Agricultural Sciences
Vol.3 No.4(2012), Article ID:20167,5 pages DOI:10.4236/as.2012.34066
Evaluation of pathogencity and race classification of Xanthomonas oryzae pv. oryzae in Guilan province—Iran
![]()
1Department of Plant Pathology, Rice Research Institute of Iran (RRII), Rasht, Iran
2Department of Plant Breeding, Rice Research Institute of Iran (RRII), Rasht, Iran; *Corresponding Author: Ebady_al@yahoo.com
3Department of Plant Breeding, Razi University, Kermanshah, Iran
Received 3 March 2012; revised 9 April 2012; accepted 16 May 2012
Keywords: Bacterial Blight; Near-Isogenic Lines; Pyramiding Lines; Race Classification; Rice; Xanthomonas oryzae pv. oryzae
ABSTRACT
To evaluation of pathogenicity and race classification of Xanthomonas oryzae pv. oryzae agent bacterial leaf blight of rice, 153 isolates of X. oryzae pv. oryzae were collected from different rice-growing cities of Guilan province—Iran. All of isolates were inoculated to assess the differential characteristics of 26 near isogenic rice lines containing a single resistance gene or two to five genes. Inoculation was done 21 days after sowing in the greenhouse. Scoring of inoculated plants was done 18 days after inoculation. The level of infection was not so clear among pyramiding lines, expect IRBB53 and IRBB61. Therefore, the pyramiding lines can not be used as differentials for pathogenicity evaluation of X. oryzae pv. oryzae The 12 rice lines with a single resistance gene were used further to establish a system of races classification of X. oryzae pv. oryzae IRBB14, IRBB21and IRBB7 were resistance to the most isolates. Whereas, IRBB1, IRBB2, IRBB4 and IRBB10 were susceptible to all isolates. Based on the interactions between the isolates X. oryzae pv. oryzae and the 12 nearisogenic rice lines, seven single-gene rice lines were chosen as differentials, and the 153 tested isolates were classified into four races. Except for cultivar types, different terrain, climate, period of rice planting and other factors may be associated with the population diversity and virulent variation of X. oryzae pv. oryzae.
1. INTRODUCTION
Bacterial leaf blight (BLB) of rice, [Oryza sativa] caused by Xanthomonas oryzae pv. oryzae [1], is a major disease in the tropics and subtropics of Asia countries [2]. The disease has become a major rice disease in last three decades because of the extensive deployment of more nitrogen-responsive modern rice varieties. The disease has also been reported in Australia, Africa, and latin America as well as in North America [3,4]. Bacterial blight can cause severe yield loss of up to 50% depending on the rice growth stage, the geographic location and seasonal conditions [5].
The disease, reported in Iran as early as in the 2007 [6], and because of the widely grown hybrid rice combinations with high yield spread rapidly. Efficacy of chemical on this disease is not significant. Therefore, planting resistance varieties provide an effective control and up to the present most breeders concentrate on the development of highly resistance varieties. Recently, thirty resistance genes for bacterial blight have been identified from rice [7-11], and most of them have been used to characterize the virulence of and differentiate among pathogenic races of X. oryzae pv. oryzae. Jeung et al. [12] demonstrated that developing resistant cultivars requires an understanding of the dynamics of the pathogen populations as well as the genetics of host resistance. Mew et al. [13] detected six virulence types (pathotypes or races) of the pathogen among 800 strains taken from around the Philippine over a 17-year period. Nine pathogenic races of X. oryzae pv. oryzae have thus been described from Nepal [14] and 10 have been described from the Philippines (C. M. vera cruz, personal communication). Liu et al. [15] worked on virulence analysis and race classification of X. oryzae pv. oryzae in china and reported that pyramided lines cannot be used as differential for the virulence analysis of X. oryzae pv. oryzae in china, so they used rice lines with a single gene further to establish a system of races classification of X. oryzae pv. oryzae in china. In Iran, 153 isolates from four rice cultivars (Hashemi-Khazar-Sadri-Alikazemi) from 2007 to 2009 had been collected. The genetic characteristic of the four rice cultivars are not completely known, probability the isolates identified in Iran did not match known resistance genes, and so they could not be compared with races identified in other countries. This study attempted for the pathogenicity analysis and races classification of X. oryzae pv. oryzae in Iran based on the interactions between the isolates of X. oryzae pv. oryzae collected from cities of Guilan province—Iran and differential lines carrying specific genes for BB resistance, provided from International Network for Genetic Evaluation of Rice (INGER), Philippines.
