainly used way of encoding the feature selection is a binary string. Here, the random values are generated for feature position. If the value of the variable is 1, then the feature is selected else feature is not selected. The egg is represented as a binary string which is shown in Figure 5.
5.2.2. Initial Population
In this work, each egg represents a possible set of features that are selected and used to classify the samples correctly. Here the initial population size is considered as 50.
5.2.3. Finding New Solutions and Levy Flight
ECS based feature selection method makes use of levy flight for finding the new solutions from Equation (4). Some of the new solutions should be generated by a levy walk around the best solution obtained so far, this will speed up the local search.
By using Levy flight, the new solution is  is produced for cuckoo i, and is given below
is produced for cuckoo i, and is given below
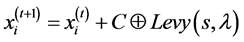 (4)
(4)
α is the step size. The step length follows the Levy distribution
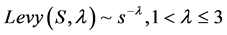
5.2.4. Fitness Function
The fitness function plays a vital role in the selection process. The performance of any classification system is measured by its classification accuracy. Classification accuracy plays a major role in the process of breast cancer classification using significant features from the feature vector. Here, the classification accuracy of Minimum distance Classifier or k-NN classifier is used as the fitness function of ECS. The fitness function fitness(f) of ECS is defined as in Equation (5)
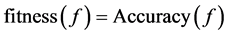 (5)
(5)
Accuracy(f) is the test accuracy of testing data f in the classifier which is built with the feature subset selection of training data. The classification accuracy of minimum distance classifier or k-NN is given in Equation (6).
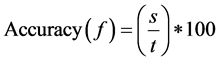 (6)
(6)
s: Number of samples that are correctly classified in test data by minimum distance classifier or k-NN Classifier
t: Total number of samples in test data
5.2.5. Parameter Pa
In enhanced cuckoo search the Pa value is changed dynamically by using the formula given in Equation (7)



Figure 5. Egg representation of feature selection using ECS.
5.2.6. Crossover and Mutation
o Cuckoo type is common cuckoo then crossover is used to create two eggs in the nest and choose the best one among them.
o Cuckoo type is European cuckoo then create two eggs using crossover with uniform mutation operator and choose the best one among them.
o Otherwise eggs (cryptic) are created with random solution.
5.2.7. K Fold Cross Validation
K fold cross validation procedure has been used to evaluate the efficiency of the proposed system. In this procedure, the feature vector is randomly divided into K disjoint parts of approximately the equal size. Classifier is trained with K-1 parts and then is tested with a single part. This process is repeated K times (K folds) with each of the K parts used exactly once as the test data. The average of K results from the folds can be calculated to produce an average accuracy of minimum Distance Classifier and K nearest neighbor classifier.
5.2.8. Similarity Measure
To find the closeness between training data set and test data set normally some distance measures are used and they are Euclidean distance, City block distance and Minkowski distance and chi-Square Distance, are given in Equation (8) to Equation (11)
Euclidean distance 
City block distance 
Minkowski distance 
Chi-Square Distance 
Table 2 shows the parameters and its values for ECS for feature selection in breast cancer classification.
6. Results and Discussions
The images in the Digital Database for Screening Mammography (DDSM) have been considered for developing breast cancer classification system. The proposed system has been implemented and the features were also extracted for further analysis using MATLAB. In this work, initially 123 features were extracted. The proposed feature selection method based minimum distance classifier selects totally 34 features and produces 98.75% average classifier accuracy by using chi-square distance, ECS with k-NN classifier selects totally 29 features and produces 99.13% average classifier accuracy using Euclidean distance. The results of various distance measures and its average classification accuracy are given in Figure 6 & Figure 7. Optimal number of features selected from CS, HS and ECS are shown in Figure 8.
Table 2. Parameter setting for ECS for breast cancer classification.
Figure 6. Classifier accuracy using ECS with minimum distance classifier.
Figure 7. Classifier accuracy using ECS with k-NN classifier.
Figure 8. Optimal feature selected from CS, HS and proposed ECS.
