American Journal of Plant Sciences
Vol.06 No.13(2015), Article ID:59217,6 pages
10.4236/ajps.2015.613216
Molecular Markers Associated with Ph-3 Gene Conferring Late Blight Resistance in Tomato
Dilip R. Panthee*, Randy G. Gardner, Ragy Ibrahem, Candice Anderson
Department of Horticultural Science, North Carolina State University, Mountain Horticultural Crops Research and Extension Center, Mills River, NC, USA
Email: *dilip_panthee@ncsu.edu
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 2 July 2015; accepted 23 August 2015; published 28 August 2015
ABSTRACT
Late blight (LB), caused by the oomycete Phytophthora infestans, is one of the most devastating diseases of tomato. Three major genes Ph-1, Ph-2 and Ph-3 conferring resistance to LB have been identified and mapped to the chromosomes 7, 10 and 9, respectively. However, PCR-based molecular markers associated with these genes are limited. Molecular markers are extremely useful in the screening and selection of tomato lines for the development of LB resistant genotypes. The objective of this study was to identify molecular markers associated with Ph-3 gene conferring LB resistance in tomato. Four co-dominant markers were found to be associated with Ph-3, all of which were sequence characterized amplified region (SCAR) type. Breeding lines and cultivars were inoculated with a field isolate of Phytophthora infestans to collect phenotypic data on disease resistance. Genotypic data from molecular markers associated with Ph-3 were in close agreement with the phenotypic data for the lines tested. With the verification of genotypic data from novel molecular markers in known genotypes supported by phenotypic data, the novel molecular markers may be useful in screening tomato populations aiming to develop LB resistant genotypes or cloning the LB resistant genes.
Keywords:
Late Blight, Molecular Breeding, Phytophthora infestans, Resistance Breeding, Tomato

1. Introduction
Late blight (LB), caused by the oomycete Phytophthora infestans (Montagne, Bary), is one of the most potentially devastating diseases of tomato in areas with high humidity and cool temperatures and can cause 100% crop loss in unprotected tomato fields or greenhouses. Because of its devastating economic impact, this disease has been the subject of intensified pathological and genetic research since the occurrence of the Irish potato famine in the 1840s. Genetic resistance to LB in tomato has been of interest for many years, and three major resistance genes have been identified in the red-fruited tomato wild species S. pimpinellifolium, including Ph-1, Ph-2 and Ph-3, which have been mapped to tomato chromosomes 7, 10, and 9, respectively. Ph-1 is a single dominant gene providing resistance to race T-0, but it was rapidly overcome by new races of the pathogen. Ph-1 was mapped to the distal end of chromosome 7 using morphological markers [1] . However, no molecular marker associated with this resistance gene has been reported. Currently, P. infestans race T-1 predominates, rendering the resistance conferred by Ph-1 ineffective. The resistance conditioned by Ph-2, a single incomplete-do- minant gene mapped to the lower end of the long arm of tomato chromosome 10 [2] , provides partial resistance to several isolates of race T-1 [1] [3] . Ph-2 slows, but does not stop the disease progress [2] . Furthermore, Ph-2 often fails in the presence of more aggressive isolates [4] [5] . Ph-2 has been mapped to an 8.4 cM interval on the long arm of chromosome 10 between RFLP markers CP105 and TG233 [2] . A much stronger resistance gene, Ph-3, was discovered in S. pimpinellifolium accessions L3707 and L3708 (a.k.a. LA 1269 or PI365957) at the Asian Vegetable Research and Development Center (AVRDC) in Taiwan [4] . Currently this gene is much more useful than Ph-1 and Ph-2 and confers incompletely dominant resistance to a wide range of P. infestans isolates of tomato, including those that overcome Ph-1 and Ph-2 [6] . Ph-3 has been mapped to the long arm of chromosome 