2. MATERIALS AND METHODS
2.1. Bacterial Isolates
One hundred and fifty-three isolates of X. oryzae pv. oryzae form 2007-2009 were collected from Guilan province in Iran. Bacteria were isolated from leaf portions showing typical bacterial blight lesions. A single colony of each isolate was selected from nutrient agar (NA) plates and incubated at 27˚C for 48 hours. For long-term storage, the purified strains were grown in peptone sucrose and frozen at –80˚C in 20% glycerol. The strains were revived on Luria Pepton (LP) (Difco) medium for experiments. All isolates were inoculated to local susceptible cultivar (Hashemi) for confirm their pathogenicity.
2.2. Rice Lines
Seeds of rice lines were obtained from the International Network for Genetic Evaluation of Rice (INGER) and used to preliminary evaluation of pathogenicity and race classification of Xanthomonas oryzae pv. oryzae in 2011 at Rice Research Institute of Iran. The 26 differential rice lines with a single gene or pyramid lines of two to five genes were tested in this study (Table 1). Rice seeds are pre-germinated for 48 hours at 28˚C in petri dishes. Germinated seeds were planted in rows 1-cm apart, in 35 × 26 × 10 cm plastic trays. Each line was planted in a 10-cm long row. The differential lines are planted in separate trays. Local resistant (Khazar) and susceptible (Hashemi) cultivars were planted at the center of each tray.
2.3. Inoculums Preparation and Inoculation
Bacterial isolates that were maintained at –80˚C were revived on NA (nutrient agar) slants at 27˚C for 48h. Each isolates was transferred to NA slants and incubated an additional 24 h at 27˚C. Each bacterial colony on the slants were suspended with sterilized distilled water and adjusted to concentrations of approximately 109 cfu/ml prior to inoculation [16]. Inoculation was done 21 days after sowing by clipping method with sterilized scissors [17]. The inoculated plants were grown in green house. To reduce the possible effects of high temperature on disease reactions, favour the entry of bacteria into infection courts in the presence of sufficient moisture on the leaf surface and maintain inoculation time consistency, the inoculation was conducted in one morning.
2.4. Disease Assessment
Disease severity was rated by measuring the lesion length from the leaf tips. Scoring was done at 18 days after inoculation (DAI). The scales to be used in scoring