7. Conclusion
In this paper, a total of 508 (288 benign and 212 malignant) mammogram images were used as case samples. For the segmentation, region growing algorithm was used, after the segmentation, each mass was represented with 123 features, including 96 texture features, 9 histogram features, 11 Shape features, 7 Radial distance features. Several feature selection methods were investigated for breast cancer classification, including the CS, HS and the proposed ECS. All these feature selection method uses a minimum distance classifier, k-nearest neighbor classifier. ECS with k-NN Classifier for breast cancer classification outperforms well when compared to other algorithms. The proposed feature selection method for breast cancer classification shows better performance in terms of accuracy with the least number of features.
Cite this paper
M. N. Sudha,S. Selvarajan, (2016) Feature Selection Based on Enhanced Cuckoo Search for Breast Cancer Classification in Mammogram Image. Circuits and Systems,07,327-338. doi: 10.4236/cs.2016.74028
References
- 1. Jemal, A., Siegel, R., Ward, E., Hao, Y., Xu, J., Murra, T. and Thun, M.J. (2008) Cancer Statistics 2008. CA: A Cancer Journal for Clinicians, 8, 71-96. http://dx.doi.org/10.3322/CA.2007.0010
- 2. Boyle, P. and Levin, B. (2008) World Cancer Report 2008. International Agency for Research on Cancer, Lyon.
- 3. Lewis, C. (1999) FDA Sets Higher Standards for Mammography. FDA Consumer, 33, 13-17.
- 4. Mandelblatt, J.S., Cronin, K.A., Bailey, S., Berry, D.A., de Koning, H.J., Draisma, G., Huang, H., Lee, S.J., Munsell, M. and Plevritis, S.K. (2009) Effects of Mammography Screening under Different Screening Schedules: Model Estimates of Potential Benefits and Harms. Ann. Intern. Med., 151, 738-747.
http://dx.doi.org/10.7326/0003-4819-151-10-200911170-00010 - 5. Winsberg, F., Elkin, M., Macy, J., Bordaz, V. and Weymouth, W. (1967) Detection of Radiographic Abnormalities in Mammograms by Means of Optical Scanning and Computer Analysis. Radiology, 89, 211-215.
http://dx.doi.org/10.1148/89.2.211 - 6. Marx, C., Malich, A., Facius, M., Grebenstein, U., Sauner, D., Pfleiderer, S.O.R. and Kaiser, W.A. (2004) Are Unnecessary Follow-Up Procedures Induced by Computer-Aided Diagnosis (CAD) in Mammography? Comparison of Mammographic Diagnosis with and without Use of CAD. European Journal of Radiology, 51, 66-72.
http://dx.doi.org/10.1016/S0720-048X(03)00144-X - 7. Doi, K., MacMahon, H., Giger, M.L. and Hoffmann, K.R. (1999) Computer-Aided Diagnosis and Its Potential Impact on Diagnostic Radiology. In: Computer-Aided Diagnosis in Medical Imaging, Elsevier Science, Amsterdam, 11-20.
- 8. Hadjiiski, L., Sahiner, B., Chan, H.P., Petrick, N., Helvie, M.A. and Gurcan, M. (2001) Analysis of Temporal Changes of Mammographic Features: Computer-Aided Classification of Malignant and Benign Breast Masses. Medical Physics, 28, 2309-2317. http://dx.doi.org/10.1118/1.1412242
- 9. Huo, Z., Giger, M.L., Vyborny, C.J., Bick, U., Lu, P., Wolverton, D.E. and Schmidt, R.A. (1995) Analysis of Spiculation in the Computerized Classification of Mammographic Masses. Medical Physics, 22, 1569-1579.
http://dx.doi.org/10.1118/1.597626 - 10. Erkang, C., Nianhua, X., Haibin, L., Bakic, P.R., Maidment, A.D.A. and Megalooikonomou, V. (2010) Mammographic Image Classification Using Histogram Intersection. IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Rotterdam, 14-17 April 2010, 197-200.