9 near RFLP marker TG591a [6] . Recently, this gene was fine-mapped in the 0.5 cM genomic region of long arm of chromosome 9 in between Indel_3 and P55 molecular markers [7] . Utilizing the fine-map information, and considering the significance of the gene in late blight resistance, it was further characterized by cloning a 24 kb region [8] . Recently, co-dominant sequence characterized amplified region (SCAR) [9] and cleaved amplified polymorphic sequence (CAPS) markers have been developed for use in marker-assisted breeding for Ph-3 (M. Mutschler, pers. comm.) [10] . Identification of these markers associated with Ph-3 has been extremely useful to transfer the Ph-3 gene into the desirable genetic background by marker-assisted selection (MAS). However, it has been suggested that the full resistance conferred by L3707 and L3708 (the original sources of Ph-3) is conferred by more than just one Ph-3 locus, and that Ph-3 alone in either homozygous or heterozygous conditions would not be highly commercially desirable as it does not confer strong resistance against aggressive isolates such as US-7 and US-17 [11] [12] . The presence of yet undetermined additional hypostatic gene(s) present in L3708 is necessary to provide full resistance. Further, it has been determined that there are new P. infestans isolates which have overcome the Ph-3 resistance [6] . However, it appears that a combination of Ph-2 and Ph-3 confers strong resistance to most such isolates. Recently, several tomato-breeding programs around the world, including the North Carolina State University, Pennsylvania State University, Cornell University (M. Mutschler, pers. comm.), and AVRDC―The World Vegetable Center (P. Hanson, pers. comm.), have succeeded in transferring LB resistance genes to fresh-market and/or processing tomato breeding lines or hybrid cultivars using a combination of phenotypic screening and MAS. For example, most recently, several fresh-market tomato breeding lines (e.g., NC1 CELBR (Ph-2 + Ph-3) and NC2 CELBR (Ph-2 + Ph-3) and hybrid cultivars Plum Regal (Ph-3), Mountain Magic (Ph-2 + Ph-3) and Mountain Merit (Ph-2 + Ph-3)) have been released by the North Carolina State Tomato Breeding Program, USA [13] [14] . Also, more breeding lines and cultivars are in the pipeline from these and other tomato breeding programs. However, the availability of more useful PCR- based markers for Ph-2 and Ph-3 will make the selection and breeding for LB resistance in tomato more expedient. The objective of this study was to identify molecular markers associated with Ph-3 gene conferring LB resistance in tomato.
2. Materials and Methods
Tomato lines used in this study are described in Figure 1. The control lines used were NC 1CELBR (Resistant homozygous), NC 123S (Susceptible homozygous), and Mountain Merit F1 hybrid (NC 1CELBR × NC 123S) (Heterozygous) for Ph-2 and Ph-3. Lines NC 25P (Resistant homozygous), NC 30P (Susceptible homozygous) and hybrid Plum Regal (NC 30P × NC 25P) (Heterozygous) for Ph-3 were also included. Additional lines with known status for Ph-2 and Ph-3 included were 63EB(2002)-1C, NC 870(2002)-1B and the F1 hybrid, NC03220, from the cross between these two lines. The original source of the Ph-3 gene used in developing NC 1CELBR (L3707) and the source of the Ph-2 gene for NC 1CELBR (Richter’s Wild Tomato) were also included. Other




Figure 1. Molecular markers associated with Ph-3 mediated late blight resistance in tomato. In the gel, the sample order is: 1: NC1CELBR-R, 2: NC25P-R, 3: NC123S-S, 4: NC30P-S, 5: Mtn. Merit-H, 6: Plum Regal-H, 7: 08135(x)-1W, 8: 08135(x)- 6W, 9: 145L-1(2008), 10: 62L-1(2008), 11: 08135(x)-11W, 12: 860-1A(2008), 13: 860-1B(2008), 14: LA3707, 15: Micro Tom, 16: 68L-1(2008), 17: 146L-1(2008), 18: 22L-2W(2008), 19: 22L-1W(2008), 20: 69L-1(2008), 21: 70L-1(2008), 22: 870(2002)-1B, 23: 63EB(2002)-1C, and 24: NC03220. (a) NCLB-9-6676 SCAR-type molecular marker associated with Ph-3 gene conferring late blight resistance in tomato. Resistant genotypes NC 