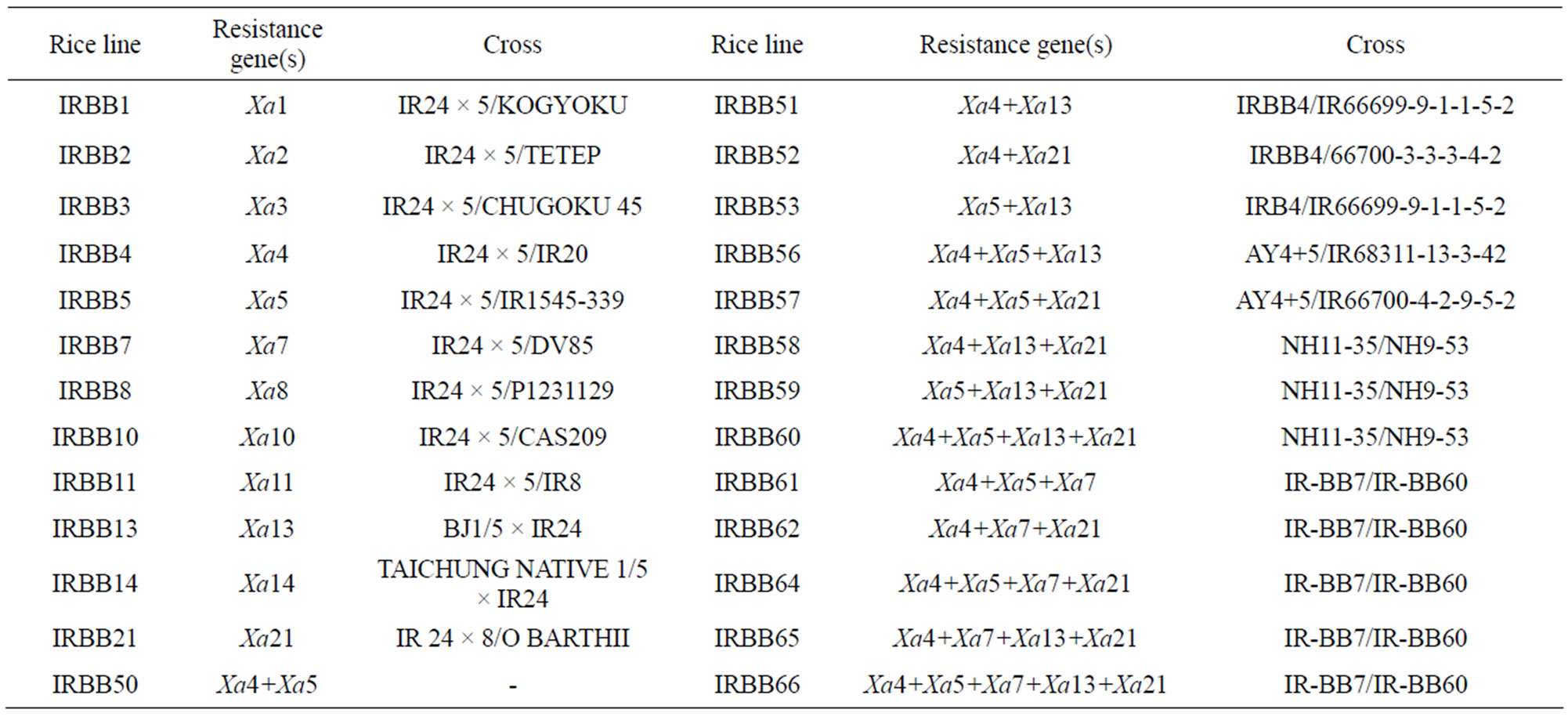
Table 1. Crosses and Resistance genes of differential rice lines used to characterize virulence of Xanthomonas oryzae pv. oryzae isolates in Iran.
were those from the 4th edition of the Standard Evaluation System for Rice [18]. The data analyzed for the main effect and interaction effect between cultivars and bacterial isolates.
3. RESULTS
3.1. Reactions of Differential Rice Lines
On each cultivar, lesions developed uniformly downward from the point of inoculation. Interaction between differential rice lines and 153 isolates were used to assess the differential characteristic of 26 near isogenic rice lines that provided from INGER. The degree of disease reaction in this experiment showed different relationship between combination of rice lines and bacterial isolates. The pyramiding lines containing two to five genes expect, IRBB53 and IRBB61 were relatively high resistant to approximately most tested isolates. IRBB50, IRBB52, IRBB57, IRBB59, IRBB60 and IRBB66 showed hypersensitive reaction to all tested isolates (Figure 1). Many isolates with high virulence on near-isogenic lines were avirulent on pyramiding lines. Therefore the most pyramiding lines could not be used as differential to virulence analysis of X. oryzae pv. oryzae. The 12 rice lines with a single-gene were selected as candidates for races classification X. o. pv. oryzae in Guilan province.
3.2. Race Classification of X. oryzae pv. oryzae
One hundred and fifty-three isolates were subjected to virulence evaluation on 12 near-isogenic lines with a single gene. All isolates were virulent on IRBB1, IRBB2, IRBB4 and IRBB10 and most isolates were avirulent on IRBB7, IRBB14 and IRBB21. Therefore, IRBB1 was chosen as a susceptible line and IRBB7 was chosen as a resistant line. IRBB3, IRBB5, IRBB13, IRBB8 and IRBB11 showed distinct resistant or susceptible reactions to different isolates. Based on the interaction between the tested isolates and the near-isogenic rice lines, seven rice lines with a single gene, IRBB1, IRBB3, IRBB5, IRBB13, IRBB8, IRBB11 and IRBB7, were chosen as the differentials for identifying pathogenic races of X. oryzae pv. oryzae in Guilan province of Iran. As a result, 153 isolates of X. oryzae pv. oryzae were classified into four pathogenic races (Table 2).