- 11. Hassanien, E.A. and Ali, J.M.H. (2003) Rough Set Approach for Classification of Breast Cancer Mammogram Images. In: Di Gesú, V., Masulli, F. and Petrosino, A., Eds., Fuzzy Logic and Applications, Springer, Berlin Heidelberg, 224- 231.
- 12. Arod, T., Kurdziel, M., Sevre, E.O.D. and Yuen, D.A. (2005) Pattern Recognition Techniques for Automatic Detection of Suspicious-Looking Anomalies in Mammograms. Computer Methods and Programs in Biomedicine, 79, 135-149.
http://dx.doi.org/10.1016/j.cmpb.2005.03.009 - 13. Zhang, P., Kumar, K. and Verma, B. (2005) A Hybrid Classifier for Mass Classification with Different Kinds of Features in Mammography. In: Wang, L.P. and Jin, Y.C., Eds., Fuzzy Systems and Knowledge Discovery, Springer, Berlin Heidelberg, 316-319. http://dx.doi.org/10.1007/11540007_38
- 14. Eddaoudi, F., Regragui, F., Mahmoudi, A. and Lamouri, N. (2011) Masses Detection Using SVM Classifier Based on Textures Analysis. Applied Mathematical Sciences, 5, 367-379.
- 15. Islam, M., Ahmadi, M. and Sid-Ahmed M. (2010) An Efficient Automatic Mass Classification Method in Digitized Mammograms Using Artificial Neural Network. International Journal of Artificial Intelligence & Applications, 1, 1-13.
- 16. Delogu, P., Fantacci, M.E., Kasae, P. and Retico, A. (2007) Characterization of Mammographic Masses Using a Gradient Based Segmentation Algorithm and a Neural Classifier. Computers in and Biology Medicine, 37, 1479-1491.
http://dx.doi.org/10.1016/j.compbiomed.2007.01.009 - 17. Kinoshita, S.K., Marques, P.M.A., Slates, A.F.F., Marana, H.R.C., Ferrari, R.J. and Villela, R.L. (1998) Detection and Characterization of Mammographic Masses by Artificial Neural Network. In: Karssemeijer, N., Thijssen, M., Hendriks, J. and van Erning, L., Eds., Digital Mammography, Springer, Netherlands, 489-490.
http://dx.doi.org/10.1007/978-94-011-5318-8_85 - 18. Rangayyan, R.M., El-Faramawy, N.M., Desautels, J.E.L. and Alim, O.A. (1997) Measures of Acutance and Shape for Classification of Breast Tumors. IEEE Transactions on Medical Imaging, 16, 799-810.
http://dx.doi.org/10.1109/42.650876 - 19. Priebe, C.E., Lorey, R.A., Marchette, D.J., Solka, J.L. and Rogers, G.W. (1994) The Application of Fractal Analysis to Mammographic Tissue Classification. Cancer Letters, 77, 183-189.
http://dx.doi.org/10.1016/0304-3835(94)90101-5 - 20. Sahiner, B., Chan, H.P., Petrick, N., Helvie, M.A. and Goodsitt, M.M. (1998) Computerized Characterization of Masses on Mammograms: The Rubber Band Straightening Transform and Texture Analysis. Medical Physics, 25, 516- 526.
http://dx.doi.org/10.1118/1.598228 - 21. Chitre, Y., Dhawan, A.P. and Moskowitz, M. (1993) Artificial Neural Network Based Classification of Mammographic Microcalcifications Using Image Structure Features. International Journal of Pattern Recognition and Artificial Intelligence, 7, 1377-1402.
http://dx.doi.org/10.1142/S0218001493000674 - 22. Furuya, S., Wei, T., Hagihara, Y., Shimizu, A., Kabotake, H. and Nawano, S. (2004) Improvement of Performance to Discriminate Malignant Tumors from Normal Tissue on Mammograms by Feature Selection and Evaluation of Features Selection Criteria. Systems and Computers in Japan, 35, 72-84.
http://dx.doi.org/10.1002/scj.10587 - 23. Bruce, L.M. and Adhami, R.R. (1999) Classifying Mammographic Mass Shapes Using the Wavelet Transform Modulus-Maxima Method. IEEE Transactions on Medical Imaging, 18, 1170-1177.