1CELBR and NC 25P produced 1000 bp PCR product whereas susceptible genotypes NC 30P and NC 123S produced 1300 bp PCR product. Both fragments were present in heterozygous genotypes Plum Regal and Mountain Merit. (b) NCLB-9-6677 SCAR-type molecular marker associated with Ph-3 gene conferring late blight resistance in tomato. Resistant genotypes NC 1CELBR and NC 25P produced 1000 bp PCR product and susceptible genotypes NC 30P and NC 123S produced 1250 bp PCR product. Both fragments were present in heterozygous genotypes Plum Regal and Mountain Merit. (c) NCLB-9-6678, SCAR-type molecular marker associated with Ph-3 gene conferring late blight resistance in tomato. Resistant genotype NC 1CELBR and NC 25P produced 600 bp PCR product whereas susceptible genotypes NC 30P and NC 123S produced 900 bp PCR product. Both fragments were present in heterozygous genotypes Plum Regal and Mountain Merit. (d) NCLB-9-6679 SCAR-type molecular marker associated with Ph-3 gene conferring late blight resistance in tomato. Resistant genotype NC 1CELBR and NC 25P produced 1000 bp PCR product whereas susceptible genotypes NC 30P and NC 123S produced 1200 bp PCR product. Both fragments were present in heterozygous genotypes Plum Regal and Mountain Merit.
lines with known status for Ph-2 and Ph-3 included 96LB, an early generation backcross line derived from L3707 and 139LB(2001)3W-8 and 139LB(2001)3W-64, both early backcross generation lines derived from “Richter’s Wild Tomato”. Other lines tested were resistant selections made from field trials where late blight was present but that had not been identified for their status of the Ph-2 and Ph-3 genes. Three grape tomato lines (08135 selfed lines from F4 generation) that had been selected for superior horticultural traits and had limited testing for late blight resistance were included. The lines were planted with two replications per line and three plants per replication in 24-cell plastic trays. Plants were maintained in a controlled condition with 25/15˚C day and night temperatures, 70% - 90% relative humidity, and 12/12 hour photoperiods in the greenhouse. Field isolate of Phytophthora infestans was collected from naturally infested susceptible tomato genotypes from the field at Mountain Horticultural Crops Research & Extension Center (MHCREC) in the summer of 2012 and inoculum was maintained on NC 123S (susceptible genotype) leaves under controlled conditions. In the greenhouse, seven-week-old plants were inoculated with the field isolate of Phytophthora infestans at the rate of 10,000/mL sporangia. Individual plants were hand misted twice at 30 minute intervals to ensure that all plants received uniform inoculum pressure. Plants were evaluated after one week using a score from 0 - 5 (where, 0 was no lesions, and 5 was for plant parts completely covered with lesions or dead).
The main basis for designing primers in this study is the mapping information available for Ph-2 and Ph-3. These two genes are mapped to chromosome 10 and 9, respectively. The latest information on functional genomics resources from these two chromosomes was utilized, which was given as follows: NC-LB-10-6395 (Solyc10g085460.1-CC-NBS-LRR, resistance protein); NC-LB-9-6676-2 (Solyc09g092280.1.1-CC-NBS-LRR, resistance protein); NC-LB-9-6677-2 (Solyc09g092290.1.1-CC-NBS-LRR, resistance protein); NC-LB-9-6678 (Solyc09g092300.2.1-CC-NBS, resistance protein fragment); NC-LB-9-6679 (Solyc09g092310.1.1-CC-NBS- LRR, resistance protein) (The downstream end of the integration depending on SNP data). Associated genome sequence was downloaded from Sol Genomics Network (http://solgenomics.net/organism/Solanum_lycopersicum/genome). Resources were also utilized from Solanaceae Coordinated Agriculture Project (http://solcap.msu.edu/). Twenty-five primer pairs were designed on the basis of this sequence using Primer 3.0 software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) with default settings. A list of informative primers associated with Ph-3 is reported in Table 1.