4. DISCUSSION
Pathogenic race of plant bacterial is usually classified based on the interaction of an avr gene in pathogen and
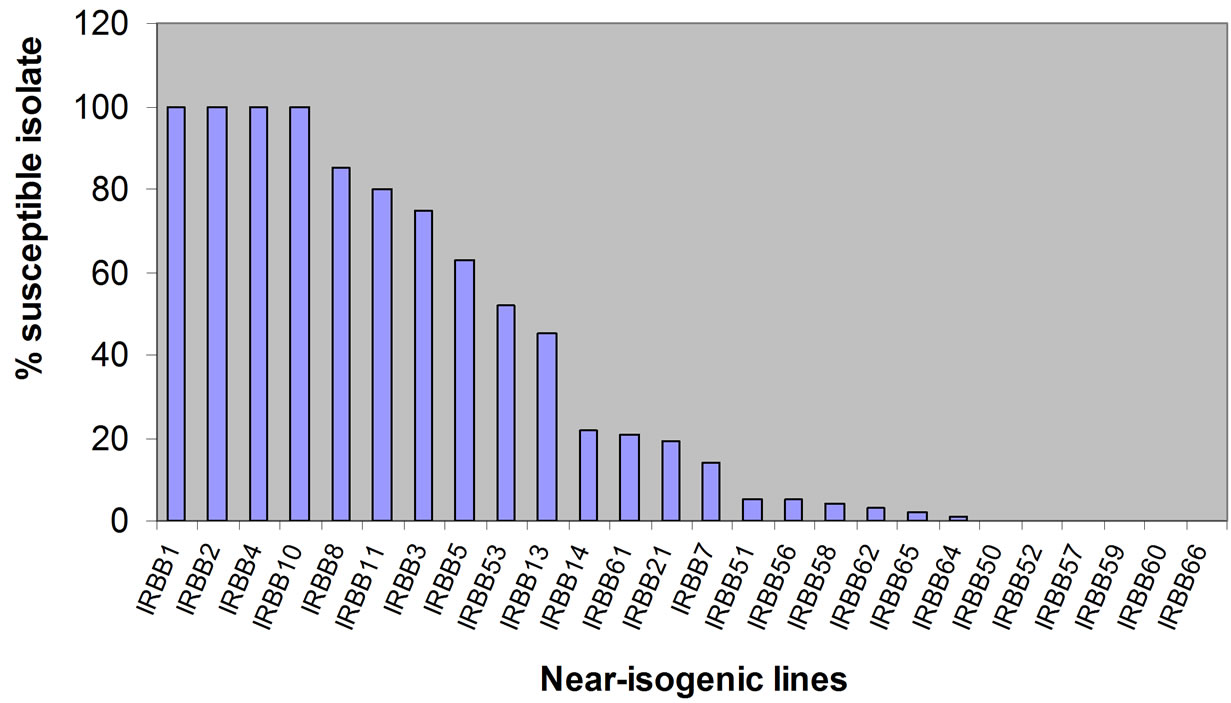
Figure 1. Percentage of susceptible isolates on 26 rice lines inoculated with 153 isolates of Xanthomonas oryzae pv. oryzae. The discrimination of 26 rice lines in X. oryzae pv. oryzae was evaluated by Standard Evaluation System for rice of IRRI, 21 days after inoculation. (Hs-S-Ms = s and HR-R-MR = R).
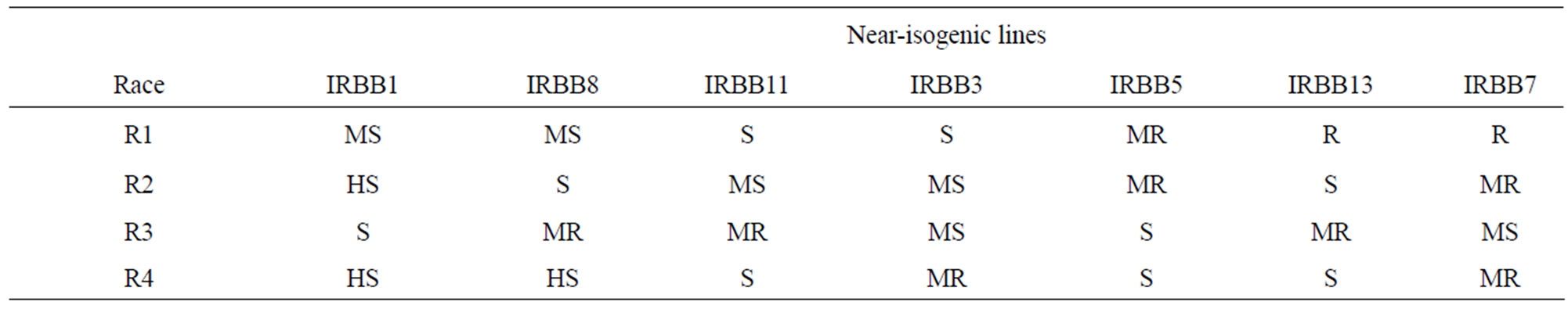
Table 2. Interaction between Xanthomonas oryzae pv. oryzae isolates and near-isogenic lines.