http://dx.doi.org/10.1109/42.819326 - 24. Pohlman, S., Powell, K.A., Obuchowshi, N.A., Chilote, W.A. and Grundfest-Broniatowski, S. (1996) Quantitative Classification of Breast Tumors in Digitized Mammograms. Medical Physics, 23, 1337-1345.
http://dx.doi.org/10.1118/1.597707 - 25. Schroeter, P., Vesin, J.M., Langenberger, T. and Meuli, R. (1998) Robust Parameter Estimation of Intensity Distributions for Brain Magnetic Resonance Images. IEEE Transactions on Medical Imaging, 17, 172-186.
http://dx.doi.org/10.1109/42.700730 - 26. Bazi, Y. and Melgani, F. (2006) Toward an Optimal SVM Classification System for Hyperspectral Remote Sensing Images. IEEE Transactions on Geoscience and Remote Sensing, 44, 3374-3385.
http://dx.doi.org/10.1109/TGRS.2006.880628 - 27. Daamouche, A., Melgani, F., Alajlan, N. and Conci, N. (2013) Swarm Optimization of Structuring Elements for VHR Image Classification. IEEE Geoscience and Remote Sensing Letters, 10, 1334-1338.
http://dx.doi.org/10.1109/LGRS.2013.2240649 - 28. Nandi, R.J., Nandi, A.K., Rangayyan, R.M. and Scutt, D. (2006) Classification of Breast Masses in Mammograms Using Genetic Programming and Feature Selection. Medical and Biological Engineering and Computing, 44, 683-694.
http://dx.doi.org/10.1007/s11517-006-0077-6 - 29. Mavroforakis, M., Georgiou, H., Dimitropoulos, N., Cavouras, D. and Theodoridis, S. (2005) Significance Analysis of Qualitative Mammographic Features, Using Linear Classifiers, Neural Networks and Support Vector Machines. European Journal of Radiology, 54, 80-89.
http://dx.doi.org/10.1016/j.ejrad.2004.12.015 - 30. Wu, Z., Yuan, J., Lv, B. and Zheng, X. (2010) Digital Mammography Image Enhancement Using Improved Unsharp Masking Approach. 3rd International Congress on Image and Signal Processing (CISP), Yantai, 16-18 October 2010, 668-672.
http://dx.doi.org/10.1109/cisp.2010.5647218 - 31. Ramos, R.P., do Nascimento, M.Z. and Pereira, D.C. (2012) Texture Extraction: An Evaluation of Ridgelet, Wavelet and Co-Occurrence Based Methods Applied to Mammograms. Expert Systems with Applications, 39, 11036-11047.
http://dx.doi.org/10.1016/j.eswa.2012.03.020 - 32. Bick, U. and Diekmann, F. (2010) Digital Mammography. Springer, Berlin.
http://dx.doi.org/10.1007/978-3-540-78450-0 - 33. Mencattini, A., Rabottino, G., Salmeri, M., Lojacono, R. and Colini, E. (2008) Breast Mass Segmentation in Mammographic Images by an Effective Region Growing Algorithm. Springer-Verlag, New York, 948-957.
- 34. Haralick, R.M., Shanmugam, K. and Distein, I. (1973) Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics, SMC-3, 610-621.
http://dx.doi.org/10.1109/TSMC.1973.4309314 - 35. Haralick, R.M. (1979) Statistical and Structural Approaches to Texture. Proceedings of the IEEE, 67, 786-804.
http://dx.doi.org/10.1109/PROC.1979.11328 - 36. Kinoshita, S.K., de Azevedo-Marques, P.M., Pereira, R.R., Rodrigues, J.A.H. and Rangayyan, R.M. (2007) Content-based Retrieval of Mammograms Using Visual Features Related to Breast Density Patterns. Journal of Digital Imaging, 20, 172-190.