Genomic DNA was extracted from 2-week-old plants of each line using a standard CTAB protocol [15] . DNA concentration was measured using a Nanodrop-1000 (Nano Drop Products, Wilmington, DE) and adjusted to a final concentration of 20 ng/µL. All markers examined in this study were PCR-based, including cleaved amplified polymorphic sequences (CAPS) and sequence characterized amplified regions (SCAR). AllPCR primers were ordered from Integrated DNA Technologies, Inc. (Coralville, IA). PCR master-mix was used from Phenix Research Products (Candler, NC) and QIAGEN (Valencia, CA). PCR was conducted in 25-µL volume consisting of 12.5 µL master mix (1.5 mM MgCl2, 0.2 mM dNTPs, 0.5 U Taq polymerase and 10× PCR buffer), 2 µL DNA (20 ng/µL), 1.5 µL (12.5 µM) primers (total of forward and reverse), and 7.5 µL of nuclease free water. DNA amplifications were performed in a thermal cycler (Eppendorf, New York) using the following cycling condition: one cycle of 92˚C for 3 min, followed by 35 cycles of 92˚C for 0.30 min, 52˚C for 1 min, and 72˚C for 0.30 min; and final extension at 72˚C for 8 min, followed by holding at 4˚C. Following PCR amplification, PCR products were either directly separated on 1.5% Agarose gel and DNA bands visualized with 0.5 µg/ml ethidium bromide staining or first digested with a restriction enzyme(Hae III) and then separated on Agarose gel (for CAPS markers). DNA fragment sizes were determined using a 100-bp DNA ladder (Phenix Research Products, Candler, NC). For all markers, PCR amplification and gel electrophoresis were carried out at least twice to confirm the results.
3. Results and Discussion
Four sequence characterized amplified region (SCAR) molecular markers were found to be associated with Ph-3 gene. The novel co-dominant SCAR marker NCLB-9-6676 associated with Ph-3 gene produced 1000 bp and 1300 bp PCR product from resistant and susceptible genotypes, respectively (Figure 1(a)). Similarly, NCLB-9- 6677 was also found to produce the PCR product of 1000 and 1250 bp from resistant and susceptible lines, respectively (Figure 1(b)).
Yet another co-dominant SCAR molecular marker NCLB-9-6678 was also capable to discriminate resistant
Table 1. List of forward and reverse primers for informative sequence characterized amplified region (SCAR) markers associated with Ph-3 gene conferring late blight resistance in tomato.
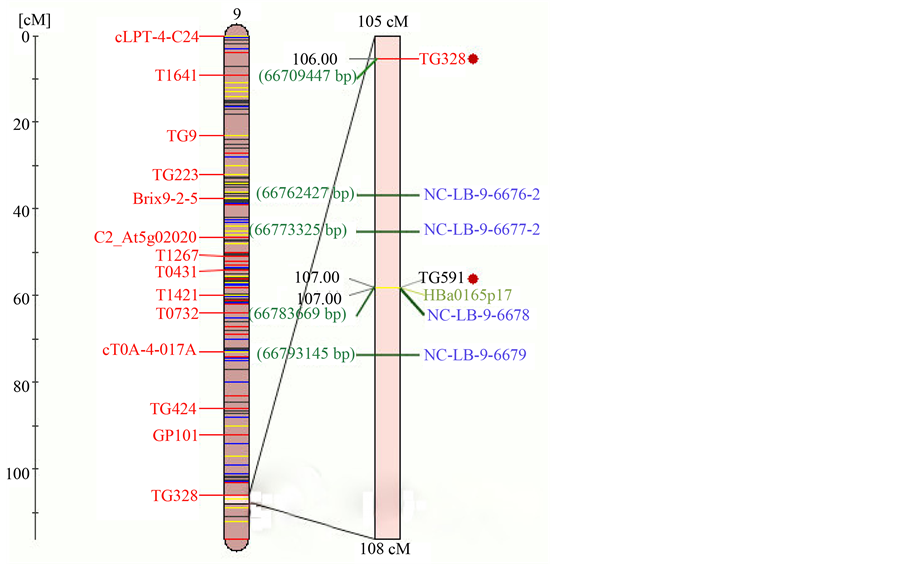
and susceptible genotypes of tomato by producing 600 and 900 bp DNA fragments (Figure 1(c)). Hybrid genotypes Plum Regal and Mountain Merit had distinct double bands. Similarly, NCLB-9-6679 produced 1000 and 1200 bp PCR product from resistant and susceptible genotypes, respectively (Figure 1(d)). Chromosomal location of these markers associated with Ph-3 gene is shown in Figure 2. Informative primers associated with Ph-3 gene, and expected DNA fragments are given in Table 1.