Xa gene in the host. Therefore, we used a set of differential lines with various resistance genes [12,19,20]. Virulence of Xanthomonas oryzae pv. oryzae races against near-isogenic rice lines with known resistance genes will give a better understanding of interactions between the host and the pathogen in genetics and a characterization of pathogen races in relation to resistance genes, as well as provide an important information that can be used in breeding programmes to develop effective resistance in rice cultivars [12,21].
In this study, we estimated the differential characteristics of 26 near-isogenic rice lines carrying different single and combinations resistance genes to 153 isolates of X. oryzae pv. oryzae in Guilan province, Iran. The results showed that approximately most pyramiding lines were resistance to the tested isolates and could not be used as differential to characterize the virulence of X. oryzae pv. oryzae. So, we selected 12 rice lines with a single gene to identification and classification the races of Xanthomonas oryzae pv. oryzae. The phenotype reactions between the single-gene lines and isolates were clear and easily classified into virulence or avirulence patterns. The degree of IRBB2, IRBB4 and IRBB10 susceptible were similar to that of IRBB1. The resistance of IRBB14 and IRBB21 were similar to that of IRBB7. Another lines showed different reactions to isolates. As a result, seven differential lines chosen for race classification and all X. oryzae pv. oryzae isolates in Guilan province were classified into four pathogenic races.
The major goals of rice-breeding programmes was to develop rice cultivars with effective resistance genes to X. oryzae pv. oryzae. In this study, we establish the level of resistance to the bacterial blight pathogen conferred by a single gene individually and multiple gene combinations. Among 12 single genes Xa1 was low resistance and Xa7 was high resistance on the basis of percentages of the compatible strains, also Xa7 was not avirulence to all tested isolates. These results indicate that Xa21, Xa14 and especially Xa7 are resistant to the most isolates and they could be effectively used for Bacterial leaf blight resistance breeding in Iran. The gene combination showed higher resistance levels than a single gene, but the Xa5 and Xa13 combination in IRBB53 was less resistant than gene Xa13. The results of this study just classified isolates of X. oryzae pv. oryzae that were collected from different rice-growing cities of Guilan province—Iran and may not reflect the diversity and differentiation of pathogen population in other countries where bacterial blight is an important constraint to rice production. There are obvious difference in the population structure of Xanthomonas oryzae pv. oryzae from different countries due to different agroecosystems [22]. Except for cultivar types, different terrain, climate, period of rice planting and other factors may be associated with the population diversity and virulent variation of Xanthomonas oryzae pv. oryzae.
Therefore, a breeding program should be initiated to transfer resistance genes from these differential lines to commercial and high quality Iranian rice varieties for controlling bacterial blight disease effectively.
5. ACKNOWLEDGEMENTS
We thank Rice Research Institute of Iran (RRII) for preparation of field, plant material and all finance to doing this project.
REFERENCES
- Swings, J., Vander Mooter, M., Vauterin, L., Hoste, B., Gillis, M., Mew, T.W. and Kersteres, K. (1990) Reclassification of the causal agents of bacterial blight (Xanthomonas campestris pv. oryzicola) of rice as pathovars of Xanthomonas oryzae (ex Ishiyama, 1922) sp. nov., nom. rev. International Journal of Systematic Bacteriology, 40, 309-311.
- Mew, T.W. (1987) Current status and future prospects of research on bacterial blight of rice. Annual Review of Phytopathology, 25, 359-382. doi:10.1146/annurev.py.25.090187.002043
- Mew, T.W. (1993) Xanthomonas oryzae pathovars on rice, cause of bacterial blight and bacterial leaf streak. in: Swings, J.G. and Civerolo, F.L., Eds., Xanthomonas, Chapman & Hall, Lpndon, 30-40.
- Ryba-White, M., Notteghem, J.L. and Leach, J.E. (1995) Comparison of Xanthomonas oryzae pv. oryzae strains from Africa, north America and Asia by RFLP analysis. International Rice research notes, 20, 25-26.
- Ou, S.H. (1985) Rice diseases. Common wealth Mycological Institute, Surrey.