http://dx.doi.org/10.1007/s10278-007-9004-0 - 37. Matsubara, T., Fujita, H., Kasai, S., Goto, M., Tani, Y., Hara, T. and Endo, T. (1997) Development of New Schemes for Detection and Analysis of Mammographic Masses. Proceedings of Intelligent Information Systems IIS’97, Grand Bahama Island, 8-10 December 1997, 63-66.
http://dx.doi.org/10.1109/IIS.1997.645180 - 38. Mencattini, A., Rabottino, G., Salicone, S. and Salmeri, M. (2009) Uncertainty Propagation for the Assessment of Tumoral Masses Segmentation. IEEE International Workshop on Advanced Methods for Uncertainty Estimation in Measurement, Bucharest, 6-7 July 2009, 39-43.
http://dx.doi.org/10.1109/amuem.2009.5207603 - 39. Georgiou, H., Mavroforakis, M., Dimitropoulos, N., Cavouras, D. and Theodoridis, S. (2007) Multi-Scaled Morphological Features for the Characterization of Mammographic Masses Using Statistical Classification Schemes. Artificial Intelligence in Medicine, 41, 39-55.
http://dx.doi.org/10.1016/j.artmed.2007.06.004 - 40. Dash, M. and Liu, H. (1997) Feature Selection for Classification. Intelligent Data Analysis, 1, 131-156.
http://dx.doi.org/10.1016/S1088-467X(97)00008-5 - 41. Leardi, R., Boggia, R. and Terrile, M. (1992) Genetic Algorithms as a Strategy for Feature Selection. Journal of Chemometrics, 6, 267-281.
http://dx.doi.org/10.1002/cem.1180060506 - 42. Wang, X., Yang, J., Teng, X., Xia, W. and Jensen, R. (2007) Feature Selection Based on Rough Sets and Particle Swarm Optimization. Pattern Recognition Letters, 28, 459-471.
http://dx.doi.org/10.1016/j.patrec.2006.09.003 - 43. Yang, X.S. (2010) A New Meta-Heuristic Bat-Inspired Algorithm. In: Cruz, C., Gonzlez, J., Krasnogor, G.T.N. and Pelta, D.A., Eds., Nature Inspired Cooperative Strategies for Optimization (NISCO 2010), Studies in Computational Intelligence Vol. 284, Springer Verlag, Berlin, 65-74.
http://dx.doi.org/10.1007/978-3-642-12538-6_6 - 44. Cai, J., Ma, X., Li, Q., Li, L. and Peng, H. (2010) A Multi-Objective Chaotic Ant Swarm Optimization for Environmental/Economic Dispatch. International Journal of Electrical Power & Energy Systems, 32, 337-344.
http://dx.doi.org/10.1016/j.ijepes.2010.01.006 - 45. Geem, Z.W. (2010) Recent Advances in Harmony Search Algorithm. Studies in Computational Intelligence. Springer-Verlag, Berlin.
http://dx.doi.org/10.1007/978-3-642-04317-8 - 46. Geem, Z.W., Kim, J.H. and Loganathan, G.V.A. (2001) New Heuristic Optimization Algorithm: Harmony Search. Simulation, 76, 60-68.
http://dx.doi.org/10.1177/003754970107600201 - 47. Yang, X.S. and Deb, S. (2009) Cuckoo Search via Lévy Flights. World Congress on Nature & Biologically Inspired Computing, Coimbatore, 9-11 December 2009, 210-214.
- 48. Payne, R.B. (2005) The Cuckoos. Oxford University Press, New York.
- 49. Tuba, M., Subotic, M. and Stanarevie, N. (2011) Modified Cuckoo Search Algorithm for Unconstrained Optimization Problems. Proceeding of the 5th European Conference on European Computing Conference, Paris, 28-30 April 2011, 263-268.
- 50. Moksnes, A. and Roskaft, E. (1995) Egg-Morphs and Host Preference in the Common Cuckoo (Calculus canorus): An Analysis of Cuckoo and Host Eggs form European Museums and Collections. Journal of Zoology, 236, 625-648.
http://dx.doi.org/10.1111/j.1469-7998.1995.tb02736.x
NOTES
*Principal.