Results were consistent for all lines for the four new SCAR markers associated with Ph-3 gene. Important point to note was that genotypic data from new molecular markers on established genotypes (such as NC1- CELBR, NC123S, Mountain Merit, NC25P, NC30P and Plum Regal) with known gene status was in perfect agreement making the finding robust.
In the phenotypic ratings for late blight incidence, NC 1CELBR and 08135(x)-1W, both of which were R for Ph-2 and Ph-3 genotypic data, and Richter’s Wild Tomato, which was R for Ph-2 but S for Ph-3 according to the genotypic data, were completely free of late blight. Two other lines, 860-1A(2008) and NC161L [16] both of which were R for both Ph-2 and Ph-3 according to the genotypic data, had a significantly higher ratings for late blight incidence than the above three lines. The hybrid “Mountain Merit” (NC 1CELBR xNC123S), which was H for both Ph-2 and Ph-3, had a higher disease rating than the resistance parent NC 1CELBR (data not shown). Also, the hybrid “Plum Regal”, which was heterozygous for the Ph-3 gene alone, had a higher disease rating than its resistant parent, NC 25P.
We identified four co-dominant molecular markers associated with Ph-3 gene. These molecular markers (NCLB-9-6676, NCLB-9-6677, NCLB-9-6678, and NCLB-9-6679) are novel and are expected to be useful for tomato breeding program in marker-assisted selection. Because Phytophthora infestans spores can travel a long distance by wind in a relatively short time, have a relatively short incubation period, and produce millions of spores before symptoms are visible, and screening is complicated [17] . Therefore, development of reliable co- dominant molecular markers is extremely important. Findings from this study are likely to pave the way towards fulfilling this necessity.
We adapted the approach of genome-based molecular marker development utilizing the currently available tomato genome (http://solgenomics.net/organism/Solanum_lycopersicum/genome). On the basis of this information, novel molecular markers NCLB-9-6676, NCLB-9-6677, NCLB-9-6678, and NCLB-9-6679 associated with Ph-3 are located on 66,762,879, 66,773,785, 66,784,000, and 66,792,551 bp, respectively. With respect to previously identified RFLP molecular marker, TG591 associated with Ph-3, distance between TG591 and these molecular markers associated with Ph-3 are within 8 to 83 kbp distance (<1 cM) in this study (http://solgenomics.net/marker/SGN-M88/details),which is within the genome sequence of Ph-3 gene (Figure 2). This is potentially within the fine-map and clonned distance of the Ph-3 gene [7] [8] . Furthermore, we utilized the annotation information including CC-NBS-LRR to select the genomic sequence to design the primers (see Materials and Methods), which was the protein found in characterizing the clonned Ph-3 gene [8] . A strong association between genotypic and phenotypic data obtained from greenhouse experiments using field isolate was found in the present experiment. The combination of Ph-2 and Ph-3 confers resistance to late blight better than with a single gene, which is also observed in the present study.
Since molecular markers were found strongly associated with Ph-3 gene conferring resistance to late blight, these molecular markers may facilitate the combination of Ph-2 and Ph-3 genes in tomato breeding lines even without field inoculation. However, testing of lines for late blight resistance in field, greenhouse, and growth chamber inoculation trials will likely be needed in addition to marker testing to achieve a high level of resistance. Some lines homozygous for the Ph-2 and Ph-3 genes separately or combined were highly resistant whereas oth- er lines indicated to be homozygous for the resistance genes by marker results had a lower level of resistance. This
Figure 2. Chromosome 9 showing the positions of molecular markers identified in the present study with respect to Ph-3 conferring resistance to late blight in tomato.