- Khoshkdaman, M., Niknejad kazmpour, M., Ebadi, A.A. and Pedramfar, H. (2009) Identification of causal agent of bacterial bight of rice in the fields of Guilan province. Plant Protection Journal, 23, 50-57.
- Chu, Z.H., Fu, B.Y., Li, Z.K., Zhang, Q.F. and Wang, S.P. (2004) Targeting the recessive rice gene, Xa13, for bacterial blight resistance to a 14.8-kb DNA fragment. The fourth international crop science congress. http://www.cropscience.org.au/icsc2004/poster/3/2/1/index.htm
- Huang, N., Angeles, E.R., Domingo, J., Magpantay, G., Singh, S., Zhang, G., Kumaravadivel, N., Bennett, J. and Khush, G.S. (1997) Pyramiding of bacterial blight resistance genes in rice, marker-assisted selection using RFLP and PCR. Theoretical and Applied Genetics, 95, 313-320. doi:10.1007/s001220050565
- Khush, G.S., Mackill, D.J. and Sindhu, G.S. (1989) Breeding rice resistance to bacterial blight. In: Ogawa, T. and Khush, G.S., eds., Bacterial Blight of Rice, Manila, Philippines. International Rice Research Institute, Los Baños, 207-217.
- Ogawa, T., Yamamoto, T., Khush, G.S. and Mew, T.W. (1991) Breeding of near-isogenic lines of rice with single genes for resistance to bacterial blight pathogen (Xanthomonas campestris pv. oryzae). Japanese Journal of Breeding, 41,523-529.
- Sun, X.L., Cao, Y.L., Yang, Z.F., Xu, C.G., Li, X.H., Wang, S.P. and Zhang, Q.F. (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. The Plant Journal, 37,517-527. doi:10.1046/j.1365-313X.2003.01976.x
- Jeung, J.U., Heu, S.K., Shin, M.S., Cruz, C.M.V. and Jena, K.K. (2006). Phytopathology, 96, 867-875. doi:10.1094/PHYTO-96-0867
- Mew, T.W., Cruz, C.M.V. and Medalla, E.S. (1992) Changes in the race frequency of Xanthomonas oryzae pv. oryzae in response to rice cultivars planted in the Philippines. Plant Disease, 76, 1029-1032. doi:10.1094/PD-76-1029
- Adhikari, T.B., Mew, T.W. and Teng, P.S. (1994) Phenotypic diversity of Xanthomonas oryzae pv. oryzae in Nepal. Plant Disease, 78, 68-72. doi:10.1094/PD-78-0068
- Liu, H., Yang, W., Hu., B. and Liu, F. (2007) Virulence analysis and race classification of Xanthomonas oryzae pv. oryzae in China. Journal of Phytopathology, 155, 129- 135. doi:10.1111/j.1439-0434.2007.01197.x
- Fang, Z.D., Xu, Z.G., Guo, C.J., Chen, Y.L., Deng, X.R., Li, H.S. and Zhu, L.M. (1981) Variability of pathogenicity of rice bacterial leaf blight organism. Journal of Nanjing Agricultural College, 1, 1-11.
- Kauffman, H.E., Reddy, A.P.K., Hsieh, S.P.Y. and Merca, S.D. (1973) An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Disease Reporter. 57, 537-541.
- International Rice Research Institute (1996) Standard Evaluation System for Rice. 4th edition, IRRI, Los Baños.
- Adhikari, T.B., Basnyat, R.C. and Mew, T.W. (1999) Virulence of Xanthomonas oryzae pv. oryzae on rice lines containing single resistance genes and gene combinations. Plant Disease, 83, 46-50. doi:10.1094/PDIS.1999.83.1.46
- Mew, T.W., Alvarez, A.M., Leach. J.E. and Swings. J. (1993) Focus on bacterial blight of rice. Plant Disease, 77, 5-12. doi:10.1094/PD-77-0005
- Mazzola, M., Leach, J.E., Nelson, R. and White, F.F. (1994) Analysis of the interaction between Xanthomonas oryzae pv. oryzae and the rice cultivars IR24 and IRBB21. Phytopathology, 84, 392-397. doi:10.1094/Phyto-84-392
- Ardales, E.Y., Leung, H., Vera Cruz, C.M., Leach, J.E., Mew, T.W. and Nelson, R.J. (1996) Hierarchical analysis of spatial variation of the rice bacterial blight pathogen across agroecosystems in the Phillippines. Phytopathology, 86, 241-252. doi:10.1094/Phyto-86-241