indicates the presence of additional modifying genes for late blight resistance or resistance gene analog (RGA) in addition to the Ph-2 and Ph-3 major genes. “Richter’s Wild Tomato” which had a high level of late blight resistance in field trials was chosen as a source of the Ph-2 gene. However, the lower resistance level in the 63EB(2002)-1C line derived from backcrossing the Ph-2 gene from “Richter’s Wild Tomato” indicates that there may have been modifying genes for late blight resistance that are lost in the backcross process. Earlier backcross generation lines (139LB(2001)3W-64 and 139LB(2001)3W-8) had a higher level of late blight resistance than the 63EB(2002)-1C line (data not shown).
Ph-2 and Ph-3 genes have been combined by conventional methods in a large-fruited tomato [18] . However, these two genes are yet to be combined in plum and grape tomatoes, which are commercially important classes of tomatoes. With the availability of these markers, the process of combining these genes in plum and grape tomatoes may be expedited. The 08135(x) selections tested in this study are grape tomato lines. The highly resistant reaction of the 08135(x)-1W selection compared to the other 08135 selection, together with the marker results indicating it to be homozygous resistant for Ph-2 and Ph-3 combined, makes this line desirable for further selection and breeding for late blight resistance in grape tomatoes. Because of the incompletely dominant control of late blight resistance with both the Ph-2 and Ph-3 genes, it would be desirable to have the genes in both parents used to make commercial F1 hybrids to enhance the resistance level of the hybrids. The use of molecular markers to identify the Ph-3 resistance genes and their status in lines will greatly facilitate the process of developing diverse lines with the resistance genes for use as parents in hybrids.
4. Conclusion
We have developed novel PCR-based molecular markers associated with Ph-3 gene. With the verification of molecular data using phenotypic data generated from field isolate, it is expected that these markers will be useful to expedite the process of introgression of Ph-3 gene into desirable genetic background. However, it is always desirable to verify the findings in populations derived from wider genetic backgrounds. SCAR markers associated with Ph-3 are expected to be more useful since they are found more consistent in the present experiment.
Acknowledgements
Support for this study was provided, in part, from the North Carolina Tomato Growers Association. The authors thank Dr. Julia Kornegay, Dr. George Allen, and Rosy Hatch for reviewing the manuscript.
Cite this paper
Dilip R.Panthee,Randy G.Gardner,RagyIbrahem,CandiceAnderson, (2015) Molecular Markers Associated with Ph-3 Gene Conferring Late Blight Resistance in Tomato. American Journal of Plant Sciences,06,2144-2150. doi: 10.4236/ajps.2015.613216
References
- 1. Peirce, L.C. (1971) Linkage Tests with Ph Conditioning Resistance to Race 0, Phytophthora infestans. Report of the Tomato Genetics Cooperative, 21, 30.
- 2. Moreau, P., Thoquet, P., Olivier, J., Laterrot, H. and Grimsley, N. (1998) Genetic Mapping of Ph-2, a Single Locus Controlling Partial Resistance to Phytophthora infestans in Tomato. Molecular Plant-Microbe Interactions, 11, 259-269.
http://dx.doi.org/10.1094/MPMI.1998.11.4.259 - 3. Gallegly, M.E. (1960) Resistance to the Late Blight Fungus in Tomato. Proceedings of Plant Science Seminar, Campbell Soup Company, Camden, USA, 113-135.
- 4. Black, L.L., Wang, T.C., Hanson, P.M. and Chen, J.T. (1996) Late Blight Resistance in Four Wild Tomato Accessions: Effectiveness in Diverse Locations and Inheritance of Resistance. Phytopathology, 86, S24.
- 5. Goodwin, S.B., Schneider, R.E. and Fry, W.E. (1995) Use of Cellulose-Acetate Electrophoresis for Rapid Identification of Allozyme Genotypes of Phytophthora infestans. Plant Disease, 79, 1181-1185.
http://dx.doi.org/10.1094/PD-79-1181 - 6. Chunwongse, J., Chunwongse, C., Black, L. and Hanson, P. (2002) Molecular Mapping of the Ph-3 Gene for Late Blight Resistance in Tomato. Journal of Horticultural Science & Biotechnology, 77, 281-286.
- 7. Zhang, C.Z., Liu, L., Zheng, Z., Sun, Y.Y., Zhou, L.X., Yang, Y.H., Cheng, F., Zhang, Z.H., Wang, X.W., Huang, S.W., Xie, B.Y., Du, Y.C., Bai, Y.L. and Li, J.M. (2013) Fine Mapping of the Ph-3 Gene Conferring Resistance to Late Blight (Phytophthora infestans) in Tomato. Theoretical and Applied Genetics, 126, 2643-2653.
http://dx.doi.org/10.1007/s00122-013-2162-1 - 8. Zhang, C.Z., Liu, L., Wang, X.X., Vossen, J., Li, G.C., Li, T., Zheng, Z., Gao, J.C., Guo, Y.M., Visser, R.G.F., Li, J.M., Bai, Y.L. and Du, Y.C. (2014) The Ph-3 Gene from Solanum pimpinellifolium Encodes CC-NBS-LRR Protein Conferring Resistance to Phytophthora infestans. Theoretical and Applied Genetics, 127, 1353-1364.
http://dx.doi.org/10.1007/s00122-014-2303-1 - 9. Park, Y., Hwang, J., Kim, K., Kang, J., Kim, B., Xu, S. and Ahn, Y. (2013) Development of the Gene-Based SCARs for the Ph-3 Locus, Which Confers Late Blight Resistance in Tomato. Scientia Horticulturae, 164, 9-16.
http://dx.doi.org/10.1016/j.scienta.2013.08.013 - 10. Robbins, M.D., Masud, M.a.T., Panthee, D.R., Gardner, C.O., Francis, D. and Stevens, M.A. (2010) Marker-Assisted Selection for Coupling Phase Resistance to Tomato Spotted Wilt Virus and Phytophthora infestans (Late Blight) in Tomato. HortScience, 45, 1424-1428.
- 11. Kim, M.J. and Mutschler, M.A. (2005) Transfer to Processing Tomato and Characterization of Late Blight Resistance Derived from Solanum pimpinellifolium L. L3708. Journal of the American Society for Horticultural Science, 130, 877-884.
- 12. Lee, S.J., Kelley, B.S., Damasceno, C.M.B., John, B.S., Kim, B.S., Kim, B.D. and Rose, J.K.C. (2006) A Functional Screen to Characterize the Secretomes of Eukaryotic Pathogens and Their Hosts in Planta. Molecular Plant-Microbe Interactions, 19, 1368-1377.
http://dx.doi.org/10.1094/mpmi-19-1368 - 13. Gardner, R.G. and Panthee, D.R. (2010) “Plum Regal” Fresh-Market Plum Tomato Hybrid and Its Parents, NC 25P and NC 30P. HortScience, 45, 824-825.
- 14. Panthee, D.R. and Gardner, R.G. (2010) “Mountain Merit”: A Late Blight-Resistant Large-Fruited Tomato Hybrid. HortScience, 45, 1547-1548.
- 15. Fulton, T.M., Chunwongse, J. and Tanksley, S.D. (1995) Microprep Protocol for Extraction of DNA from Tomato and Other Herbaceous Plants. Plant Molecular Biology Reporter, 13, 207-209.
http://dx.doi.org/10.1007/bf02670897 - 16. Panthee, D.R. and Gardner, R.G. (2014) “Mountain Rouge”: A Pink-Fruited, Heirloom-Type Hybrid Tomato and Its Parent Line NC 161L. HortScience, 49, 1463-1464.
- 17. Foolad, M.R., Merk, H.L. and Ashrafi, H. (2008) Genetics, Genomics and Breeding of Late Blight and Early Blight Resistance in Tomato. Critical Reviews in Plant Sciences, 27, 75-107.
- 18. Gardner, R.G. and Panthee, D.R. (2010) NC 1 CELBR and NC 2 CELBR: Early Blight and Late Blight Resistant Fresh Market Tomato Breeding Lines. HortScience, 45, 975-976.
NOTES

*Corresponding author.